Abstract
Translocon-associated protein complex (TRAP) is thought to be required for efficient protein-specific translocation across the endoplasmic reticulum membrane. We created a mutation in the Trapα gene that leads to the synthesis of a truncated TRAPα protein fused to ShBle-β-galactosidase. Analysis of Trapα cDNAs reveals that among three different messenger RNAs expressed in the mouse, one of them encodes a slightly larger protein that differs in its C-terminal end. This mRNA, specific for skeletal muscle and heart, is only expressed after birth. Homozygous Trapα mutant pups die at birth, likely as a result of severe cardiac defects. Indeed, the septation of the proximal part of the outflow tract is absent, resulting in a double-outlet right ventricle. Studies of protein secretion in transfected embryonic fibroblasts reveal that the TRAP complex does not function properly in homozygous mutant cells and confirm, in vivo, the involvement of TRAP in substrate-specific translocation. Our results provide the first in vivo demonstration that a member of the TRAP complex plays a crucial role in mammalian heart development and suggest that TRAPα could be involved in translocation of factors necessary for maturation of endocardial cushions.
In eucaryotes, most secretory and membrane proteins are cotranslationally transported across or integrated into the endoplasmic reticulum (ER) membrane. These proteins are synthesized on membrane-bound ribosomes and are transported through a protein-conducting channel in the ER membrane. In the process, they undergo various modifications, including folding and maturation events (19, 32). The elements that are involved in the translocation machinery have been characterized by biochemical studies of cell-free systems and constitute the translocon. The major component is the Sec61p complex, which is conserved among all eucaryotes. This heterotrimeric complex is thought to be the main constituent of the translocation channel (3, 12, 28). Translocation experiments using proteoliposomes with translocons reconstituted from purified elements have shown the Sec61 complex to be sufficient for the formation of a membrane channel (20). However, other components, such as the TRAM protein (15, 36) or the heterotetrameric translocon-associated protein (TRAP) complex (10, 13), are generally required for efficient translocation. Other proteins can interact with the nascent chain during its transport, for example, to cleave the signal sequence or to catalyze other posttranslational modifications. Despite the important body of information on the different steps of the translocation process, the large amount of biochemical information available for the different elements involved in that process, and recent crystallographic data, some aspects of the organization of the translocon and of the functions of the different associated complexes are still unclear (19, 27, 34). The exact role of the TRAP complex, previously called the signal sequence receptor, remains to be clarified. Previous studies have suggested that TRAP is not essential for protein translocation (12, 29). However, the efficiency of protein translocation in reconstituted systems deprived of TRAP varied considerably according to the protein (12). Structural studies of ribosome channel complexes engaged in protein translocation have revealed that the translocon channel contains TRAP or OST complexes (28). More recently, TRAP was identified as a ubiquitous component of the mammalian translocon, and a model was proposed, according to which every native channel may contain two TRAP complexes, suggesting that in higher eucaryotes TRAP may have a general role in protein translocation (27). In previous functional studies, Fons and collaborators have demonstrated that TRAP is indeed required for the initiation of the translocation of a subset of specific proteins, depending on the functional features of signal sequences. The same authors have shown that, in a reconstituted system, the absence of TRAP results in an important decrease in translocation efficiency for those proteins (10).
Most of the studies on the translocon and translocon-associated proteins have been carried out using in vitro systems, mainly with microsomes from dog or pig pancreas. There are only two reports about in vivo studies of TRAP subunits. The first one described the expression of the beta subunit during zebra fish embryogenesis (26), while the other one showed the involvement of the gamma subunit in pronephros development in Xenopus laevis (25).
During the analysis of embryonic stem (ES) cell clones generated by gene trapping (5), we identified a clone, TL5, that carries an insertion in the gene encoding one of the TRAP subunits (TRAPα). This results in a hybrid gene in which the β-galactosidase coding sequence is inserted towards the end of the Trapα coding region. Phenotypic analysis of homozygous (HZ) embryos carrying such an insertion mainly revealed cardiovascular defects, namely malformation of the septum of the outflow tract (OFT). In mammals, heart development involves complex cellular interactions and morphogenetic movements. In mice, the primitive heart tube forms by embryonic day 8 (E8). The separation of the four chambers occurs between E10.5 and E14.5 and proceeds first with the development of the ventricular septum, followed by the development of the endocardial cushions and the reorganization of the inflow and outflow tracts (14, 30). The single outflow vessel, the truncus arteriosus, which connects the heart with the aortic arches, is divided by a septum that grows from the aortic arches towards the ventricles and divides the truncus into two independent arteries: the pulmonary artery and the aorta. As it forms, the outflow septum rotates in such a way that the aorta communicates with the left ventricle while the pulmonary artery emerges from the right ventricle (9). Three kinds of cells participate in the formation of the OFT septum: myocardial cells, endocardial cells, and neural crest cells (NCC). Upon myocardial signaling, a subpopulation of endocardial cells undergoes an endothelial-mesenchymal transition (EMT), which results in the formation of the cardiac cushions. NCC migrating from the trunk through the artery arches contribute to the mesenchyme of the cushions, as well as to the formation of the walls of the great vessels (22, 39). The specific functions of each of these cell populations in the establishment of the outflow septum are still unclear, as are the underlying molecular mechanisms.
In the present article, we report on a detailed study of ES cells carrying an insertional mutation in the Trapα gene, as well as on the analysis of the mouse line established from these cells, TL5. Our data revealed that, in vivo, TRAPα is involved in the secretion of specific proteins and that the TL5 mutation affects that function. In addition, they show that three different Trapα messengers are expressed. Two different proteins are encoded, one of which is constitutively expressed while the other, which is muscle specific, is developmentally regulated. Intriguingly, the mutation in Trapα results in specific defects in heart formation. HZ mutant embryos display an absence of septation of the proximal part of the OFT due to an abnormal development of the endocardial cushions. Our results suggest a critical role for TRAPα-mediated protein translocation during endocardial cushion maturation.
MATERIALS AND METHODS
5′ and 3′ RACE-PCR.
In order to identify the gene trap insertion, 5′ rapid amplification of cDNA ends (RACE)-PCR was performed on ES mRNA using the MARATHON kit (Clonetech) according to the instructions of the manufacturer. To increase the specificity of the RACE, two nested primers were designed from the ShBle gene: 5′GGGATCATCGGATCTGTCCT3′ and 5′ACCACTCGGCGTACAGCTCGT3′. The amplified cDNA was cloned and sequenced. The entire set of Trapα cDNAs were obtained using the 5′GGTTGTTGTTGGCCTTCATCAGCTCCTGG3′ primer designed from the 5′-amplified cDNA after 3′ RACE-PCR on either testis or muscle MARATHON Ready CDNA libraries (Clontech). In order to control the 5′ end of the Trapαm, 5′ RACE-PCR was performed using the 5′TTTCGGGGCTGCCTTTTGGGAGAAG3′ primer. The different PCR products were cloned and sequenced.
In order to identify the exact position of the vector in the seventh intron, the region between exon 7 and the construct was amplified. Using primer 5′GGTTGTTGTTGGCCTTCATCAGCTCCTGG3′ in exon 7 and 5′TGAGCACCGGAACGGCACTGGTCAACTTGG3′ in the construct, the PCR product (2.3 kb) was sequenced starting from the construct and aligned on the genomic sequence (ENSMU SG00000021427).
RNA isolation and Northern blotting.
Poly(A)+ RNAs were isolated from ES cells, E15.5 whole embryos, or different tissues from newborn or adult mice using a Dynabeads mRNA DIRECT kit (Dynal). Northern blot analysis was performed using 2 μg of poly(A)+ RNA per sample and a Northern Max kit (Ambion) until transfer onto a Hybond membrane. To study Trapα adult mRNA expression, commercial Northern blot membranes (Clonetech) were used. In both cases, membranes were hybridized with a 638-bp probe obtained by PCR amplification of the cDNA located between nucleotides 97 and 735.
RT-PCR.
Two hundred to three hundred nanograms of mRNA was reverse transcribed (RT) with SuperscriptII (Invitrogen) according to the manufacturer's instructions, and one-twentieth of the products were amplified using either primers P1 (5′-GTCAGAATGATGTTGACATGAGCTGG-3′) and P2 (5′-GCGTTTTGCTGAGTATGCTAAAGGT-3′) to detect Trapαg mRNA (PCR product length, 400 bp) or P3 (5′-GGTTGTTGTTGGCCTTCATCAGCTCCTGG-3′) and P4 (5′-CGCAGAAAGAGAGAGGCGCGCTGAG-3′) to detect Trapαm mRNA (PCR product length, 200 bp).
