Abstract
AML1 (RUNX1) regulates hematopoiesis, angiogenesis, muscle function, and neurogenesis. Previous studies have shown that phosphorylation of AML1, particularly at serines 276 and 303, affects its transcriptional activation. Here, we report that phosphorylation of AML1 serines 276 and 303 can be blocked in vivo by inhibitors of the cyclin-dependent kinases (CDKs) Cdk1 and Cdk2. Furthermore, these residues can be phosphorylated in vitro by purified Cdk1/cyclin B and Cdk2/cyclin A. Mutant AML1 protein which cannot be phosphorylated at these sites (AML1-4A) is more stable than wild-type AML1. AML-4A is resistant to degradation mediated by Cdc20, one of the substrate-targeting subunits of the anaphase-promoting complex (APC). However, Cdh1, another targeting subunit used by the APC, can mediate the degradation of AML1-4A. A phospho-mimic protein, AML1-4D, can be targeted by Cdc20 or Cdh1. These observations suggest that both Cdc20 and Cdh1 can target AML1 for degradation by the APC but that AML1 phosphorylation may affect degradation mediated by Cdc20-APC to a greater degree.
The AML1 proteins, including AML1a, AML1b, and AML1c (AML1c is also known as AML1B), are generated from one gene by alternative splicing (22). This gene has been given the names RUNX1, AML1, CBFA2, and PEBP2αB (37). The AML1 protein is composed of a DNA binding runt homology domain located in the amino terminus followed by a transcriptional activation domain and a negative regulatory C-terminal domain (15, 20, 29). AML1 was initially identified during the study of breakpoint t(8;21), which is a common chromosomal translocation in acute myeloid leukemia (23). The association of AML1 with blood cell development is shown by the disruption of the AML1 gene through multiple chromosomal translocations, deletions, point mutations, or amplification in approximately 30% of human myeloid leukemias and myelodysplastic syndrome patients and a significant number of lymphoid leukemia patients (21, 24, 27, 32). Furthermore, no detectable definitive hematopoiesis is observed in Aml1 knockout mice (26, 40). The importance of AML1 in nonhematopoietic cells has also been recognized in angiogenesis, muscle function, and neurogenesis (6, 12, 14, 35, 41).
AML1 is detected as a serine and threonine phosphorylated protein (9). Previous work has suggested that AML1 activity may be regulated by phosphorylation (34, 45). Phosphorylation at specific serine-proline or threonine-proline sites in AML1 appears to be necessary for normal activity (45). It has also been suggested that phosphorylation releases AML1 from an association with the nuclear matrix mediated by sin3A, in turn leading to both increased activity and an increased rate of degradation (11).
We have now shown that AML1 phosphorylation by cyclin-dependent kinases (CDKs) affects the overall stability of AML1 as well as the ability of certain ubiquitin ligase complexes, such as Cdc20-anaphase-promoting complex (APC), to target AML1 for degradation.
MATERIALS AND METHODS
Cell culture and treatment.
293T and NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin-streptomycin at 37°C in 5% CO2. Stable NIH 3T3 cell pools expressing wild-type or mutant AML1 were established by cloning AML1 coding sequences into pMSCV-puro vectors. NIH 3T3 cells were spin infected with empty mouse stem cell virus (MSCV) vector as a control or with vector expressing wild-type or mutant AML1. Two days after infection, the cells were subjected to 5 days of selection in 2 μg/ml puromycin. 293T cells were transfected using Polyfect (QIAGEN, Valencia, CA). AEL-ΔR1 endothelial lines and 293T lines expressing AML1 were established in the same manner. Nocodazole (Sigma-Aldrich, St. Louis, MO) was added to the 293T cells 8 to 16 h after transfection at a concentration of 1 μg/ml. Roscovitine, alsterpaullone, and Cdk2 inhibitor II (Calbiochem, San Diego, CA) were added to 293T cells at a final concentration of 50 μM 8 to 16 h after transfection. Cdk4 inhibitor (Calbiochem) was added to a final concentration of 5 μM. To label proteins with [32P]orthophosphate, 24 h after transfection cells were placed in phosphate-free Dulbecco's modified Eagle's medium (Cellgro, Herndon, VA) supplemented with 10% dialyzed fetal bovine serum and HEPES buffer, pH 7.5. A total of 100 μCi per ml of [32P]orthophosphate was added to the medium, and the cells were incubated for 4 to 6 h.
Plasmids.
Full-length AML1B was cloned into the HindIII site of pFLAG-CMV2 (Invitrogen, Carlsbad, CA) to generate FLAG-tagged AML1. To generate the glutathione S-transferase (GST)-AML1(267-315) mammalian expression construct, AML1B coding sequences from amino acids 267 to 315 were amplified by PCR and then cloned into the EcoRI and NotI sites of pGEX-4T-2 (Promega, Madison, WI). A BstBI-NotI fragment from this plasmid was then cloned into the pEBG vector, which contains the human elongation factor EF-1α promoter to drive expression (a gift from B. Mayer). BstBI cuts within the GST coding sequence. pcDNA3-AML1 (wild type and 4A) have been described previously (45). Similar expression vectors for AML1 containing two mutations (serines 276 and 303 or serine 293 and threonine 300) or single mutations were generated in the same manner as described previously (45). To construct GST-AML1(267-315) mammalian expression vectors with mutations, PCR was used to amplify amino acids 267 to 315 coding the sequence from full-length AML1 DNA carrying the appropriate mutations. These sequences were fused to GST and cloned into pEBG as described above. The pcDNA3-HA-Cdh1 (where HA is hemagglutinin) expression plasmid was received from Michele Pagano (New York University), the pCS2-HA-Cdc20 plasmid was received from Guowei Fang (Stanford University), and the pcDNAB-Myc-Skp2 plasmid was from Xiaohua Wu (The Scripps Research Institute).
Cell labeling, immunoprecipitation, Western blotting, and Northern blotting.
Labeling of cells with [32P]orthophosphate and immunoprecipitation of FLAG-tagged AML1 were performed as described previously (45) and as above. GST-AML1(267-315) protein was purified from the lysate of 32P-labeled cells in a similar fashion, but glutathione-S agarose was used. Western blotting was performed as described previously (4). Phospho-AML1 antibodies were raised against AML1 peptides containing either phosphor-S276 or phosphor-S303 (Biosource) (39). Total RNA was prepared from cells using RNA-Bee (Tel-Test, Inc.) and the manufacturer's protocol. Northern blotting was performed as described previously (45).
In vitro kinase assays.
GST-AML1(267-315) constructs (wild type and mutants) were expressed in Escherichia coli from pGEX-4T-2 (Pfizer-Pharmacia, New York, NY) and bound to glutathione agarose. The GST-AML1 substrate was incubated in kinase buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 2 mM dithiothreitol, 10 μM ATP) with [γ-32P]ATP and purified active Cdk1/cyclin B (Calbiochem), Cdk2/cyclin A (Upstate Biotechnologies), Cdk6/cyclin D3 (Upstate Biotechnologies), or Cdk4/cyclin D1 (Biaffin GmbH and Co. KG, Kassel, Germany). The retinoblastoma fusion protein used as a control for Cdk4/cyclin D1 activity was purchased from Santa Cruz Biotechnology. After incubation for 10 min at 30°C, the in vitro kinase reaction mixtures were placed on ice and then washed three times with phosphate-buffered saline (PBS). The GST-AML1(267-315) on glutathione beads was then prepared for analysis on sodium dodecyl sulfate (SDS) gels by boiling in SDS sample buffer.
Coimmunoprecipitation.
Lysates were prepared following transfection of 293T cells expressing either AML1, AML1-2xDBM (AML1 with a mutation in two D box sequences), AML1-4A, or AML1-4D with or without myc-Skp2, HA-Cdc20, or HA-Cdh1 in 400 μl of PBS-1 mM EDTA-0.5% Triton X-100; samples were sonicated twice for 6 s, and debris was removed by centrifugation at 17.000 × g for 30 min at 4°C. A total of 200 μg of lysate was immunoprecipitated overnight with either the anti-myc or anti-HA antibody with 15 μl of a 50% slurry of protein G-Sepharose in PBS. Following washes with the lysis buffer, the beads were boiled in 10 μl of 2× Laemmli buffer, resolved on an 8 to 10 to 12% gradient SDS-PAGE gel, electroblotted to nitrocellulose, and blocked in 4% milk-PBS-0.2% Tween 20. The blots were hybridized first with rabbit anti-AML1 and donkey anti-rabbit antibody-horseradish peroxidase, and detection was performed by chemiluminescence (NEN). Following stripping with 2% SDS-PBS-0.2% Tween 20 for 20 min at 50°C, the blots were washed for 30 min with various exchanges of wash buffer PBS-0.2% Tween 20 and blocked as above; the blots were then hybridized with either mouse anti-myc (clone 9E10) or anti-HA (Babco) and donkey anti-mouse antibody-horseradish peroxidase, and detection was performed as above. The input samples were 10 μg of total protein of each sample resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and manipulated as described above for the detection of AML1, myc-Skp2, HA-Cdc20, or HA-Cdh1.
RESULTS
CDK inhibitors block AML1 phosphorylation in vivo.
Since we used various AML1B expression constructs in this study, the numbering of AML1 amino acids in this report is that used for the AML1B protein. Numerous AML1 phosphorylation sites have been identified previously (34, 45), but the kinases that phosphorylate AML1 in vivo have not been definitively identified. A previous report suggested that AML1 was phosphorylated by extracellular signal-related kinase (ERK) upon serum starvation followed by epidermal growth factor stimulation (34). However, our initial experiments (in which the MEK inhibitor U0156 blocked ERK activity but failed to inhibit AML1 phosphorylation) suggested that not all AML1 phosphorylation was due to ERK activity (data not shown). We were especially interested in the identification of the kinase that phosphorylates AML1 serines 276, 293, and 303 and threonine 300. These phosphorylation sites have been shown to affect AML1 transcriptional activity (34, 45). All 13 known AML1 phosphorylation sites are serine or threonine residues followed by a proline residue (45). (S/T)P is the core consensus sequence for both ERK and CDKs (8, 25, 36). This suggests the possibility that CDKs may be responsible for some AML1 phosphorylation.
To determine whether CDKs phosphorylate AML1 in vivo, we first transfected 293T cells with AML1 and then treated the cells with the CDK inhibitor roscovitine. Roscovitine inhibits the activity of Cdk1, Cdk2, and Cdk5 but not ERK (1). As shown in Fig. 1A, roscovitine inhibits in vivo phosphorylation of full-length AML1.
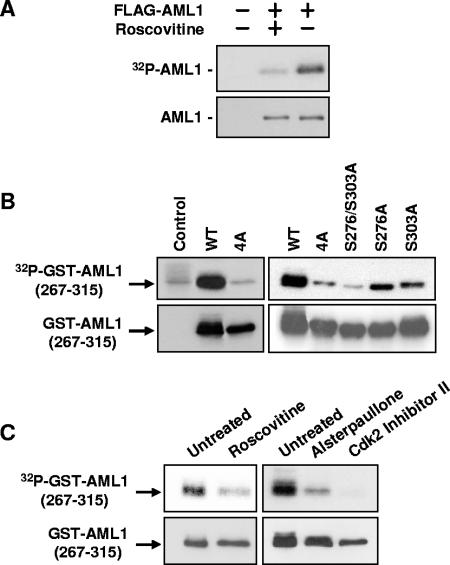
FIG. 1.
Phosphorylation of AML1 is inhibited by CDK inhibitors. (A) 293T cells were transfected with full-length FLAG-AML1 and then split into two samples. One sample was treated for approximately 20 h with 30 μM roscovitine. Nontransfected cells served as a control. During the last 4 to 6 h of roscovitine treatment, all samples were labeled with [32P]orthophosphate. The FLAG-AML1 was then immunoprecipitated with anti-FLAG agarose and used for Western blotting with anti-AML1 antibodies and for autoradiography. (B) Wild-type GST-AML1(267-315), GST-AML1(267-315)-4A (serines 276, 293, and 303 and threonine 300 mutated to alanine), GST-AML1(267-315)-S276/S303A (serines 276 and 303 mutated to alanine), or GST-AML1(267-315) with single mutations (serine 276 to alanine and serine 303 to alanine) were transfected into 293T cells, as indicated above the lanes. The far left lane shows nontransfected control cells. All cells were labeled with 32P, and the GST-AML1(267-315) was isolated using glutathione agarose. The GST-AML1(267-315) was then used for Western blotting with anti-GST antibodies and for autoradiography. (C) 293T cells were transfected with GST-AML1(267-315), split into separate samples, and then treated with CDK inhibitors as indicated above the lanes. Sixteen hours after treatment with CDK inhibitors, all samples were labeled using [32P]orthophosphate (CDK inhibitor concentrations were maintained during labeling). The GST-AML1(267-315) was pulled down using glutathione agarose and subjected to Western blotting with anti-GST antibodies and for autoradiography.
To examine the effect of roscovitine and other CDK inhibitors on phosphorylation of AML1 serines 276, 293, and 303 and threonine 300, a GST-AML1 fusion protein containing AML1 amino acids 267 to 315 was transfected into 293T cells. Figure 1B shows that mutation of serines 276, 293, and 303 and threonine 300 eliminates all in vivo phosphorylation of GST-AML1(267-315). The control lane, without GST-AML1(267-315), shows the presence of a small amount of phosphorylated contaminant that migrates at the same position in the gel as GST-AML1(267-315). The vast majority of the in vivo phosphorylation that occurs on GST-AML1(267-315) occurs on serines 276 and 303. Mutation of these two sites reduces phosphorylation to background levels. The presence of either serine 276 or serine 303 restores some phosphorylation but not to full wild-type levels. To test the CDK inhibitors, 293T cells were transfected with wild-type GST-AML1(267-315). Twenty-four hours after transfection, the cells were split into separate samples and treated with roscovitine, alsterpaullone (Cdk1-specific inhibitor), or a Cdk2-specific inhibitor (Fig. 1C). All inhibitors blocked the phosphorylation of GST-AML1(267-315).
The data shown in Fig. 1 suggest that CDK inhibitors block phosphorylation of AML1 serines 276 and 303. To confirm this, Western blotting was performed using antibodies that specifically recognize AML1 phosphorylated on serine 276 or serine 303 (see Materials and Methods). The phosphospecific AML1 antibodies were used to examine phosphorylation of wild-type AML1 (FLAG-tagged). FLAG-AML1 was immunoprecipitated from transfected 293T cells; the immunoprecipitates were then used for Western blotting with anti-AML1 (P-303) or anti-AML1 (P-276). As shown in Fig. 2, treatment with CDK inhibitors blocks phosphorylation of both serine 279 and serine 303.
FIG. 2.
Phosphorylation of AML1 serines 276 and 303 is blocked by CDK inhibitors. 293T cells were transfected with FLAG-AML1 and then divided. One sample was left untreated, while the others were treated with 100 μM roscovitine, 50 μM alsterpaullone, or 50 μM Cdk2 inhibitor II, as indicated above the lanes. Nontransfected 293T cells were used as a control. Twenty hours after treatment, all samples were lysed and FLAG-AML1 was immunoprecipitated with anti-FLAG-agarose. Each sample of immunoprecipitated FLAG-AML1 was then used for Western blotting with anti-serine 276-phosphorylated AML1 (PS278-AML1) antibodies, anti-serine 303-phosphorylated AML1 (PS303-AML1) antibodies, and anti-AML1 antibodies.
Cdk1/cyclin B, Cdk2/cyclin A2, and Cdk6/cyclin D3 phosphorylate AML1 in vitro, but Cdk4/cyclin D1 does not.
The fact that inhibitors of Cdk1 and Cdk2 block AML1 phosphorylation at serines 276 and 303 strongly indicates that these residues of AML1 are substrates for these kinases in vivo. To confirm that these residues of AML1 can serve as substrates for CDKs, GST-AML1(267-315) was used as a substrate for in vitro kinase reactions with purified active Cdk1/cyclin B, Cdk2/cyclin A2, Cdk6/cyclin D3, or Cdk4/cyclin D1. Figure 3A shows that the first three CDK/cyclin complexes were able to phosphorylate wild-type GST-AML1(267-315) in vitro, but when GST-AML1(267-315)-4A was used as a substrate, only slight background phosphorylation was observed.