Protein expression.
For Western blotting, ES cells or E15.5 embryos were homogenized after freezing in liquid nitrogen in buffer containing 0.1 M NaCl, 0.01 M Tris-HCl (pH 7.6), 0.001 M EDTA (pH 8.0), and 0.5% Triton X-100. The protein suspension was quantified by Bio-Rad protein assay, and 30 μg of protein was used for Western blot analysis (Bio-Rad system). Two different polyclonal antibodies were used to reveal TRAPα protein: one was raised against the C-terminal region (gift of Tom A. Rapoport) and the other was directed against the N-terminal region, produced by Neosystem, and purified using HiTrap affinity columns (Amersham Pharmacia). A monoclonal antibody was used to reveal β-galactosidase protein (40-1A; Developmental Studies Hybridoma Bank [DSHB]).
Cellular localization.
E13.5 embryos were collected from TL5 heterozygous (HT) intercrosses to prepare embryonic fibroblasts. Yolk sacs were used for genotyping. The embryos were chopped with a sterile scalpel, and cells were dissociated by 15 min of incubation in trypsin/EDTA (Invitrogen) at room temperature. After centrifugation, the cells were plated on tissue culture dishes and cultured in Dulbecco's minimal essential medium (Invitrogen) complemented with 10% fetal calf serum for 1 or 2 days. For each sample, 4 × 105 cells were used. They were plated on circular glass slides, cultured overnight, and fixed for 20 min in 4% paraformaldehyde at pH 7.2. After two washes in phosphate-buffered saline (PBS)-0.1 M glycine, the slides were incubated for 20 min in PBS complemented with 0.2% bovine serum albumin and 0.05% saponin before the first antibody was added. Three polyclonal antibodies were used; they were raised against either the C-terminal or N-terminal region of TRAPα (see “Protein expression”) and against calnexin. In addition, a mouse monoclonal antibody (40-1A; DSHB) was used to reveal β-galactosidase. All of the antibodies were used at 1/200. After 1 h of incubation, cells were washed five times in PBS for 5 min each and stained with a second antibody that was either raised against rabbit and marked with fluorescein isothiocyanate or directed against mouse and marked with rhodamine (1/200; DAKO). After 30 min of incubation, cells were washed in PBS. DAPI (4′,6′-diamidino-2-phenylindole) at 1/50,000 was added in one of the five washes.
Ex vivo study of protein secretion.
Gamma interferon (IFN-γ), atrial natriuretic peptide (ANP), and preprolactin cDNA were amplified from mouse IFN-γ cDNA (primers 5′CCCAAGCTTCTGAGACAATGAACGCTAC3′ and 5′CGGGGTACCGAGCGACCCTTTTCCGCTTC3′), heart cDNA library from a mouse newborn (primers: 5′CCCAAGCTTACCCACGCCAGCATGGGCT3′ and 5′TCTTCGGTACCCGGAAGTGTTG3′), and brain cDNA library from an adult mouse (primers 5′CCCAAGCTTAGTCACCATGACCATGAAC3′ and 5′CGGGGTACCGAGTTGTTTTGATGGGCAATT3′), respectively. All PCR products were subcloned into the expression vector peGFP-N1 (Stratagene), in which enhanced green fluorescent protein (GFP) was replaced by the hemagglutinin tag. The peGFP-N1 vector was cotransfected with each of the vectors in order to get a control of transfection efficiency.
Embryonic fibroblasts were grown in modified Eagle's minimal essential medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin at 37°C in 8% CO2. Transient transfection was performed with Lipofectamine 2000 (Invitrogen, Inc.) in 24-well clusters according to the manufacturer's instructions. Each transfection was carried out in triplicate. Five hours later, the transfection medium was replaced by Optimem-1 (Life Technologies) supplemented with 0.5% fetal calf serum, and both supernatants and cells were harvested 48 h later. Proteins from the supernatant were extracted in 80% cold acetone and resuspended in loading buffer. Western blots were performed using a monoclonal anti-hemagglutinin antibody (Covance, Inc.). GFP from pellet samples was revealed using a polyclonal anti-GFP antibody (Biovalley SA). Quantitation was performed by chemifluorescent detection (ECLplus; Amersham) using a phosphorimager (Molecular Dynamics Inc.). For each protein, after normalization to the GFP content, data were expressed as the percentage of secretion by the wild-type (WT) cells. Statistical significance of difference was assessed by Student's t testing.
Histological analyses.
After PFA fixation or in situ revelation, complete embryos or heart/lung ensembles were treated and embedded in resin according to the instructions of the manufacturer (Historesin; Leica), and sectioned at 10 μm. The sections were counter-colored with 1% safranin.
Cell proliferation was evaluated by bromodeoxyuridine (BrdU) incorporation or phosphohistone H3 analysis. E12.5 pregnant females were injected intraperitoneally with 10 μM of BrdU (Roche) per 100 g of body weight 2 h prior to embryo harvest. TL5 HZ embryos and WT littermates were sectioned, followed by in situ immunochemical detection of BrdU-incorporated cells. Sections were incubated with a 1:200 dilution of monoclonal anti-BrdU (Becton Dickinson) revealed with an AlexaFluor 594 anti-mouse antibody (1/500). Phosphohistone H3 was detected on paraffin sections of E12.5 embryos using anti-phosphohistone H3 (1/200; Upstate) and revealed with an AlexaFluor 488 goat anti-rabbit antibody. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) was performed on paraffin-embedded sections using an in situ cell death detection kit (Roche) according to the manufacturer's instructions, and the fluorescein isothiocyanate signal was enhanced using an AlexaFluor 488 signal amplification kit for fluorescein (Molecular Probes). Sections were counterstained with Hoechst. Data are expressed as a percentage of positive cells among cushion cells.
Myocardialization.
Paraffin sections were incubated with MF20 antibody (1/200; DSHB) and revealed with AlexaFluor 594 anti-mouse antibody (1/500; Molecular Probes).
In situ hybridization was performed in toto on opened trunks of embryos of different genotypes collected between E11.5 and E13.5. PCR fragments were obtained from mouse plexin A2 (912 bp: nucleotides 2121 to 3033) or mouse periostin (679 bp: nucleotides 22 to 701) cDNAs and cloned using pCRII TOPO vector dual promoter (Invitrogen). Sense and antisense RNA probes were synthesized using either SP6 or T7 RNA polymerase (Boeringer). In situ hybridization was performed as described by de Robertis et al. (http://www.hhmi.ucla.edu/derobertis/protocol_page/mouse.PDF).
Nucleotide sequence accession numbers. Trapαg and Trapαm cDNA sequences were deposited in the GenBank database under the accession numbers AF326229 and AY993942, respectively.
RESULTS
Isolation of mouse Trapα cDNA from TL5 ES cells.
The TL5 ES cell clone was obtained during a gene trapping experiment. The construct inserted into the CK35 ES cell genome comprised a fusion between ShBle and lacZ genes framed by a splice acceptor sequence in the 5′ end and an SV40 poly(A)+ sequence in the 3′ terminus. The ShBle gene allowed selection on phleomycin, while the lacZ reporter gene permitted monitoring of the expression of the trapped endogenous gene (5). Using 5′ RACE PCR, an 879-bp cDNA was amplified from RNA purified from TL5 ES cells. Analysis of the sequence revealed a high similarity with dog and human Trapα cDNA (accession numbers gi 505979075 and gi 6552340, respectively). Alignment with the mouse genome sequence (ENSMU SG00000021427) revealed that the insert is in the seventh intron, leading, after splicing of the construct, to the removal of the eighth exon as well as the 3′-noncoding region (Fig. 1B).
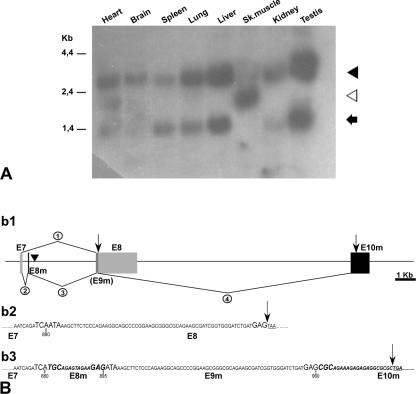
FIG. 1.
Characterization of Trapα messengers. (A) Northern blot analysis of mouse adult tissues (commercial membrane; Clontech) using a 638-bp probe encompassing the 5′ region of Trapα cDNA. Three different messengers of 2.7 kb (black arrowhead), 1.8 kb (white arrowhead), and 1.2 kb (black arrow) are shown. (B) 3′ end of the three different Trapα cDNAs. Sk.muscle, skeletal muscle. (b1) Schematic representation of 3′-genomic region encompassing the last exons of the Trapα gene. Angular lines indicate splicing events, E7 and E8 (gray) correspond to the last two exons of both the 2.7-kb and 1.2-kb messengers; E8m and E10m (black) are the two exons specific to the 1.8-kb mRNA. E9m is identical to the E8 coding region. A filled inverted triangle indicates the position of the vector insertion. (b2 and b3) Sequences of the 3′-end coding region of the different mRNAs. (b2) Sequence of 2.7-kb and 1.2-kb mRNAs resulting from splicing event 1 in b1. (b2) Sequence of 1.8 kb resulting from splicing events 2, 3, and 4 shown in b1. Black arrows indicate the ends of the coding regions. In b2 and b3 the three nucleotides at each extremity of the exons are in large uppercase letters and the stop codons are underlined. Numbers below the sequence indicate nucleotide positions in the cDNA sequences.
Three different Trapα messengers are expressed in the adult mouse.
Northern blot analysis revealed that three different mRNAs of 1.2 kb, 1.8 kb, and 2.7 kb were expressed in the adult (Fig. 1A). Both the 1.2-kb and 2.7-kb messengers were seen in most of the tissues examined, to varying extents. They were not detected in skeletal muscle, where a different 1.8-kb mRNA which was also detected in the heart was expressed. Complete coding sequences were obtained after RACE-PCR on either testis or muscle cDNA libraries. Figure 1B shows a schematic representation of the alternative splicing events that lead to three different Trapα messengers RNA (b1) as well as the sequence differences at the 3′ ends of the coding regions (b2 and b3). The 2.7-kb and 1.2-kb mRNAs differed only by the length of their 3′-noncoding regions due to an alternative polyadenylation site. The 1.2-kb form stopped at position 1172. The 1.8-kb mRNA differed in its 3′-noncoding sequence (not shown) and also, through alternative splicing, in the presence of two additional small exons. The first one is located 5′ of and close to the insertion site of the construct (Fig. 1B, b1 and b3).
The mouse TRAPα protein sequences confirmed a high degree of conservation among vertebrates.
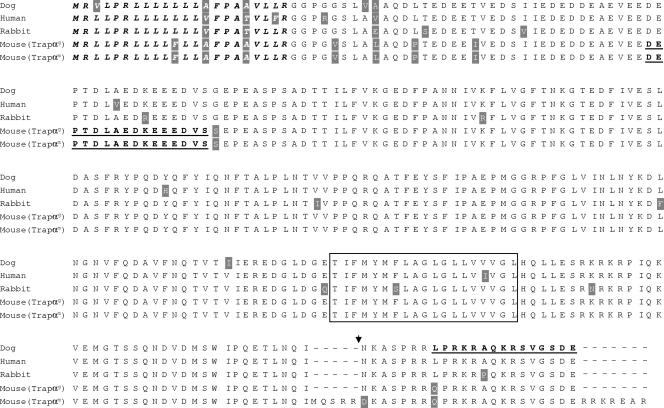
Analysis of the Trapα messengers reveals the presence of two open reading frames. The first one, common to the 2.7-kb and 1.2-kb Trapα mRNAs, encodes a protein of 286 amino acids (hereafter referred to as TRAPαg). The second one corresponds to the 1.8-kb mRNA and encodes a protein that comprises 12 supplementary residues (hereafter referred to as TRAPαm). This protein is exclusively found in skeleton muscles and in the heart. Figure 2 displays the mouse protein sequences that were deduced from the cDNAs by comparison with those of dog, human, and rabbit. TRAPα is highly conserved: the percentage of identity between the TRAPαg protein sequence and those of other species varies from 89% (human) to 96% (dog). Most differences are concentrated in the N-terminal part, in or close to the signal sequence. The mouse protein expressed in skeletal muscle (TRAPαm) differs from the TRAPαg at the C-terminal end. Five amino acids are inserted at position 264, the asparagine at position 265 is replaced by an aspartic acid, and seven amino acids, of which four are arginine, are added at the C-terminal extremity. It is interesting that 11 of the 31 C-terminal residues (35%) are arginine.
FIG. 2.
Sequence comparison between mouse TRAPα proteins and their human, dog, and rabbit orthologs. Shaded amino acids display some differences according to the species. The signal peptide region is in italics, while the transmembrane region is boxed. Underlined sequences correspond to the immunopeptides used to produce N- and C-terminal antibodies. The point of junction between TRAPα and ShBle-β-galactosidase in the hybrid protein synthesized in TL5 is indicated by a filled inverted triangle. Mouse TRAPαg corresponds to the protein expressed in all tissues but muscles; mouse TRAPαm corresponds to the muscle-specific protein. Protein sequence accession numbers from the SwissProt database: human, P43307; dog, P16967; rabbit, PS3815.
In TL5 ES cells, the insertion of the vector at position 880 of the Trapα gene should result, after splicing of the trap construct, in the synthesis of a fusion protein between TRAPα and ShBle-β-galactosidase. TRAPαg protein would be truncated by removal of the last 22 C-terminal amino acids, while the TRAPαm protein would lose its last 29 C-terminal residues (Fig. 2).
Mice, homozygous for the modified TRAPα, die around birth.
In order to check for an effect of TRAPα modification on mouse development, we generated a mouse line (TL5) carrying the vector insertion. HT animals did not display any abnormality and were fertile. Crosses between HT animals for the Trapα mutated allele never gave rise to HZ live animals. However, during development, HZ embryos were detected at E19.5, and careful checking of the deliveries revealed that most homozygotes were born but died very soon after birth.
Analysis of Trapα mRNA during embryonic development revealed that only the two mRNAs encoding TRAPαg are expressed before birth.
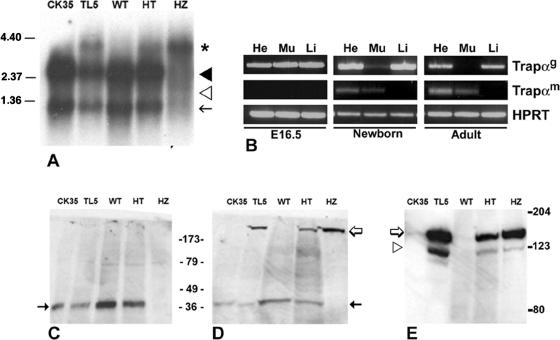
To try to understand the origin of perinatal death, we first investigated the expression of endogenous Trapα mRNA during embryonic development. Poly(A)+ RNAs were isolated from either ES cells or WT, HT, and HZ E15.5 embryos and analyzed by Northern blotting. Hybridization with the 638-bp Trapα cDNA probe (Fig. 1A) revealed that the 2.7- and 1.2-kb mRNAs were expressed in WT and HT embryos as well as in CK35 (WT) and TL5 (HT) ES cells (Fig. 3A). The 1.8-kb muscle-specific messenger was missing in all of the samples. A new 4.2-kb form was found in HT and HZ mutants as well as in TL5 ES cells (Fig. 3A). This mRNA corresponds to the fusion between the first 800 Trapα nucleotides and the ShBle/LacZ messenger. It was the only mRNA expressed in the HZ embryos (Fig. 3A), demonstrating that the vector insertion has totally interrupted the endogenous message.
FIG. 3.