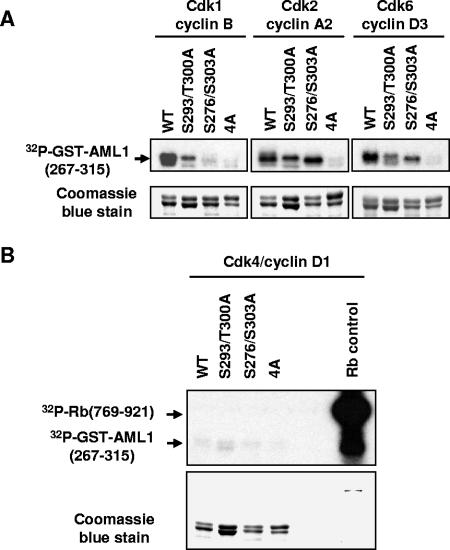
FIG. 3.
Cdk1/cyclin B, Cdk2/cyclin A, and Cdk6/cyclin D phosphorylate GST-AML1(267-315) in vitro, but Cdk4/cyclin D1 does not. (A) Wild-type GST-AML1B(267-315), GST-AML1B(267-315) with serine 293 and threonine 300 mutations, GST-AML1B(267-315) with serine 276 and 303 mutations, and GST-AML1B(267-315) with mutations of serines 276, 293, and 303 and threonine 300(4A) were expressed in 293T cells and bound to glutathione agarose. The agarose was incubated in kinase buffer with [γ-32P]ATP and either purified active Cdk1/cyclin B, Cdk2/cyclin A2, or Cdk6/cyclin D, as indicated above the lanes. After incubation for 10 min at 30°C, the GST-AML1(267-315) was boiled off the agarose in SDS sample buffer and run on an SDS polyacrylamide gel, which was stained with Coomassie blue to verify loading of proteins. The gel was then dried and used for autoradiography. (B) GST-AML1 substrates were incubated as above with purified active Cdk4/cyclin D1 along with a fusion protein containing amino acids 769 to 921 of the retinoblastoma protein as a positive control for Cdk4/cyclin D1 activity. All substrates were then analyzed as described for panel A.
When GST-AML1(267-315) proteins with a subset of sites mutated (serines 276 and 303 to alanine or serine 293 and threonine 300 to alanine) were used as substrates, more variable results were obtained. Incubation with Cdk1/cyclin B resulted in some phosphorylation of the S293/T300A substrate but much less than that observed for wild-type substrate. Very little phosphorylation of the S276/303A substrate was detected. This indicates that serines 276 and 303 are the preferred substrates for Cdk1/cyclin B but that all four residues must be present to achieve maximum levels of phosphorylation. Cdk2/cyclin A utilized all substrates except the 4A protein, although it appeared to exhibit some preference for the S293/T300 sites. Cdk6/cyclin D also utilized all substrates except 4A, although again the presence of all four sites was required for maximal phosphorylation.
In contrast to the first three CDK/cyclin complexes tested, Cdk4/cyclin D1 phosphorylation of wild-type GST-AML1(267-315) was not above the background phosphorylation observed for GST-AML1(267-315)-4A. This was not due to lack of Cdk4/cyclin D1 activity, because a control substrate (a fusion protein containing amino acids 769 to 921 of the retinoblastoma protein) was highly phosphorylated (Fig. 3B).
Although all four residues (serines 276, 293, and 303 and threonine 300) appear to be sites for phosphorylation by some CDK/cyclin complexes in vitro, different CDK/cyclin complexes exhibit subtle preferences for particular substrates. The presence of all four sites in the wild-type substrate often results in the highest level of phosphorylation, suggesting the possibility of cooperative interaction between sites. Our observations also suggest that serines 276 and 303 are much more highly phosphorylated in vivo. This may reflect the fact that in vitro phosphorylation conditions are more permissive than in vivo conditions. As discussed below, serine 293 and/or threonine 300 do appear to be weakly phosphorylated in vivo.
Phosphorylation of AML1 as cells progress through the cell cycle and cross talk between phosphorylation sites.
As cells progress from the G1 phase of the cell cycle to S phase, Cdk2/cyclin E is replaced by Cdk2/cyclin A, while in G2 Cdk1/cyclin B and Cdk1/cyclin A are active (25). We examined phosphorylation of GST-AML1(267-315) as cells progressed through the cell cycle to determine whether specific CDK/cyclin complexes have greater AML1 phosphorylation activity.
To synchronize cells expressing GST-AML1(267-315), 293T cells were transfected with GST-AML1(267-315) and treated for 16 to 20 h with hydroxyurea (G1/S block) or with nocodazole (G2/M block). The cells were released from the block when the medium containing hydroxyurea or nocodazole was removed and replaced by fresh medium. Samples were collected 0, 4, and 8 h after removal of the blocks, and synchronization was confirmed by labeling with propidium iodide and by flow cytometry (Fig. 4A). To examine GST-AML1(267-315) phosphorylation, cells were labeled with [32P]orthophosphate for 4 h before the collection of time points. GST-AML1(267-315) was then isolated from the 32P-labeled cells and analyzed by Western blotting and autoradiography. As shown in Fig. 4B, there is a slight decrease in GST-AML1(267-315) phosphorylation in cells arrested with hydroxyurea and a slight increase when they reenter the cell cycle. This is consistent with the observation that hydroxyurea activates a cell cycle checkpoint which involves down-regulation of Cdk2 activity (7). The relatively small decrease in GST-AML1(267-315) phosphorylation in hydroxyurea-treated cells may be due to the fact that some Cdk2 activity remains (7) or to low levels of phosphatases which might dephosphorylate GST-AML1(267-315). Overall, the changes in GST-AML1(267-315) phosphorylation as cells progress from late G1 to G2/M are not drastic. This relatively steady level of phosphorylation over the cell cycle is similar to that observed for other transcription factors believed to be CDK/cyclin substrates (19, 31) and involved in the regulation of cell proliferation (19). It is thus possible that cells maintain a fairly even level of total CDK/cyclin activity during the cell cycle, even though individual CDK/cyclin complexes appear and disappear.
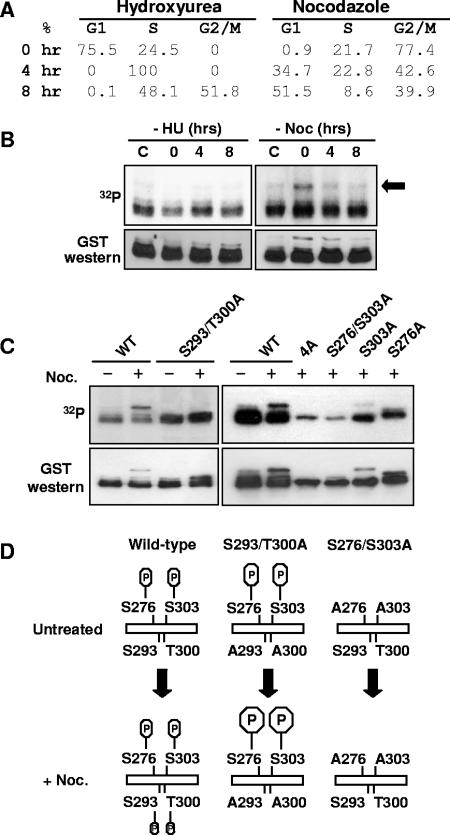
FIG. 4.
Cross talk between AML1 phosphorylation sites. (A) Cell cycle state of 293T cells synchronized with hydroxyurea or nocodazole. 293T cells were treated for 16 to 24 h with either 2 mM hydroxyurea or 0.1 mg/ml nocodazole or left untreated. Cells were collected immediately after removal of the hydroxyurea (HU) or nocodazole (Noc) (0 h) and after 4 and 8 h of culture. All cell samples were then fixed in ethanol, stained with propidium iodide, and analyzed by flow cytometry. The percentages of cells in the different phases of cell cycle are indicated. (B) Cells were treated as above and labeled with [32P]orthophosphate for 4 h before collection. After labeling, the GST-AML1(267-315) was isolated from cell lysates using glutathione agarose and used for Western blotting with anti-GST antibodies and for autoradiography. (C) 293T cells were transfected with wild-type or mutant GST-AML1(267-315) as indicated above the lanes. Some samples were treated for 16 to 20 h with 1 μg/μl nocodazole, as indicated above the lanes. During the last 4 to 6 h of nocodazole treatment, all samples were labeled with [32P]orthophosphate. After labeling, samples were analyzed as described for panel B. (D) A diagram of AML1 phosphorylation in 293T cells arrested in G2/M by nocodazole is shown. The upper left part of the diagram indicates that in unsynchronized cells, phosphorylation is detected only on serines 276 and 303 of GST-AML1(267-315). When the cells are arrested at G2/M by treatment with nocodazole, additional phosphorylation on serine 293 and/or threonine 300 is observed. The deletion of serine 293 and threonine 300 phosphorylation sites greatly enhances the phosphorylation at serines 276 and 303 of AML1. If serines 276 and 303 are mutated to alanine, the G2/M-specific phosphorylation at 293/300 does not occur (indicated at the right).