Trapα mRNA and protein expression in ES cells, embryos, or adult mouse cells. (A) Northern blot analysis of poly(A)+ RNAs isolated from ES cells (CK35, TL5) and from E15.5 embryos (WT, HT, HZ) hybridized with a 638-bp Trapα probe (see the legend to Fig. 1). Two messengers of 2.7 kb (black arrowhead) and 1.2 kb (black arrow) are expressed in WT and HT forms, both in ES cells and embryos; they are absent in the HZ embryos. A larger band of 4.2-kb mRNA (asterisk) is observed in HT or HZ embryos. It represents the fusion between TRAPα and ShBle-β-galactosidase mRNAs. The 1.8-kb RNA (white arrowhead) is expressed neither in ES cells nor in embryos. (B) RT-PCR of poly(A)+ RNA purified from heart (He), muscle (Mu), and liver (Li) from E16.5, newborn, or adult mouse cells. Trapαg corresponds to both 2.7- and 1.2-kb RNAs, while Trapαm is specific for the 1.8-kb RNAs. HPRT, hypoxanthine phosphoribosyltransferase. (C to E) Western blot analysis of ES cells and E15.5 embryos. Antibodies raised against C- or N-terminal TRAPα (C and D) or against β-galactosidase (E) were used. (C) C-terminal antibody revealed a protein of 34 kDa (black arrow) in wild-type (CK35, WT) and heterozygous (TL5, HT) samples but not in HZ samples. (D) N-terminal antibody binds the 34-kDa bands (black arrow) in WT and HT samples; in addition, it reveals a larger band of >123 kDa (white arrow) present in TL5, HT, and HZ samples but not in the WT. That larger band is the only one present in the HZ embryos. (E) Antibody directed against β-galactosidase binds to an identical band of >123 kDa (white arrow) in TL5, HT, and HZ samples, demonstrating that it corresponds to the TRAPα-ShBle-β-galactosidase fusion protein. The fainter band of 123 kDa, recognized by β-galactosidase antibody, corresponds to ShBle-β-galactosidase translated from the ATG present in the gene trap construct (white arrowhead).
Analysis of Trapα transcript expression at different developmental stages (ranging from E7 to E17) revealed that the 2.7-kb and 1.2-kb transcripts are expressed throughout embryogenesis. In contrast, the 1.8-kb mRNA was never found (not shown).
To determine the onset of 1.8-kb mRNA expression, RT-PCR was performed on mRNA prepared from heart, muscle, or liver obtained from E16.5 embryos, newborn mice, and adult mice. Trapαg (2.7- and 1.2-kb) transcripts were found in the three samples at E16.5 and in the newborn; however, their expression decreased in muscle at birth. In the adult, Trapαg transcripts were not expressed in muscle, while they remained in heart and liver (Fig. 3B, adult). The muscle-specific form (Trapαm, 1.8 kb) was completely absent in the embryonic samples (Fig. 3B, E16.5), confirming the results obtained by Northern blotting (Fig. 3A). Faint expression was observed in the heart and muscle of the newborn (Fig. 3B) and increased in the adult. It remained absent in the liver samples (Fig. 3B). Thus, at birth there is a switch between the Trapαg and Trapαm forms in muscle.
The TRAPα- ShBle-β-galactosidase-TRAPα fusion protein is the only Trapα protein expressed in homozygous embryos.
Protein extracts obtained from CK35 and TL5 ES cells or from WT, HT, and HZ E16.5 embryos were analyzed by Western blotting using antibodies directed against either the last 15 residues at the C-terminal end or a polypeptide in the N-terminal region (Fig. 2). In WT and HT samples, the C-terminal antibody revealed a band of 34 kDa, which corresponds to the size of TRAPα (Fig. 3C). In contrast, there was no TRAPα protein present in the extracts from the HZ embryos (Fig. 3C). In addition to the 34-kDa protein, the N-terminal revealed a protein larger than 123 kDa in the heterozygotes. It was the only protein found in HZ samples (Fig. 3D). Moreover, this protein is absent from the WT samples (Fig. 3D, CK35 and WT). Antibody directed against β-galactosidase stained a band of the same size (Fig. 3E); we conclude that this band corresponds to the fusion protein between TRAPα and ShBle-β-galactosidase. A small amount of a slightly shorter protein that must correspond to translation initiated at the ATG present in the 5′ terminus of the gene trap construct, giving rise to a ShBle-β-galactosidase fusion protein, was also observed in TL5, HT, and HZ embryos. It is interesting that this translation start site seems to be used more frequently in ES cells than in embryos (Fig. 3E).
TRAPα and TRAPα-ShBle-β-galactosidase fusion protein have the same subcellular localization.
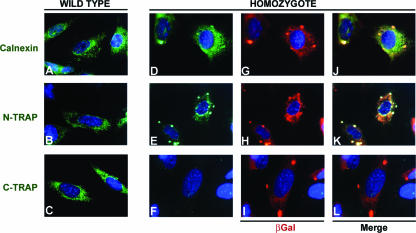
Analysis of β-galactosidase activity in preimplantation embryos revealed subcellular localization of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining (see Fig. S1 in the supplemental material), which suggests that the TRAPα-ShBle-β-galactosidase protein depends on the TRAPα signal sequence and thus follows endogenous TRAPα localization. To verify this hypothesis, we compared the localization of TRAPα-ShBle-β-galactosidase fusion protein with that of endogenous TRAPα protein by using embryonic fibroblasts prepared from WT, HT, and HZ E13.5 embryos. In WT cells (Fig. 4A to C), the distribution of TRAPα was studied by using antibodies directed against either the N- or C-terminal region (Fig. 4B and C). The staining spread around the nucleus in a way compatible with localization in the ER. Comparison with the staining obtained using an antibody directed against calnexin, which is an ER integral membrane protein, reinforced that conclusion (Fig. 4A). In HT fibroblasts (not shown), antibody directed against β-galactosidase revealed that the fusion protein had a distribution similar to that of WT TRAPα molecules, suggesting that the fusion protein integrates into the ER membrane. In WT fibroblasts, Trapα C-terminal antibody revealed homogenous distribution of the protein in the plane of the ER membrane (Fig. 4C), while no staining was observed in HZ fibroblasts (Fig. 4F). These observations demonstrate that in HZ cells all the TRAPα proteins lack the C-terminal region. In HT fibroblasts, the C-terminal antibody staining of WT TRAPα subunits (not shown) revealed that their distribution is homogenous and, thus, not perturbed by the presence of the fusion protein. On the contrary, both N-terminal and β-galactosidase antibodies showed aggregates formed by the fusion proteins (Fig. 4E, G, and H). Those aggregates are more numerous and larger in HZ than in HT cells. Most probably they are due to tetramerization of β-galactosidase. Interestingly, some calnexin seems to be localized in these aggregates, at least in HZ cells, where costaining with β-galactosidase is observed (Fig. 4J). These observations suggest that calnexin could still interact with TRAPα within the fusion protein (37).
FIG. 4.
Subcellular localization of TRAPα and TRAPα-ShBle-β-galactosidase fusion protein in WT and HZ embryonic fibroblasts. Immunofluorescence staining was performed using Calnexin antibody (green) (A, D, and J), TRAPα N-terminal antibody (N-TRAP; green) (B, E, and K), TRAPα C-terminal antibody (C-TRAP; green) (C, F, and L), and β-galactosidase antibody (βGal; red) (G to I). TRAPα N-terminal antibody stains both WT (B) and HZ (E) samples and reveals that TRAPα is localized in the same region as calnexin (A and D), thus, in the ER membrane. TRAPα C-terminal antibody stains WT cells (C) but does not recognize any protein in the HZ cells (F). β-Galactosidase colocalizes with calnexin (G and J) and TRAPα (H and K), revealing that the fusion protein is also inserted in the ER membrane. However, in HZ cells, the distribution of TRAPα-ShBle-β-galactosidase fusion protein in the membrane, revealed by either the N-terminal (E) or β-galactosidase antibody (G and H), is different from the distribution of TRAPα in wild-type cells (B); indeed, aggregates are clearly visible.
TRAPα-ShBle-β-galactosidase fusion protein does not function properly.
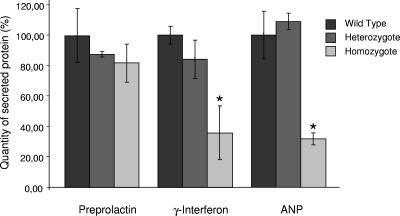
In order to test the activity of the TRAPα-ShBle-β-galactosidase fusion protein, WT, HT, or HZ embryonic fibroblasts were transfected with vectors expressing secreted proteins whose translocation has been determined to be independent (preprolactin) or dependent (IFN-γ and ANP) on TRAP (10). A vector expressing a GFP cytoplasmic protein was cotransfected with each of the vectors. The proteins secreted in the culture medium or the GFP present in the cells was analyzed by Western blotting 48 h after transfection (see Fig. S2 in the supplemental material). Quantitation of the data after normalization to the GFP content is shown in Fig. 5; it revealed that, while the secretion of preprolactin by either HT or HZ fibroblasts is not significantly changed compared to that by WT cells, secretion of both IFN-γ and ANP is considerably decreased in HZ mutants. Indeed, in TL5 HZ fibroblasts, the percentage of secretion of IFN-γ or ANP is reduced to, respectively, 36% or 32% of that of the WT cells. These results are reminiscent of those obtained in a reconstituted system deprived of TRAP (10), suggesting that (i) in vivo IFN-γ and ANP translocation is also dependent on TRAP and (ii) in TL5 HZ cells, the TRAP complex is not functional due to the replacement of TRAPα by TRAPα-ShBle-β-galactosidase fusion protein. It is worth noting that the secretion of any of the proteins is not perturbed in HT cells, demonstrating that the fusion proteins do not prevent normal functioning of the endogenous TRAP complex.
FIG. 5.
Secretion of proteins by embryonic fibroblasts after cotransfection of vectors expressing enhanced GFP and either preprolactin, IFN-γ, or ANP. After Western blot analysis, quantitation of secreted proteins was normalized to GFP content. The quantities of proteins secreted by either WT, HT, or HZ cells were plotted as, respectively, black, gray, and light gray bars (each bar corresponds to the mean for three independent samples). For each protein the secretion by wild-type fibroblasts was defined as 100%. Error bars represent the standard errors of the means; *, P < 0.05.
Trapα−/− mutant displayed abnormal heart morphogenesis.
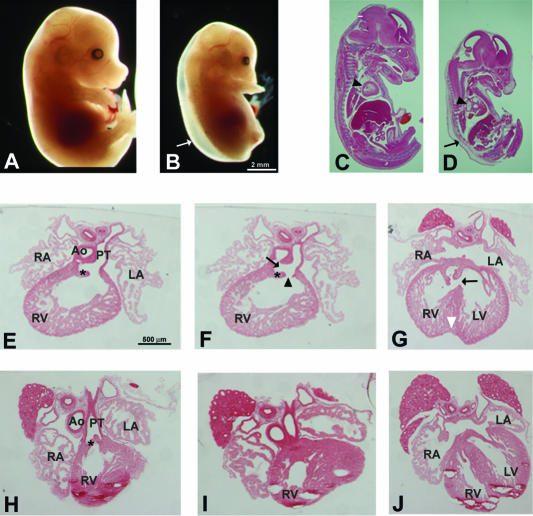
At E15.5, embryos homozygous for the modified Trapα gene displayed an abnormal phenotype, including smaller size and development of severe subcutaneous edema (Fig. 6B and D). The organs were proportionally smaller than those of the WT. In contrast, the heart appeared bigger and presented some abnormalities, in particular, a prominent enlargement of the ventricles (Fig. 6C and D). Edema and ventricular dilatation were suggestive of poor cardiac function.
FIG. 6.
Trapα mutants are retarded and exhibit a strong cardiac phenotype. (A and B) The E14.5 HZ TL5 mutant (B) is growth retarded compared to a WT littermate (A). In addition, the HZ embryo displays an important subcutaneous edema (B, arrow). (C and D) Sagittal sections of E15.5 embryos reveal that in the TL5 mutant (D), the organs are proportionally smaller compared to those of the WT (C), except for the heart (arrowheads), which displays an important dilatation. (E to J) In serial transverse sections through E15.5 HZ (E to G) and WT (H to J) hearts, a partial separation of the aorta (Ao) and the pulmonary trunk (PT) is evident in the mutant embryo (F, arrow). This defect gives rise to a DORV (F, arrowhead) and an interventricular septal defect (G, arrow). RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle. Asterisks in panels E, F, and H show the semilunar valves. The white arrowhead in panel G points to the indentation in the ventricle apex.
The general aspect of the heart was normal in the mutant until E12.5; however, later the heart was enlarged with a clear indentation at the apex of the ventricles (Fig. 6G), which generally signals incomplete ventricle septation. At E15.5, the aorta and pulmonary artery had separated distally, but the two large arteries remained side by side, indicating abnormal rotation of the aorta (not shown). Comparison of sections through either HZ (Fig. 6E to G) or HT (Fig. 6H to J) hearts clearly revealed morphogenetic defects in the mutant. The incomplete ventricle septation (Fig. 6G) was due to the absence of septum formation in the proximal part (conus) of the OFT (Fig. 6F). This absence of septum resulted in the persistence of a common trunk connected to the right ventricle and thus to the formation of a double-outlet right ventricle (DORV) (Fig. 6F). Semilunar valves were abnormally developed: both their size and their orientation were incorrect (Fig. 6 E and F). The valve leaflets pointed down to the ventricle, and in most cases they retained a thick and cushion-like appearance.
Mutant neural crest cells migrate as far as the wild type.
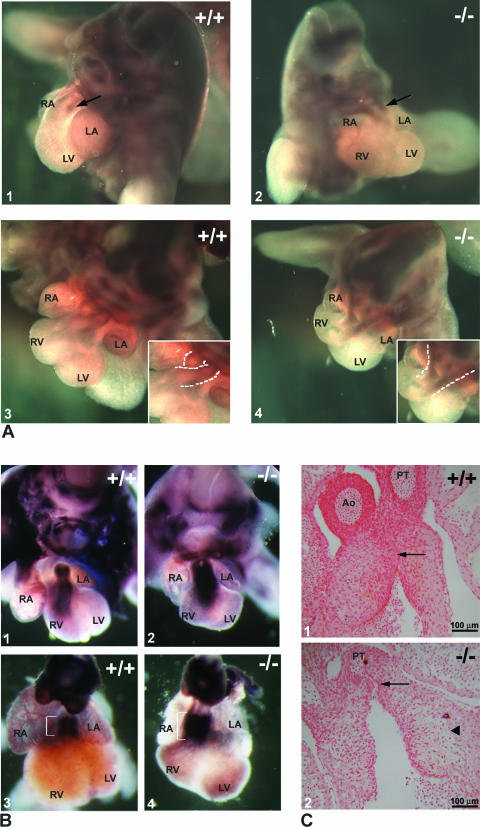
Cardiac NCC migrate through the pharyngeal region and the aortic arches into the endocardial cushions of the OFT. Monitoring plexin A2 expression in the cardiac NCC during their migration through the endocardial ridges (4) allows us to study their localization by whole-mount in situ hybridization. The results, obtained on WT or HZ E12.5 embryos, are shown in Fig. 7. Prongs of NCC extended into the OFT of the Trapα mutant as far as in the WT embryos (Fig. 7A1 and 2). However, in the mutant, the route followed by the cells was straight instead of spiraling, suggesting abnormal architecture of the endocardial cushions into which the NCC migrate (Fig. 7A3 and 4, insets). We therefore studied the formation and development of the cushions between E10.5 and E13.5.
FIG. 7.
Development of the outflow tract. (A) Neural crest cells visualized after whole-mount in situ hybridization on the trunk of E12.5 embryos using a plexin A2 riboprobe. Comparison between a TL5 mutant (A2 and 4) and a WT littermate (A1 and 3) reveals that the extent of migration of the crest cells is similar (A1 and 2, arrows). However, the route followed by the cells is very different in TL5 HZ embryos (compare the dashed lines in the insets in A3 and 4). (B) Endocardial cushion maturation as indicated by expression of periostin, which marks endocardial cells after EMT. Periostin expression in E12.5 WT (B1) and mutant (B2) or E13.5 WT (B3) and mutant (B4) embryos shows that endocardial cushions of the mutant had undergone EMT but that the staining is broader in the mutant at E13.5, suggesting an absence of maturation of the cushions (compare brackets). (C) Sections through OFT of E12.5 embryos revealed that the cushions have already fused in the WT (arrow in C1), while they are still separated in the mutant (arrow in C2). In addition, detachment of the cushions from the myocardial layer is frequently observed in the mutant (arrowhead in C2). Ao, aorta; PT, pulmonary trunk; RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle.
Maturation of endocardial cushions.
Periostin starts to be expressed in the mesenchyme of endocardial cushions after EMT at E10.5. Its expression decreases after myocardialization at E12.5 and remains confined to the valves from E14 to the adult stage (23). We used periostin as a marker of the mesenchymal transformation of the endocardial cells and therefore of the development of the cushions. Whole-mount in situ hybridization on E11.5 (not shown), E12.5 (Fig. 7B1 and 2), and E13.5 embryos (Fig. 7B3 and 4) revealed that Trapα mutant endocardial cells expressed periostin (Fig. 7B2 and 4), indicating that EMT had taken place. However, the extent of the signal is broader in the mutant (Fig. 7B). These observations suggested either an increased number of mesenchymal cells due to an excess in proliferation and/or a decrease of apoptosis within this cell population. It could also reflect a failure in the differentiation of the endocardial mesenchymal cells, which continued to express mesenchymal markers at stages when they are no longer expressed in WT cells. The absence of fusion of endothelial cushions in the mutant was evident on frontal sections made in OFTs (Fig. 7C1 and 2). In addition, the mutant's endocardial cushions were frequently detached from the myocardium (Fig. 7C2), while this was rarely observed in the WT. Observation of the distribution of the mesenchymal cells in the cardiac jelly suggested that in the mutant this tissue was looser or that there were fewer cells invading the jelly.