In contrast to cells blocked by hydroxyurea, nocodazole-treated cells display a specific phosphorylated species of GST-AML1(267-315) (Fig. 4B, arrow), which disappears as the cells move out of the nocodazole block.
Subsequent analysis (Fig. 4C) revealed that mutation of serine 293 and threonine 300 to alanine in GST-AML1(267-315) results in the loss of the nocodazole-specific phosphorylated species and results in increased phosphorylation at serines 276 and 303. Interestingly, when serines 276 and 303 are both mutated to alanine, this nocodazole-specific phosphorylation on serine 293/threonine 300 cannot occur (Fig. 4C, right). These results suggest a complex interaction between AML1 phosphorylation at serines 276, 293, and 303 and threonine 300, which is diagrammed in Fig. 4D.
The results described above indicate that mutation of serines 276 and 303 is equivalent to mutation of all four sites (serines 276, 293, and 303 and threonine 300) on AML1 phosphorylation. We have used AML1-4A, the full-length AML1 containing mutations in all four sites, for subsequent analysis.
Mutations in AML1 phosphorylation sites affect the stability of the protein and targeting by the APC.
To determine whether expression of AML1 with mutations in phosphorylation sites had different in vivo effects compared to wild-type AML1, AEL-ΔR1 endothelial cells or NIH 3T3 cell lines stably expressing three types of AML1 were established. The AEL-ΔR1 cell line was established by the immortalization of endothelial cells derived from the aorta-gonad-mesonephros region of a Runx1-null embryo (12). Wild-type AML1, AML1-4A, or AML1-4D were cloned into MSCV retroviral vectors, which were then used to produce virus and to infect cells. The AML1-4D protein is a phospho-mimic mutant, with serines 276, 293 and 303 and threonine 300 mutated to glutamic acid. Three pools of AEL-ΔR1 and two to three pools of NIH 3T3 cells were infected with empty MSCV vector or vector expressing wild-type (WT) AML1, AML1-4A, or AML1-4D. After each infected pool was subjected to puromycin selection, the pools were tested for AML1 expression. Figure 5A and B show the expression of AML1 protein and mRNA in two pools each of empty vector AEL-ΔR1 controls or cells expressing AML1-WT, AML1-4A, or AML1-4D. Figure 5A clearly shows that AML1-4A accumulates to a higher level than AML1-WT, while the AML1-4D protein does not. The Northern blot (Fig. 5B) shows that the level of AML1 mRNA expressed from the MSCV construct bears no relationship to the level of protein present in the various lines. Figure 5C and D show a similar pattern of expression in NIH 3T3 cell lines (AML1-4D pool not shown).
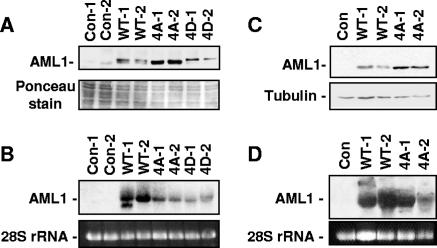
FIG. 5.
Mutation of AML1 phosphorylation sites to alanine increases cellular levels of AML1. (A) Lysates from control AEL-ΔR1 endothelial cells infected with empty MSCV-puro vector and cells stably expressing wild-type AML1, AML1B-4A mutant, or the phospho-mimic AML1B-4D mutant protein were used for Western blotting with anti-HA antibodies. Two independently infected pools expressing each type of AML1 were analyzed. Samples were stained with Ponceau solution after transfer to membranes to confirm approximately equal loading. (B) Total RNA was prepared from the pools of cells used to make the protein lysates analyzed above in panel A. The RNA was run on an agarose gel, and the 28S rRNA was stained with ethidium bromide to determine relative amounts of RNA in each lane. The RNA was then transferred to a membrane for Northern blotting with an AML1 probe. Protein lysates (C) and RNA (D) were prepared from NIH 3T3 cells infected with empty MSCV-puro vector and cells stably expressing wild-type AML1 and AML1B-4A mutant and analyzed as described for panels A and B. Equal loading of the NIH 3T3 protein lysate samples was confirmed by staining with Ponceau solution after transfer (not shown) and by Western blotting with antitubulin antibodies. Con, control.
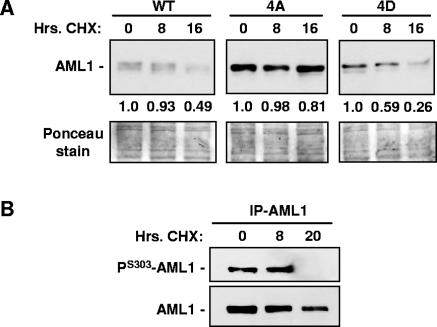
The difference in protein levels observed for wild-type AML1, AML1-4A, and AML1-4D suggest that AML1-4A may be more stable (have a longer half-life) than the other forms. To compare the half-lives of the various forms of AML1, the endothelial cell lines expressing wild-type AML1, AML1-4A, or AML1-4D were treated with cycloheximide as shown in Fig. 6A. After inhibition of protein synthesis with cycloheximide, AML1-4D degradation proceeded at the fastest rate, AML1-4A was the most stable of the proteins, and the rate of wild-type AML1 degradation was intermediate. This result suggests that AML1 phosphorylation does, in fact, promote degradation of the protein. To confirm that phosphorylated wild-type AML1 disappears from cycloheximide-treated cells more rapidly than the nonphosphorylated form, endothelial cells expressing wild-type AML1 were treated for 0, 8, or 20 h with cycloheximide. The HA-tagged AML1 was then immunoprecipitated from each sample and analyzed by Western blotting with anti-serine 303-phosphorylated AML1 antibodies or anti-AML1 antibodies. Figure 6B shows that the anti-serine 303 phosphorylated AML1 antibodies detect no protein after 20 h of cycloheximide treatment, but anti-AML1 antibodies still detect AML1 lacking S303 phosphorylation, indicating the that S303-phosphorylated AML1 is less stable than the nonphosphorylated form.
FIG. 6.
Wild-type and phospho-mutant AML1 display different stabilities after cycloheximide treatment. (A) Endothelial cell lines expressing wild-type AML1, AML1-4A, or AML1-4D were treated for 0, 8, or 16 h with 20 μg/ml cycloheximide (CHX) as indicated above the lanes. Before lysis, the number of cells in each sample was determined, after which whole-cell lysates were prepared from each sample. Lysate from an equal number of cells was loaded into each lane, and the level of AML1 in each sample was determined by Western blotting with anti-AML1 antibodies. Ponceau staining of the membrane after transfer from the SDS gel was used to determine the amount of total protein in each sample. The relative amount of wild-type AML1, AML1-4A, or AML1-4D in each sample was calculated by densitometry, and the value was normalized to the amount of total protein (values are given beneath the upper panel). (B) Endothelial cells expressing HA-tagged wild-type AML1 were treated for 0, 8, or 20 h with 20 mg/ml cycloheximide (CHX) as indicated above the lanes. AML1 was then immunoprecipitated with anti-HA antibodies and analyzed by Western blotting with anti-serine 303-phosphorylated AML1 antibodies and anti-AML1 antibodies.