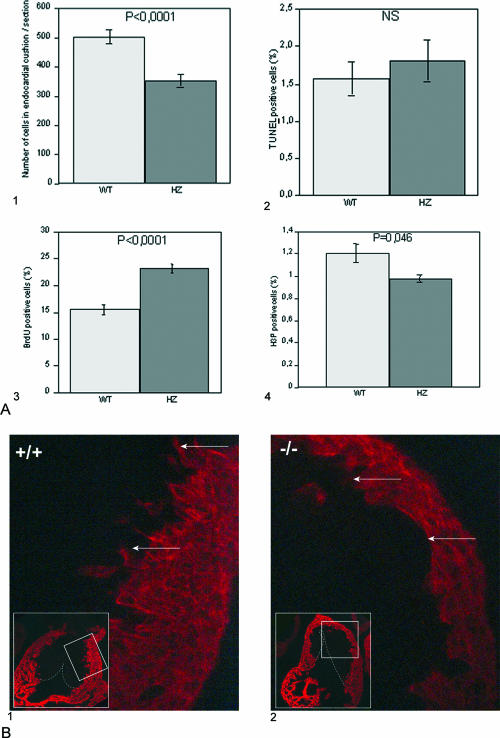
In order to analyze proliferation and apoptosis in the endocardial cushions, we examined serial sections of the OFT at E12.5. We estimated the total number of mesenchymal cells present in OFT cushions (13 sections by genotype), and we observed a clear reduction in the total number of cells in HZ mutants compared to their WT counterparts (Fig. 8A1). No significant difference in the number of TUNEL-positive cells between wild-type and mutant endocardial cushions was observed (Fig. 8A2). Proliferation was monitored by use of PCNA antibody, which revealed increased proliferation in the mutant samples (not shown). This was confirmed by BrdU incorporation analysis (Fig. 8A3), indicating that the number of cells in S phase is increased in the endocardial cushions of the mutants. On the contrary, anti-phosphohistone H3 showed a small but significant decrease in the number of cells in M phase in the mutant cushions (Fig. 8A4). Taken together, these results suggest that the cell cycle is lengthened in the mutant cells, probably due to an elongated S phase, and thus that mesenchymal cells in the cushions of the OFT of TL5 mutants divide more slowly than the wild-type cells.
FIG. 8.
Endocardial cushion development. (A) Proliferation and apoptotic death of mesenchymal cells in endocardial cushions. (A1) Total number of mesenchymal cells in endocardial cushion of WT or HZ TL5 E12.5 OFT. Cells were counted on sagittal paraffin sections stained by hematoxylin (13 sections per genotype). Apoptotic death was analyzed using TUNEL (A2), while proliferation was evaluated by either BrdU (A3) or phosphohistone H3 (H3P; A4) immunostaining. In each case, results were expressed as the mean of the ratios of the number of positive cells to the total number of mesenchymal cells per section. NS, no significant difference between WT and HZ embryos was observed by TUNEL assay (six sections per genotype). An important increase in the number of BrdU-positive cells (five sections per genotype) and a small but significant (P < 0.05) decrease in the number of phosphohistone H3-positive cells (four sections per genotype) were revealed in HZ embryos. These results were confirmed with two different WT or HZ Trapα mutant embryos. Statistical significance of difference was assessed by Student's t testing. (B) Myocardialization of the endocardial cushions. Myosin heavy chain immunostaining with MF20 antibody on frontal sections of OFT in E12.5 embryos revealed that while migration of myocardial cells is clearly observed in the WT (B1, arrows), it has not yet started in the mutant (B2, arrows).
The myocardialization of the cushions was investigated using the anti-myosin heavy chain antibody MF20. In the OFT from wild-type embryos, myocardialization was observed as soon as E12.5, when myocardial cells from the periphery were seen to invade the cushions (Fig. 8B1). In contrast, in the mutant embryos there were no or very few cells protruding from the myocardium towards the cushions (Fig. 8B2). At E13.5 (not shown), the differences between the two genotypes were accentuated. These results clearly show an important delay in myocardialization of the endocardial cushions in TL5 mutants.
DISCUSSION
The efficiency of the translocation of specific proteins across the endoplasmic reticulum membrane has been shown to depend on the involvement of the TRAP complex in a substrate-specific manner (10), providing the first demonstration of a direct role of TRAP in protein transport. Moreover, the authors of that study suggested the specific involvement of Trapα. This demonstration, as well as the majority of studies on protein translocation, was achieved using an artificial in vitro system. We present the first in vivo study of the impact of TRAPα deficiency. We show that (i) in mammalian cells, TRAPα is involved in the secretion of specific proteins; (ii) during mouse development, perturbing the function of TRAPα blocks specific events crucial for the formation of the cardiac outflow tract; and (iii) in skeletal muscle a slightly different TRAPα protein is translated after birth, when it replaces the ubiquitous one in that tissue.
Trapα expression in the mouse.
We have characterized the mouse Trapα cDNA and shown that, like β and γ subunits (25, 26), it is highly conserved among vertebrates, the main differences being in the signal sequence. Trapα is maternally supplied until the eight-cell stage and then constitutively expressed during mouse embryogenesis, as well as in the adult (see S1 in the supplemental material). In keeping with data obtained in human studies (16), we identified three different transcripts. However, in the mouse, one of them encodes a slightly different protein, which is specific for skeletal muscles and also present in the heart. This transcript starts to be expressed after birth, which suggests a requirement for a specific TRAPα protein in functional muscles. This specific protein has never been described. It would be interesting to study the other TRAP subunits in muscles. The level of expression of the ubiquitous form of TRAPα reported by the β-galactosidase varies among tissues, which is particularly obvious in the central nervous system (not shown). Similar observations had been reported, in zebra fish, for Trapβ, whose messengers are enriched in the notochord and polster (26).
The insertion of the construct in the Trapα gene leads to the synthesis of a protein that is lacking the 22 C-terminal residues and is fused to a hybrid ShBle-β-galactosidase protein. HT mice do not display any phenotype and reproduce normally, while HZ mice die around birth. Based on the studies of Fons et al. (10) and using transfected embryonic fibroblasts, we show that in TL5 mutant cells, secretion of preprolactin, whose translocation is TRAP independent, is not perturbed. On the contrary, we demonstrate that the exportation of two factors dependent on TRAP, γ-INF and ANP, was strongly impaired in these cells. Based on prolactin excretion, these results reveal that the presence of the large fusion protein probably does not interfere with the functioning of the translocon. On the other hand, they established that the modification of TRAPα is sufficient for preventing the functioning of the TRAP complex during translocation of specific proteins. Importantly, they confirmed in vivo the results obtained in reconstituted systems (10), mainly that TRAP improves the translocation efficiency of some substrates but not of others. We know that in HT mice, the presence of the fusion protein per se does not prevent the wild-type TRAP complex from functioning adequately since we do not observe any dominant negative phenotype. We have shown that the fusion protein in embryonic fibroblasts is located similarly to the wild-type TRAPα in the ER membrane. However, the mutant proteins form aggregates, probably due to the formation of β-galactosidase tetramers, and may thus segregate at least partially from the endogenous complexes. That could be enough to explain why there is no dominant negative phenotype, normal complexes being able to form in the HT cells at a level high enough to ensure TRAPα function.
Cardiac malformation in Trapα mutants.
Surprisingly, while Trapα expression is ubiquitous, the HZ embryos displayed an apparent phenotype only in the heart. The proximal outflow tract septum is always absent, which results in a persistent truncus arteriosus. In addition, the semilunar valves are often abnormal and present a reverse orientation. Although weak variations could be observed, all of the mutants displayed an identical phenotype resulting in a DORV.
In humans, congenital heart defects represent the largest class of birth defects, and abnormalities in septation and cardiac valve formation are associated with high mortality (6, 17). The mechanisms involved in the formation of the septum in the proximal part of the OFT are not well understood (see references 24 and 40). Endocardial cells as well as NC cells participate in the septation of the conotruncus (21). The endocardium undergoes EMT at the onset of endocardial cushion formation, when the endocardial cells invade the cardiac jelly, which is followed by migration of the NCC into the conotruncal ridges. Proliferation of the mesenchymal cells and cell apoptosis precede compaction and fusion of the ridges before myocardialization (35, 41). While the essential role of NCC in the distal septation of the OFT is very well established, their participation in septation of the proximal OFT is still controversial even though the use of transgenic mice expressing markers in NCC has shown their presence in endocardial ridges (18, 38). The loss of NCC by apoptosis has been proposed to be important for release or mobilization of factors involved in myocardialization (31). We observe that migration of NCC occurs in both the Trapα mutant and wild-type embryos, which is consistent with normal formation of the distal OFT septum. However, the proximal septum and the semilunar valves do not develop properly. Abnormal NCC migration clearly cannot account for such a phenotype. Defective interactions with adjoining endothelial and mesenchymal cells are probably an important feature in this phenotype.
Endocardial cushion maturation.
The molecular cues that determine the precise anatomical position and extent of the endocardial cushions are as yet unknown. In the mouse, endocardial cushion formation is complete by E12.5, whereas in TL5 mutants maturation of the cushions is still ongoing at E13.5 and valve formation is compromised. Moreover, a dysplasia of the cushions, especially of the dorsal cushion, is always observed in E12.5 mutants. We have shown that apoptosis is not perturbed in the mutant but that the proliferation of mesenchymal cells is affected. In the WT, myocardialization starts at E12.5, when cushions are fusing in the proximal part of the OFT and while proliferation decreases. At the same stage, in TL5 mutants, mesenchymal cells continue to proliferate within the cushions. However, the total number of these cells is lower in the mutant. The lengthening of the S phase of the cell cycle suggested by the comparison of results obtained with either BrdU or anti-histone H3 could explain that apparent discrepancy. Maybe, due to the small number of mesenchymal cells in the E12.5 OFT, timely fusion of the cushions does not happen, resulting in impairment of inductive interactions between mesenchymal cells and the myocardium. Developmental mistiming thus could be the main reason for the abnormal phenotype. How is this observation linked to the putative function of TRAPα? One plausible hypothesis is that the abnormal proliferation rate of the mesenchymal cells is the consequence of an inadequate secretion of signal(s) during cushion formation and/or maturation.
Signaling pathways involved in morphogenesis of the OFT have begun to be identified as a result of in vitro experiments using explants from chick embryos as well as analyses of mouse mutants. The latter have revealed that transforming growth factor β (TGF-β) families play complex roles in EMT as well as in cell migration, extension, and differentiation of the endocardial cushions in birds or mammals (see reviews in references 1 and 7).
In contrast to that of most mutants displaying a cardiac phenotype, the phenotype of Trapα mutants is fully penetrant, and the defects are limited to the proximal part of the OFT. Periostin expression in these mutants indicates that EMT does occur but that maturation of the cushion is clearly retarded. Similarly to TL5 embryos, TGF-β2 null mutants display cushions that fail to condense and to thin out to form mature valve leaflets; these defects have been related to delayed myocardialization (2, 33). In addition, a case of common arterial trunk in TGF-β2-deficient mice, with abundant NCC present in the region, has already been described (11). We examined the level of expression of TGF-β2 in TL5 embryos as well as the rate of excretion of this factor by embryonic fibroblasts (not shown); we did not find any difference from the wild type.
TRAPα and heart phenotype.
The mutation introduced in TL5 modified both the Trapαg and the Trapαm messengers; however, we can exclude that the muscle- and heart-specific TRAPαm mutated proteins play a role in the TL5 phenotype as it is only expressed after birth. The fact that the mutation, introduced in the ubiquitous TRAPαg protein, results in a very specific phenotype limited to the heart is in agreement with the proposition that TRAPα is involved in facilitating the translocation of specific proteins rather than playing an obligatory role in translocation (10, 27). We cannot exclude some other function of TRAPα linked to the C-terminal truncated part; however, such a function has never been described. It has been reported that both TRAPα and Calnexin are calcium-binding proteins (37). We cannot exclude the possibility that, despite the fact that the luminal part of TRAPα is still present, the fixation of calcium and whatever function it may have could be perturbed, e.g., because of the formation of TRAPα aggregates in the ER membrane. The involvement of TRAPα in calcium homeostasis has never been established. As regards calnexin, which seems partly aggregate with TRAPα in the mutant cells, it has been proposed to be essentially involved in retaining defective proteins in the ER, and calnexin knockout mice do not display any cardiac phenotype (8). Perturbation of secretion or integration of some unknown excreted factors or membrane proteins necessary for maturation of the endocardial cushions seems to us the more probable cause of the phenotype of the Trapα mutants.
The large number of such proteins involved in OFT cushion development makes it very difficult to identify those affected by the Trapα mutation. In addition, differences in the level of integration or secretion could be subtle enough to alter the phenotype while escaping our current technical tools. Recently, the expression of Trapγ has been abolished in Xenopus by using morpholinos (25), causing failure of tubulogenesis during pronephros development. These observations, taken together with our results, could suggest that the TRAP subunits play individual roles during development, with TRAPα being required during heart morphogenesis while TRAPγ is necessary for kidney development. However, perturbation of either one of the subunits could result in the decrease or absence of function of the TRAP complex. The studies have been carried out in different species; it cannot be excluded that the defective translocation of factors dependent on TRAP is more detrimental in the pronephros in Xenopus, while it would be in the mouse heart. Additional experiments, such as inactivating another TRAP subunit in the mouse, should be carried out to address that question.
Identifying precisely the cells responsible for the TL5 phenotype could help to identify candidates whose translocation is decreased in the absence of a fully functional TRAP complex. Conditional mutagenesis using the Cre/Lox system is in progress and should yield information about the cell population involved in producing the signal(s) absent in Trapα mutants. In addition, it will permit us to address the role of muscle-specific TRAPα.
In conclusion, our results show that the α subunit of the TRAP complex is constitutively expressed during mouse development as well as in the adult. They reveal that a messenger encoding a different TRAPα protein is translated specifically in muscles after birth, while the general form is turned off in this tissue. Our observations establish that a fully functional TRAPα subunit is required for remodeling the endocardial cushions in the OFT, providing the first in vivo demonstration that TRAPα plays a crucial role during mammalian heart development. In addition, TL5 mice provide a model in which to study late events in the patterning of the proximal outflow tract.
Supplementary Material
Acknowledgments
We acknowledge Tom A. Rapoport for the gift of the C-terminal TRAPα antibody and Claire Soudais for the IFN-γ cDNA. We are very grateful to Robert Kelly, Stéphane Zaffran, and Fanny Bajolle, for precious advice and helpful discussions. We thank Laurence Fiette and Yves Pierre for technical advice. The monoclonal antibodies 40-1A and MF20, developed by J. R. Sanes and D. A. Fischman, respectively, were obtained from the DSHB, University of Iowa, Iowa City. We thank M. Buckingham, R. Kelly, and J.-L. Popot for critical reading of the manuscript.