The results shown in Fig. 5 and 6 suggest that phosphorylation of AML1 may regulate the interaction between AML1 and some ubiquitin ligase complex and that loss of phosphorylation results in stabilization of the protein. This is consistent with previous observations that AML1 is ubiquitinated (4, 10) and that phosphorylation affects AML1 stability in transient transfection experiments (4, 11). The level of AML1 protein, but not AML1 mRNA, has been shown to change as cells progress through the cell cycle (3). Two ubiquitin ligase complexes, the APC and the Skp1/cullin/F-box protein (SCF) complex, are involved in cell cycle regulation through targeted protein degradation (reviewed in references 38 and 42). Since phosphorylation appears to regulate AML1 degradation in cycling cells, we decided to investigate the ability of the APC and SCF complexes to degrade phosphorylated and nonphosphorylated AML1. Two different proteins, Cdh1 and Cdc20, can associate with the APC and promote the interaction between the APC and specific target proteins. Many different proteins can perform this targeting function when associated with the SCF complex, but only a few, such as Skp2, appear to be involved in regulation of the cell cycle.
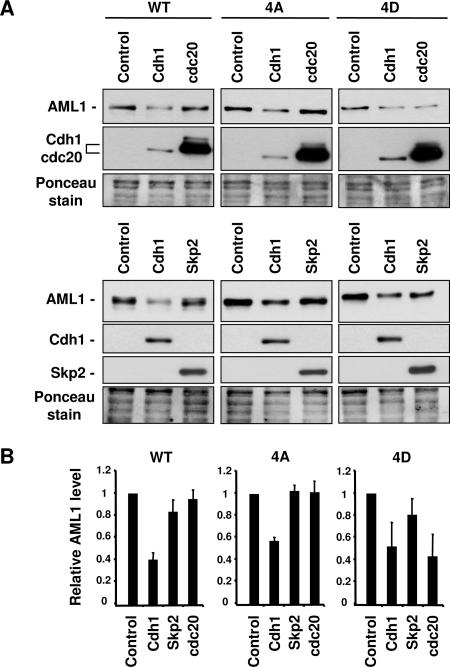
To determine whether Chd1, Cdc20, or Skp2 could promote the degradation of AML1, we first constructed 293T cell lines stably expressing wild-type AML1, AML1-4A, or AML1-4D using the MSCV vectors described above. These 293T-AML1 cell lines were then transfected with either empty expression plasmid (control) or with plasmids expressing Cdh1, Cdc20, or Skp2. An example of such an experiment is shown in Fig. 7A, where the 293T-AML1 cell lines were transfected with empty vector, the Cdh1 expression vector, or the Cdc20 expression vector. The transfected cells were then used to prepare lysates for Western blot analysis with anti-AML1 antibodies to determine the level of AML1 present. Three independent transfection experiments were performed with Cdh1, Cdc20, and Skp2 expression vectors, and the level of AML1 present in each sample was measured by Western blotting followed by densitometry. The results of these experiments are summarized in the graphs shown in Fig. 7B. These experiments show that Cdh1 can promote the degradation of all three forms of AML1 to approximately the same extent. Cdc20, on the other hand, is unable to promote degradation of AML1-4A but is active against AML1-4D. This suggests that Cdc20 preferentially targets the phosphorylated form of AML1. The pattern displayed by Skp2 is similar to that seen with Cdc20, although Skp2 is less active in promoting the degradation of AML1-4D.
FIG. 7.
APC targeting subunit Cdc20 promotes the degradation of AML1 in a phosphorylation-dependent manner. (A) 293T cells stably expressing wild-type AML1, AML1B-4A mutant, or the phospho-mimic AML1B-4D mutant protein were transfected with either empty vector (control) or vector expressing Cdh1, Cdc20, or Skp2. At 24 to 48 h after transfection, cell lysates were prepared and used for Western blotting with anti-AML1 antibodies, followed by anti-HA antibodies to detect HA-tagged Cdh1 or Cdc20. Samples were stained with Ponceau solution after transfer to membranes to confirm approximately equal loading. (B) Three independent transfection experiments were performed as described for panel A using Cdh1, Cdc20, and Skp2. The quantity of wild-type AML1, AML1-4A, or AML1-4D was determined by densitometry of the bands visualized with the anti-AML1 antibodies. Differences in sample loading were corrected by measuring the intensity of the protein bands stained with Ponceau solution. The resulting values are presented graphically, with the amount of AML1 in the sample transfected with empty vector set at 1.0.
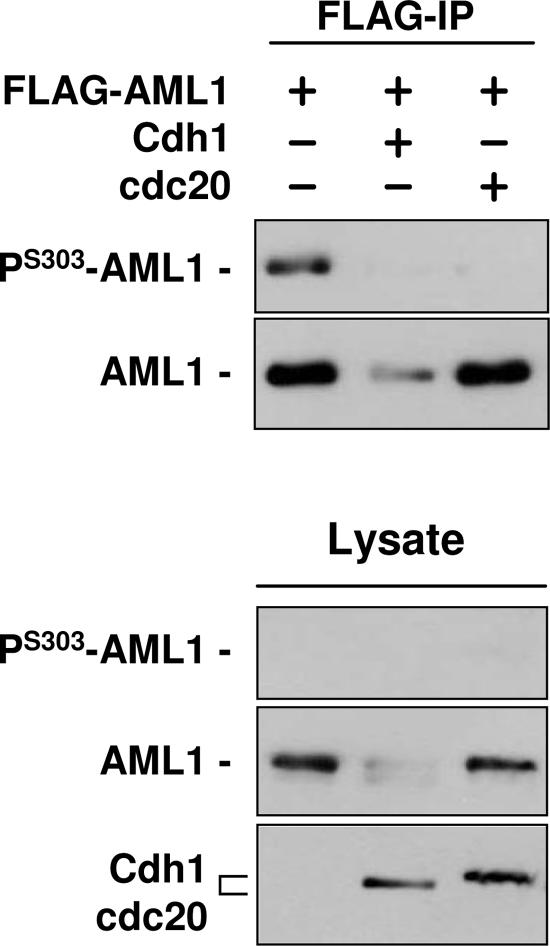
To confirm that Cdc20 targets only the phosphorylated form of wild-type AML1, 293T cells were cotransfected with FLAG-tagged wild-type AML1 and either empty vector, Cdh1 expression vector, or Cdc20 expression vector. At 48 h after transfection, FLAG-AML1 was immunoprecipitated from sample lysates and used for Western blotting with anti-serine 303-phosphorylated AML1 antibodies or anti-AML1 antibodies (Fig. 8). The results confirm that Cdh1 induces degradation of both phosphorylated and nonphosphorylated AML1. In contrast, Cdc20 promotes the disappearance of phosphorylated AML1 but has little effect on total AML1 levels, which presumably consist mostly of nonphosphorylated AML1. The level of wild-type AML1 degradation mediated by Cdc20 (or Skp2) presumably depends on the ratio of phosphorylated to nonphosphorylated wild-type AML1 present in the cell. This may vary from cell type to cell type, depending on the level of CDK (or phosphatase) activity; it would appear that in 293T cells only a small proportion of AML1 is phosphorylated.
FIG. 8.
Cdc20 promotes the degradation of phosphorylated wild-type AML1 but not nonphosphorylated AML1. 293T cells were cotransfected with FLAG-tagged wild-type AML1 and either empty vector, Cdh1 expression vector, or Cdc20 expression vector, as indicated above the lanes. At 48 h after transfection, FLAG-AML1 was immunoprecipitated, and the immunoprecipitate was used for Western blotting with anti-serine 303-phosphorylated AML1 antibodies and anti-AML1 antibodies. Western blotting was also performed with samples of lysate, and anti-HA antibodies were used to detect Chd1 and Cdc20 expression.
Cdh1, Cdc20, and Skp2 interact with AML1.
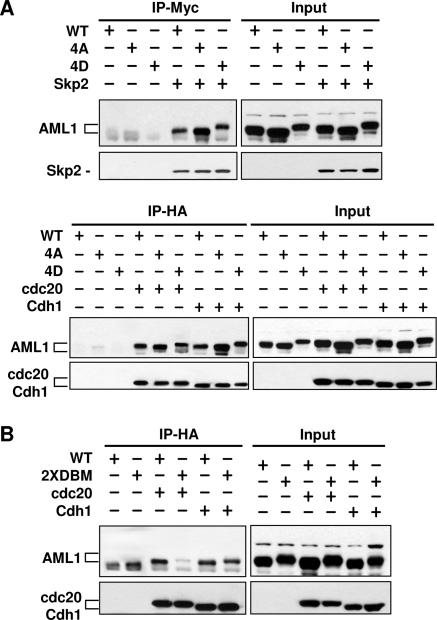
To determine whether Cdh1, Cdc20, and Skp2 physically interact with AML1, 293T cells were cotransfected with AML1 (wild-type, 4A, or 4D) and HA-Cdh1, HA-Cdc20, or myc-Skp2. Excess AML1 expression vector was used for cotransfections to ensure overexpression of AML1 relative to Cdh1, Cdc20, or Skp2. This was done to make sure that induced degradation of AML1 was not observed and confused with any effects phosphorylation might have on the association of AML1 with Cdh1, Cdc20, or Skp2. Immunoprecipitation of HA-Cdh1, HA-Cdc20, or myc-Skp2 was followed by Western blotting for the presence of AML1 in the immunoprecipitate. All three forms of AML1 can associate with Cdh1, Cdc20, or Skp2 (Fig. 9). This result suggests that the phosphorylation state of AML1 does not directly affect the interaction of Cdh1, Cdc20, or Skp2 with AML1 but affects some other aspect of the process of degradation. Past studies (4, 11) indicate that the phosphorylation state of AML1 affects nuclear localization and association with the nuclear matrix. Immunoprecipitation studies are necessarily carried out with AML1 which has been detached from the nuclear matrix by the lysis process. Cdh1, Cdc20, and Skp2 may all bind equally to AML1 after detachment from the matrix but may not find matrix-associated AML1 equally accessible.
FIG. 9.
AML1 associates with Cdh1, Cdc20, and Skp2, and the AML1 destruction boxes are required for efficient association with Cdc20 but are not required for Cdh1. (A) 293T cells were transfected with the indicated expression plasmids and lysed in PBS-1 mM EDTA-0.5% Triton X-100. Excess AML1 expression vector was used for cotransfections to ensure overexpression of AML1 relative to Cdh1, Cdc20, or Skp2. This was done to make sure that induced degradation of AML1 was not observed and confused with any effects phosphorylation might have on the association of AML1 with Cdh1, Cdc20, or Skp2. A total of 200 μg of each sample was immunoprecipitated (IP) with the indicated antibodies, and immunoblotting was performed with anti-AML1 followed by anti-myc or anti-HA. Ten micrograms of each lysate was also resolved by SDS-PAGE and immunoblotting was performed as described above to determine the level of protein expression in every sample. (B) 293T cells were transfected with the indicated plasmids and the experiment performed as described above. DBM, destruction box mutant.
We have also used coprecipitation experiments to determine whether the destruction box (D box) consensus sequences present in AML1 affect the interaction between AML1 and Cdh1 or Cdc20. The D box is one of several protein motifs that can be used by Cdh1 and Cdc20 to recognize substrates (42). AML1 contains two copies of the invariant portion of the D box consensus sequence (RXXL) beginning at amino acids 207 and 250 (amino acid numbering of the AML1B/AML1c form); AML1 does not contain any of the other known recognition motifs. Both D box consensus sequences were mutated to AXXA to create AML1-2xDBM, which was tested for coprecipitation with Cdh1 and Cdc20 (Fig. 9B). Mutation of the D boxes had little effect on the coprecipitation of AML1 and Cdh1 but greatly decreased the interaction between AML1 and Cdc20. The data presented above suggest that Cdh1 can interact with and target all forms of AML1 (phosphorylated or nonphosphorylated) for degradation and does not require the presence of the D box. In contrast, Cdc20 does require the AML1 D box motif to interact with AML1. Cdc20 cannot target nonphosphorylated AML1 (or AML1-4A) for degradation in vivo but can interact with AML1-4A by coimmunoprecipitation from a cell lysate. This suggests that phosphorylation may prevent the association of AML1 and Cdc20 in vivo by an indirect mechanism; alternatively, phosphorylation may be required for Cdc20-mediated degradation of AML1 irrespective of AML1 association with Cdc20.
DISCUSSION
In this report, we have identified specific serine residues in AML1 which are phosphorylated in vivo by Cdk1, Cdk2, and possibly Cdk6 (Cdk6 will phosphorylate AML1 in vitro, but it is not yet known whether in vivo inhibition of Cdk6 activity would affect AML1 phosphorylation). Mutation of these serines to alanines, creating a form of AML1 which cannot be phosphorylated (AML1-4A), increases the stability of AML1. We have also observed that AML1-4A is resistant to degradation mediated by the Cdc20-containing APC (Cdc20-APC). On the other hand, the phospho-mimic AML1 mutant AML1-4D can be targeted for degradation by Cdc20-APC. A similar pattern is observed for the Skp2-SCF complex, although this complex is less active than Cdc20-APC in promoting the degradation of AML1. These observations suggest that AML1 phosphorylation may affect the ability of some ubiquitin ligase complexes, such as Cdc20-APC, to target AML1 and promote degradation.
Not all ubiquitin ligase complexes that target AML1 recognize the phosphorylation state of AML1. The Cdh1-APC complex is able to promote the degradation of AML1-4A almost as efficiently as AML1-4D or wild-type AML1. This suggests that AML1 may contain more than one recognition element for ubiquitin ligase complexes and that AML1 phosphorylation could control access to some elements but not others. The Cdc20 and Cdh1 proteins both serve as substrate-targeting subunits for the APC (38, 42). Cdc20-APC mediates the degradation of proteins such as cyclin B1 and securin, allowing cells to exit mitosis. Cdh1-APC is then activated and is believed to maintain the G1 state by continuing the degradation of cyclin B1 and also proteins necessary for the initiation of S phase, such as Skp2. When S phase is initiated, Cdh1 is phosphorylated and inactivated, allowing the accumulation of Skp2 and the formation of the SCF-Skp2 complex (38). These facts are summarized in Fig. 10 and suggest that AML1 phosphorylation could be more important for the regulation of AML1 stability at specific points during the cell cycle, depending upon which ubiquitin ligase complexes are active. In addition to APC/C-Cdc20, APC/C-Cdh1, and SCF-Skp2, AML1 may be targeted for degradation by other as yet unidentified ubiquitin ligase complexes. If most ubiquitin ligase complexes which target AML1 require AML1 phosphorylation, this would explain why AML1-4A is more stable than wild-type AML1 and accumulates to higher levels. If APC/C-Cdh1 is exceptional in its ability to promote the degradation of both phosphorylated and nonphosphorylated forms of AML1, this fact might explain the observation that cellular AML1 levels are lowest during the G1 phase of the cell cycle, when APC/C-Cdh1 is active (3).
FIG. 10.
Diagram depicting cell cycle-dependent AML1 degradation by the APC/C or SCF complexes. Our data support the idea that AML1 phosphorylated at positions 276, 293, 300, and 303 is targeted for degradation by the APC-Cdc20 complex at early M phase. The APC-Cdh1 complex active during the late M and G1 phases is able to degrade AML1 independent of phosphorylation status. During reentry of the cells into S phase, the SCF-Skp2 complex slightly degrades phosphorylated AML1. This suggests a mechanism for the regulation of AML1 protein levels (and activity) during cell cycle progression.
Our observations also suggest that the manner in which AML1 levels vary with the cell cycle might depend on the level of AML1 phosphorylation, which may vary from cell type to cell type, depending on the level of CDK/cyclin activity or the activity of other unidentified kinases. Higher levels of AML1 phosphorylation may lead to a higher rate of AML1 degradation at many cell cycle phases. Where the cellular level of AML1 phosphorylation is low, substantial AML1 degradation may occur only when ubiquitin ligase complexes, which can promote the degradation of nonphosphorylated AML1, are active. The rate of AML1 degradation could also depend on the levels of activity of APC/C, SCF, or other ubiquitin ligase complexes in a particular cell type.
Cdc20 recognizes a substrate amino acid motif known as the destruction box (reviewed in references 42 and 38), while Cdh1 can interact with a variety of motifs, including the destruction box (D box), the KEN box, the GEN box, and the A box (16, 38, 42). AML1 contains two potential D box motifs, beginning at amino acids 207 and 250 (AML1B/AML1c amino acid numbering). AML1 contains no matches for the other motifs known to assist in substrate recognition by Cdh1 and Cdc20. We have determined that mutation of both putative AML1 D box sequences does not greatly affect coprecipitation of Cdh1 with AML1. Mutation of the D boxes also fails to inhibit Cdh1-mediated degradation of AML1 (J. Biggs, unpublished data). In contrast, mutation of the D boxes greatly reduces the interaction between AML1 and Cdc20. This observation suggests that while Cdc20 recognizes AML1 through the D box motifs, Cdh1 utilizes some other motif to target AML1 for degradation.