This work was supported by the Pasteur Institute and CNRS.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Azhar, M., J. Schultz Jel, I. Grupp, G. W. Dorn II, P. Meneton, D. G. Molin, A. C. Gittenberger-de Groot, and T. Doetschman. 2003. Transforming growth factor beta in cardiovascular development and function. Cytokine Growth Factor Rev. 14:391-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartram, U., D. G. M. Molin, L. J. Wisse, A. Mohamad, L. P. Sandford, T. Doestchman, C. P. Speer, R. E. Poelmann, and A. C. Gittenberger-de Groot. 2001. Double-outlet right ventricle and overriding tricuspid valve reflect disturbances of looping, myocardialization, endocardial cushion differentiation, and apoptosis in TGFbeta(2)-knockout mice. Circulation 103:2745-2752. [DOI] [PubMed] [Google Scholar]
- 3.Beckmann, R., D. Bubeck, R. Grassucci, P. Penczek, A. Verschoor, G. Blobel, and J. Frank. 1997. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science 278:2123-2126. [DOI] [PubMed] [Google Scholar]
- 4.Brown, C. B., L. Feiner, M. M. Lu, J. Li, X. Ma, A. L. Webber, L. Jia, J. A. Raper, and J. A. Epstein. 2001. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development 128:3071-3080. [DOI] [PubMed] [Google Scholar]
- 5.Camus, A., C. Kress, C. Babinet, and J. Barra. 1996. Unexpected behavior of a gene trap vector comprising a fusion between the Sh ble and lacZ genes. Mol. Reprod. Dev. 45:255-263. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C.-P., J. R. Neilson, J. H. Bayle, J. E. Gestwicki, A. Kuo, K. Stankunas, I. A. Graef, and G. R. Crabtree. 2004. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell 118:649-663. [DOI] [PubMed] [Google Scholar]
- 7.Délot, E. C. 2003. Control of endocardiac cushion and cardiac valve maturation by BMP signaling pathways. Mol. Genet. Metab. 80:27-35. [DOI] [PubMed] [Google Scholar]
- 8.Denzel, A., M. Molinari, C. Trigueros, J. E. Martin, S. Velgurman, S. Brown, G. Stamp, and M. J. Owen. 2002. Early postnatal death and motor disorders in mice congenitally deficient in calnexin expression. Mol. Cell. Biol. 22:7398-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fananapazir, K., and M. H. Kaufman. 1988. Observations on the development of the aortico-pulmonary spiral septum in the mouse. J. Anat. 158:157-172. [PMC free article] [PubMed] [Google Scholar]
- 10.Fons, R. D., B. A. Bogert, and R. S. Hegde. 2003. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 160:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gittenberger-de Groot, A. C., M. M. Bartelings, A. J. J. C. Bogers, M. J. Boot, and R. E. Poelmann. 2002. The embryology of the common arterial trunk. Prog. Pediatr. Cardiol. 15:1-8. [Google Scholar]
- 12.Görlich, D., and T. A. Rapoport. 1993. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75:615-630. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann, E., D. Görlish, S. Kostka, A. Otto, and R. Kraft. 1993. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur. J. Biochem. 214:375-381. [DOI] [PubMed] [Google Scholar]
- 14.Harvey, R. P. 2002. Patterning the vertebrate heart. Nat. Rev. Genet. 3:544-556. [DOI] [PubMed] [Google Scholar]
- 15.Hegde, R. D., S. Voigt, T. A. Rapoport, and V. R. Lingappa. 1998. TRAM regulates the exposure of nascent secretory proteins to the cytosol during translocation into the endoplasmic reticulum. Cell 92:621-631. [DOI] [PubMed] [Google Scholar]
- 16.Hirama, T., C. W. Miller, and H. P. Koeffler. 1999. Translocon-associated protein alpha transcripts are induced by granulocyte-macrophage colony-stimulating factor and exhibit complex alternative polyadenylation. FEBS Lett. 455:223-227. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman, J. I. 1995. Incidence of congenital heart disease. II. Prenatal incidence. Pediatr. Cardiol. 16:155-165. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, X., D. H. Rowitch, P. Soriano, A. P. McMahon, and H. M. Sucov. 2000. Fate of the mammalian cardiac neural crest. Development 127:1607-1616. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, A. E., and M. A. van Weas. 1999. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15:799-842. [DOI] [PubMed] [Google Scholar]
- 20.Jungnickel, B., and T. A. Rapoport. 1995. A posttargeting signal sequence recognition event in the reticulum membrane. Cell 82:261-270. [DOI] [PubMed] [Google Scholar]
- 21.Kinazuki, Y. Y., R. E. Hammer, J.-I. Miyazaki, S. C. Williams, J. A. Richardson, and M. Yanagisawa. 2001. Tie2-cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230:230-242. [DOI] [PubMed] [Google Scholar]
- 22.Kirby, M. L., T. Gale, and D. Stewart. 1983. Neural crest cells contribute to normal aorticopulmonary septation. Science 220:1059-1061. [DOI] [PubMed] [Google Scholar]
- 23.Kruzynska-Frejtag, A., M. Machnicki, R. Rogers, R. R. Markwald, and S. J. Conway. 2001. Periostin (an osteoblast-specific factor) is expressed within the embryonic mouse heart during valve formation. Mech. Dev. 103:183-188. [DOI] [PubMed] [Google Scholar]
- 24.Lamers, W. H., and A. F. M. Moorman. 2002. Cardiac septation. A late contribution of the embryonic primary myocardium to heart morphogenesis. Circ. Res. 91:93-103. [DOI] [PubMed] [Google Scholar]
- 25.Li, D.-H., T. Chan, R. Satow, S. Komazaki, K. Hashizume, and M. Asashima. 2005. The role of XTrap-γ in Xenopus pronephros development. Int. J. Dev. Biol. 49:401-408. [DOI] [PubMed] [Google Scholar]
- 26.Mangos, S., R. Krawetz, and G. M. Kelly. 2000. The translocon-associated protein β (TRAPβ) in zebrafish embryogenesis. I. Enhanced expression of transcripts in notochord and hatching gland precursors. Mol. Cell. Biochem. 215:93-101. [DOI] [PubMed] [Google Scholar]
- 27.Ménétret, J.-F., R. S. Hedge, S. U. Heinrich, P. Chandramouli, S. J. Ludtke, T. A. Rapoport, and C. W. Akey. 2005. Architecture of the ribosome-channel complex derived from native membranes. J. Mol. Biol. 348:445-457. [DOI] [PubMed] [Google Scholar]
- 28.Ménétret, J.-F., A. Neuhof, D. G. Morgan, K. Plath, M. Radermacher, and C. W. Akey. 2000. The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell 6:1219-1232. [DOI] [PubMed] [Google Scholar]
- 29.Migliaccio, G., C. V. Nicchita, and G. Blobel. 1992. The signal sequence receptor, unlike the signal recognition particle receptor, is not essential for protein translocation. J. Cell Biol. 117:15-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moorman, A. F. M., and V. M. Christoffels. 2003. Cardiac chamber formation: development, genes, and evolution. Physiol. Rev. 83:1223-1267. [DOI] [PubMed] [Google Scholar]
- 31.Poelmann, R. E., T. Mirakawa, and A. C. Gittenberger-de Groot. 1998. Neural crest cells in outflow tract septation of the embryonic chick heart: differentiation and apoptosis. Dev. Dyn. 212:373-384. [DOI] [PubMed] [Google Scholar]
- 32.Rapoport, T. A., B. Jungnickel, and U. Kutay. 1996. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem. 65:271-303. [DOI] [PubMed] [Google Scholar]
- 33.Sanford, L. P., I. Ormsby, A. C. Gittenberger-de Groot, H. Sariola, R. Friedman, G. P. Boivin, E. L. Cardell, and T. Doestchman. 1997. TGFβ2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development 124:2659-2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Berg, B., W. M. Clemons, Jr., I. Collinson, Y. Modis, E. Hartmann, S. C. Harrison, and T. A. Rapoport. 2003. X-ray structure of a protein-conducting channel. Nature 427:36-44. [DOI] [PubMed] [Google Scholar]
- 35.van den Hoff, M. J. B., A. F. M. Moorman, J. M. Ruijter, W. H. Lamers, R. W. Bennington, R. R. Markwald, and A. Wessels. 1999. Myocardialization of the cardiac outflow tract. Dev. Biol. 212:477-490. [DOI] [PubMed] [Google Scholar]
- 36.Voigt, S., B. Jungnickel, E. Hartmann, and T. A. Rapoport. 1996. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J. Cell Biol. 134:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wada, I., D. Rindress, P. H. Cameron, W. J. Ou, J. J. Doherty, D. Louvard, A. W. Bell, D. Dignard, D. Y. Thomas, and J. J. M. Bergeron. 1991. SSRα and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J. Biol. Chem. 266:19599-19610. [PubMed] [Google Scholar]
- 38.Waldo, K., C. W. Lo, and M. L. Kirby. 1999. Connexin 43 expression reflects neural crest patterns during cardiovascular development. Dev. Biol. 208:307-323. [DOI] [PubMed] [Google Scholar]
- 39.Waldo, K., S. Miyagawa-Tomita, D. Kumiski, and M. L. Kirby. 1998. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Dev. Biol. 196:129-144. [DOI] [PubMed] [Google Scholar]
- 40.Webb, S., S. R. Qayyum, R. H. Anderson, W. H. Lamers, and M. K. Richardson. 2003. Septation and separation within the outflow tract of the developing heart. J. Anat. 202:327-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ya, J., M. J. B. van den Hoff, P. A. J. de Boer, S. Tesink-Taekama, D. Franco, A. F. M. Moorman, and W. H. Lamers. 1998. Normal development of the outflow tract in the rat. Circ. Res. 82:464-472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.