The precise mechanism by which AML1 phosphorylation promotes degradation by APC/C-Cdc20 is not yet clear. Phosphorylation does not appear to be necessary for Cdc20 or Skp2 to coprecipitate with AML1, which may suggest that lack of phosphorylation does not necessarily prevent physical interaction between AML1 and Cdc20. Phosphorylation may promote proper association of the complete APC/C-Cdc20 complex with AML1 or some other step of the degradation process. Alternatively, previous studies have suggested that the nonphosphorylated form of AML1 interacts more strongly with the nuclear matrix (4, 11). It is possible that AML1 interaction with the matrix might block access of some (but not all) ubiquitin ligase complexes to AML1 or hinder placement of ubiquitin on AML1 lysine residues. This model might explain why all forms of AML1 (wild-type, 4A, and 4D) coimmunoprecipitate with Cdh1, Cdc20, and Skp2. Immunoprecipitation experiments must be performed with AML1 that has been detached from the nuclear matrix by the lysis process. Cdh1, Cdc20, and Skp2 may all bind equally to AML1 after detachment from the matrix but may not find matrix-associated AML1 equally accessible; this may especially be true for Cdc20 or Skp2. It is possible that matrix association hinders access by Cdc20 to the AML1 D box motif, but the unidentified motif recognized by Cdh1 is still accessible. Other proteins which bind to AML1 in this region, such as CBFβ (10), sin3A (11), and cyclin D (28), might also regulate access to AML1 by Chd1 or Cdc20.
Expression of AML1 may either enhance cell growth (33) or suppress cell growth (44), depending on the cell type. AML1 suppresses mouse embryonic fibroblast proliferation in the presence of a functional p53-p19ARF pathway but promotes growth in the absence of p53 (44). AML1 also inhibits transcriptional elongation by binding to the elongation factor P-TEFb (13). Inhibition of elongation is usually associated with suppression of growth. Loss of AML1 function is associated with the development of acute myeloid leukemia, but amplification of AML1 is associated with B-cell acute lymphoblastic leukemia (reviewed in reference 21). Clearly, AML1 has both growth-enhancing and growth-inhibitory effects, and which effect is predominant depends on the cellular environment.
The level of AML1 in cells may also be critical for normal development. The Notch/AML1 pathway has been identified as a key component of the process of generating adult hematopoietic stem cells (5). These observations suggest that AML1 is required at a specific time during development for the appearance of adult hematopoietic stem cells. Recently, evidence has emerged that AML1 may also be required for proliferation of selected populations of neural progenitors (6, 14, 35), for the prevention of skeletal muscle atrophy (41), and for angiogenesis by epithelial cells from the aorta-gonad-mesonephros region (12).
As might be expected for a protein which can affect cell proliferation, AML1 expression can alter the way cells move through the cell cycle. AML1 has been shown to affect the transition through G1 (2, 33). It has also recently been shown that AML1 forms a complex with cyclin D (28). AML1 has been shown to activate the cyclin D promoter (17) and to repress the p21 (WAF1/Cip1) promoter (18, 35). It has been suggested that AML1 may shorten the G1 phase of the cell cycle by the up-regulation of proteins such as cyclin D (33). The fact that AML1 can alter cell cycle progression while the CDKs which are sequentially activated during the cell cycle phosphorylate AML1 and affect AML1 stability suggests that phosphorylation by CDKs may be a way to fine-tune AML1 activity over the course of the cell cycle, ensuring that a precise level of AML1 activity is present at the required time.
The RUNX2/AML3 protein has substantial amino acid sequence homology to RUNX1/AML1, but the reported phosphorylation patterns of the two proteins appears somewhat different (34, 43, 45). One RUNX2/AML3 phosphorylation site, serine 451, was recently shown to be a target of Cdk1/cyclin B (30). Mutation of this site resulted in loss of RUNX2/AML3 DNA binding and reduced stimulation of anchorage-independent growth in endothelial cells (30). Previous studies (4) have indicated that mutation of the RUNX1/AML1 phosphorylation sites in AML1-4A has little effect on DNA binding; this might suggest that CDK phosphorylation of different sites in the RUNX proteins can regulate different processes.
The overall effect of altering AML1 stability on AML1 target gene expression remains to be determined. AML1 may act as both a transcriptional activator and repressor (21) and may also inhibit transcriptional elongation by binding to cyclin T (13). Experiments using promoter-luciferase reporter constructs indicate that AML1-4A has lower transcriptional activation activity than wild-type AML1 on most, but not all, tested AML1 target promoters (45). The variable effect of AML1 phosphorylation on its transactivation may also dependent on its cooperation with other transcription factors, such as C/EBPα, in different regulatory elements of gene expression. Further investigation is necessary to determine precisely which set of AML1 target genes is affected by mutations that alter AML1 stability and what effect this has on overall cell properties.
Acknowledgments
We thank Guowei Fang, Michele Pagano, and Xiaohua Wu for providing Cdc20, Cdh1, and Skp2 expression constructs. We thank Takahiko Hara for endothelial cell lines.
This work was supported by National Institutes of Health grants CA42533 (to A.S.K.) and CA72009 (to D.E.Z.). The Stein Endowment Fund has partially supported the MEM departmental molecular biology service laboratory for DNA sequencing and oligonucleotide synthesis.
Footnotes
This is paper number 17930-MEM from The Scripps Research Institute.
REFERENCES
- 1.Bain, J., H. McLauchlan, M. Elliott, and P. Cohen. 2003. The specificities of protein kinase inhibitors: an update. Biochem. J. 371:199-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernardin, F., and A. D. Friedman. 2002. AML1 stimulates G1 to S progression via its transactivation domain. Oncogene 21:3247-3252. [DOI] [PubMed] [Google Scholar]
- 3.Bernardin-Fried, F., T. Kummalue, S. Leijen, M. I. Collector, K. Ravid, and A. D. Friedman. 2004. AML1/RUNX1 increases during G1 to S cell cycle progression independent of cytokine-dependent phosphorylation and induces cyclin D3 gene expression. J. Biol. Chem. 279:15678-15687. [DOI] [PubMed] [Google Scholar]
- 4.Biggs, J. R., Y. Zhang, L. F. Peterson, M. Garcia, D. E. Zhang, and A. S. Kraft. 2005. Phosphorylation of AML1/RUNX1 regulates its degradation and nuclear matrix association. Mol. Cancer Res. 3:391-401. [DOI] [PubMed] [Google Scholar]
- 5.Burns, C. E., D. Traver, E. Mayhall, J. L. Shepard, and L. I. Zon. 2005. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19:2331-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, C. L., D. C. Broom, Y. Liu, J. C. de Nooij, Z. Li, C. Cen, O. A. Samad, T. M. Jessell, C. J. Woolf, and Q. Ma. 2006. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron 49:365-377. [DOI] [PubMed] [Google Scholar]
- 7.Chow, J. P., W. Y. Siu, H. T. Ho, K. H. Ma, C. C. Ho, and R. Y. Poon. 2003. Differential contribution of inhibitory phosphorylation of CDC2 and CDK2 for unperturbed cell cycle control and DNA integrity checkpoints. J. Biol. Chem. 278:40815-40828. [DOI] [PubMed] [Google Scholar]
- 8.Davis, R. J. 1993. The mitogen-activated protein kinase signal transduction pathway. J. Biol. Chem. 268:14553-14556. [PubMed] [Google Scholar]
- 9.Erickson, P. F., G. Dessev, R. S. Lasher, G. Philips, M. Robinson, and H. A. Drabkin. 1996. ETO and AML1 phosphoproteins are expressed in CD34+ hematopoietic progenitors: implications for t(8;21) leukemogenesis and monitoring residual disease. Blood 88:1813-1823. [PubMed] [Google Scholar]
- 10.Huang, G., K. Shigesada, K. Ito, H. J. Wee, T. Yokomizo, and Y. Ito. 2001. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 20:723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai, Y., M. Kurokawa, Y. Yamaguchi, K. Izutsu, E. Nitta, K. Mitani, M. Satake, T. Noda, Y. Ito, and H. Hirai. 2004. The corepressor mSin3A regulates phosphorylation-induced activation, intranuclear location, and stability of AML1. Mol. Cell Biol. 24:1033-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwatsuki, K., K. Tanaka, T. Kaneko, R. Kazama, S. Okamoto, Y. Nakayama, Y. Ito, M. Satake, S. Takahashi, A. Miyajima, T. Watanabe, and T. Hara. 2005. Runx1 promotes angiogenesis by downregulation of insulin-like growth factor-binding protein-3. Oncogene 24:1129-1137. [DOI] [PubMed] [Google Scholar]
- 13.Jiang, H., F. Zhang, T. Kurosu, and B. M. Peterlin. 2005. Runx1 binds positive transcription elongation factor b and represses transcriptional elongation by RNA polymerase II: possible mechanism of CD4 silencing. Mol. Cell. Biol. 25:10675-10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer, I., M. Sigrist, J. C. de Nooij, I. Taniuchi, T. M. Jessell, and S. Arber. 2006. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron 49:379-393. [DOI] [PubMed] [Google Scholar]
- 15.Levanon, D., R. E. Goldstein, Y. Bernstein, H. Tang, D. Goldenberg, S. Stifani, Z. Paroush, and Y. Groner. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA 95:11590-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Littlepage, L. E., and J. V. Ruderman. 2002. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 16:2274-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lou, J., W. Cao, F. Bernardin, K. Ayyanathan, F. J. RauscherIII, and A. D. Friedman. 2000. Exogenous Cdk4 overcomes reduced Cdk4 RNA and inhibition of G1 progression in hematopoietic cells expressing a dominant-negative CBF—a model for overcoming inhibition of proliferation by CBF oncoproteins. Oncogene 19:2695-2703. [DOI] [PubMed] [Google Scholar]
- 18.Lutterbach, B., J. J. Westendorf, B. Linggi, S. Isaac, E. Seto, and S. W. Hiebert. 2000. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 275:651-656. [DOI] [PubMed] [Google Scholar]
- 19.Major, M. L., R. Lepe, and R. H. Costa. 2004. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol. Cell. Biol. 24:2649-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyers, S., J. R. Downing, and S. W. Hiebert. 1993. Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol. Cell. Biol. 13:6336-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikhail, F. M., K. K. Sinha, Y. Saunthararajah, and G. Nucifora. 2005. Normal and transforming functions of RUNX1: a perspective. J. Cell Physiol. [DOI] [PubMed]
- 22.Miyoshi, H., M. Ohira, K. Shimizu, K. Mitani, H. Hirai, T. Imai, K. Yokoyama, E. Soeda, and M. Ohki. 1995. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 23:2762-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi, H., K. Shimizu, T. Kozu, N. Maseki, Y. Kaneko, and M. Ohki. 1991. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA 88:10431-10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulloy, J. C., J. Cammenga, K. L. MacKenzie, F. J. Berguido, M. A. Moore, and S. D. Nimer. 2002. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood 99:15-23. [DOI] [PubMed] [Google Scholar]
- 25.Murray, A. W. 2004. Recycling the cell cycle: cyclins revisited. Cell 116:221-234. [DOI] [PubMed] [Google Scholar]
- 26.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 27.Osato, M., N. Asou, E. Abdalla, K. Hoshino, H. Yamasaki, T. Okubo, H. Suzushima, K. Takatsuki, T. Kanno, K. Shigesada, and Y. Ito. 1999. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2alphaB gene associated with myeloblastic leukemias. Blood 93:1817-1824. [PubMed] [Google Scholar]
- 28.Peterson, L. F., A. Boyapati, V. Ranganathan, A. Iwama, D. G. Tenen, S. Tsai, and D. E. Zhang. 2005. The hematopoietic transcription factor AML1 (RUNX1) is negatively regulated by the cell cycle protein cyclin D3. Mol. Cell. Biol. 25:10205-10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrovick, M. S., S. W. Hiebert, A. D. Friedman, C. J. Hetherington, D. G. Tenen, and D. E. Zhang. 1998. Multiple functional domains of AML1: PU. 1 and C/EBPα synergize with different regions of AML1. Mol. Cell. Biol. 18:3915-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao, M., P. Shapiro, M. Fosbrink, H. Rus, R. Kumar, and A. Passaniti. 2006. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J. Biol. Chem. 281:7118-7128. [DOI] [PubMed] [Google Scholar]
- 31.Shuman, J. D., T. Sebastian, P. Kaldis, T. D. Copeland, S. Zhu, R. C. Smart, and P. F. Johnson. 2004. Cell cycle-dependent phosphorylation of C/EBPβ mediates oncogenic cooperativity between C/EBPβ and H-RasV12. Mol. Cell. Biol. 24:7380-7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song, W. J., M. G. Sullivan, R. D. Legare, S. Hutchings, X. Tan, D. Kufrin, J. Ratajczak, I. C. Resende, C. Haworth, R. Hock, M. Loh, C. Felix, D. C. Roy, L. Busque, D. Kurnit, C. Willman, A. M. Gewirtz, N. A. Speck, J. H. Bushweller, F. P. Li, K. Gardiner, M. Poncz, J. M. Maris, and D. G. Gilliland. 1999. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 23:166-175. [DOI] [PubMed] [Google Scholar]
- 33.Strom, D. K., J. Nip, J. J. Westendorf, B. Linggi, B. Lutterbach, J. R. Downing, N. Lenny, and S. W. Hiebert. 2000. Expression of the AML-1 oncogene shortens the G(1) phase of the cell cycle. J. Biol. Chem. 275:3438-3445. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka, T., M. Kurokawa, K. Ueki, K. Tanaka, Y. Imai, K. Mitani, K. Okazaki, N. Sagata, Y. Yazaki, Y. Shibata, T. Kadowaki, and H. Hirai. 1996. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol. Cell. Biol. 16:3967-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theriault, F. M., H. N. Nuthall, Z. Dong, R. Lo, F. Barnabe-Heider, F. D. Miller, and S. Stifani. 2005. Role for Runx1 in the proliferation and neuronal differentiation of selected progenitor cells in the mammalian nervous system. J. Neurosci. 25:2050-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ubersax, J. A., E. L. Woodbury, P. N. Quang, M. Paraz, J. D. Blethrow, K. Shah, K. M. Shokat, and D. O. Morgan. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425:859-864. [DOI] [PubMed] [Google Scholar]
- 37.van Wijnen, A. J., G. S. Stein, J. P. Gergen, Y. Groner, S. W. Hiebert, Y. Ito, P. Liu, J. C. Neil, M. Ohki, and N. Speck. 2004. Nomenclature for Runt-related (RUNX) proteins. Oncogene 23:4209-4210. [DOI] [PubMed] [Google Scholar]
- 38.Vodermaier, H. C. 2004. APC/C and SCF: controlling each other and the cell cycle. Curr. Biol. 14:R787-R796. [DOI] [PubMed] [Google Scholar]
- 39.Wang, S., Y. Zhang, J. Soosairajah, and A. S. Kraft. Regulation of AML1 during the G2/M transition. Leukemia Res., in press. [DOI] [PubMed]
- 40.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 93:3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, X., C. Blagden, J. Fan, S. J. Nowak, I. Taniuchi, D. R. Littman, and S. J. Burden. 2005. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 19:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wasch, R., and D. Engelbert. 2005. Anaphase-promoting complex-dependent proteolysis of cell cycle regulators and genomic instability of cancer cells. Oncogene 24:1-10. [DOI] [PubMed] [Google Scholar]
- 43.Wee, H. J., G. Huang, K. Shigesada, and Y. Ito. 2002. Serine phosphorylation of RUNX2 with novel potential functions as negative regulatory mechanisms. EMBO Rep. 3:967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wotton, S. F., K. Blyth, A. Kilbey, A. Jenkins, A. Terry, F. Bernardin-Fried, A. D. Friedman, E. W. Baxter, J. C. Neil, and E. R. Cameron. 2004. RUNX1 transformation of primary embryonic fibroblasts is revealed in the absence of p53. Oncogene 23:5476-5486. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y., J. R. Biggs, and A. S. Kraft. 2004. Phorbol ester treatment of K562 cells regulates the transcriptional activity of AML1c through phosphorylation. J. Biol. Chem. 279:53116-53125. [DOI] [PubMed] [Google Scholar]