Abstract
PGC-1-related coactivator (PRC) was initially characterized as a transcriptional coactivator that shares structural and functional features with PGC-1α. Both coactivators interact with nuclear respiratory factor 1 (NRF-1) and activate NRF-1 target genes required for respiratory chain expression. Here, we establish that PRC belongs to the class of immediate early genes that are rapidly induced in the transition from quiescence to proliferative growth. As observed for other members of this class, the rapid serum induction of PRC mRNA does not require de novo protein synthesis and inhibition of protein synthesis stabilizes PRC mRNA, leading to its superinduction. Previous work indicated that PRC activation of cytochrome c expression occurs through cis-acting elements that bind both NRF-1 and CREB. Here, we demonstrate that, like NRF-1, CREB binds PRC in vitro and exists in a complex with PRC in cell extracts. Both CREB and NRF-1 bind the same sites on PRC, and the interaction with CREB requires the CREB b-Zip DNA binding domain. Moreover, a CREB/NRF-1 interaction domain on PRC is required for its trans activation of the cytochrome c promoter and a PRC subfragment containing this domain inhibits respiratory growth on galactose when expressed in trans from a lentivirus vector. Finally, PRC associates with the cytochrome c promoter in vivo and its occupancy of the promoter is markedly elevated in response to serum induction of quiescent fibroblasts. The results establish that PRC is an immediate early gene product that can target key transcription factors as an early event in the program of cellular proliferation.
Mitochondrial biogenesis relies upon the integrated expression of both the nucleo-cytosolic and the mitochondrial genetic system (7). Although mitochondria have their own DNA (mtDNA), which, in vertebrates, is a covalently closed circular molecule of approximately 16.5 kilobases, the protein coding capacity of this genome is limited to 13 polypeptide subunits of respiratory chain complexes I, III, IV, and V. The only other products of mtDNA expression are the tRNAs and rRNAs of the mitochondrial translation system. This arrangement necessitates that nuclear genes encode the majority of respiratory chain subunits as well as all of the gene products required for the transcription and replication of mtDNA.
In recent years, there have been significant inroads into understanding the transcriptional mechanisms that contribute to nucleo-mitochondrial interactions in mammalian systems (15). Key components of the mitochondrial transcriptional machinery have been characterized (8). These include a single mitochondrial RNA polymerase (POLRMT); a stimulatory factor (Tfam), which binds DNA and is required for maintenance of the mitochondrial genome; specificity factors (TFB1M and -2M) that interact with Tfam and the polymerase; and a termination factor (mTERF), which may help regulate the rRNA/mRNA ratio. In addition, several nuclear transcription factors that act on nuclear genes required for mitochondrial function have been identified (28). These include the nuclear respiratory factors (NRF-1 and NRF-2); peroxisome proliferator-activated receptor α (PPARα), which functions in fatty acid oxidation; and, most recently, ERRα, which is thought to act as a primary biogenesis factor. Many promoter studies over the years have pointed to the involvement of NRF-1 and -2 in the expression of the majority of respiratory chain subunits as well as in the expression of mitochondrial transcription factors (15). This work is supported by recent observations that NRF-1 occupies the promoters of many of these nuclear genes in vivo (2).
An important issue concerns the mechanisms governing the coordination of multiple transcription factors into a program of mitochondrial biogenesis. This has been resolved in part by the discovery of the PGC-1 family of regulated coactivators (17, 22). The founding member of this family, PGC-1α, directs the expression of genes involved in energy metabolism in response to signaling pathways that mediate thermogenesis, gluconeogenesis, muscle fiber type switching, and mitochondrial biogenesis. The control of mitochondrial biogenesis by PGC-1α occurs at least partly through its interaction with NRF-1, as evidenced by the finding that a dominant negative allele of NRF-1 can block mitochondrial biogenesis directed by ectopically expressed PGC-1α (37). A second family member, PGC-1-related coactivator (PRC), shares key structural motifs with PGC-1α, including a potent activation domain, an LXXLL coactivator signature motif, and both an R/S domain and an RNA recognition motif (1). However, PRC mRNA is not induced during adaptive thermogenesis but is upregulated during the serum-induced G0-to-G1 transition from quiescence to proliferative growth in fibroblasts. Similarly, PRC mRNA is downregulated upon exit from the cell cycle, mediated either by serum withdrawal or by contact inhibition, suggesting that PRC is a growth-regulated coactivator. These differences in regulation between PRC and PGC-1α are intriguing in light of the fact that the two family members are indistinguishable in their abilities to interact with NRF-1 and to trans activate the promoters of NRF-1 target genes required for respiratory chain expression (1, 9). The ability of PRC to work through NRF-1, coupled with its induction in proliferating cells, suggests that it may be involved in coordinating cell growth with mitochondrial expression and function.
Cytochrome c has served as a model for understanding the genetic mechanisms regulating respiratory chain expression in both Saccharomyces cerevisiae and mammalian cells (27). Analysis of the rodent cytochrome c promoter led to the identification of NRF-1 as an activator of respiratory gene expression (5, 6). In addition, the cyclic AMP (cAMP) response element binding protein, CREB, activates the promoter through distinct recognition sites and mediates cytochrome c transcription in response to both cAMP (11) and serum (13). Serum-induced cytochrome c expression at both the mRNA and the protein level in the G0-to-G1 transition has been associated with a specific elevation of heme c-plus-c1 absorbance and enhanced mitochondrial respiration (13). Activation of both NRF-1 and CREB by phosphorylation was implicated in the transcriptional induction of the cytochrome c promoter. Moreover, NRF-1 and CREB recognition sites are required for maximal trans activation of the cytochrome c promoter by PRC, suggesting a potential relationship between growth-regulated cytochrome c expression and promoter activation by PRC (1). Here, we explore this possibility and find that PRC has the characteristics of an immediate early gene and can complex with CREB in a manner that is identical to that of its interactions with NRF-1. In addition, PRC occupies the cytochrome c promoter in vivo and promoter occupancy by the coactivator is enhanced upon serum stimulation of quiescent cells. These results suggest that PRC can target key transcription factors as an early event in the genetic program of cell growth.
MATERIALS AND METHODS
Plasmids.
The plasmids FL-PRC/pBSII, FL-PRC/pSV-Sport, pET32b/PRC(1-221), pET32c/PRC(1047-1379), and pET32c/PRC(1379-1664) have been described previously (1). pET32a/PRC(400-698) was constructed using template FL-PRC/pBSII and primers PRC/BamHI400S (AAAAAAGGATCCGCTGCTGTGCCCAAGGTA) and PRC/XhoI698AS (AAAAAACTCGAGCACTGCACCACGTCTGGG) to amplify a 900-bp fragment that was inserted into BamHI/XhoI-digested pET32a. Similarly, pET32a/PRC(400-450), pET32a/PRC(400-467), pET32a/PRC(400-604), pET32a/PRC(433-467), pET32a/PRC(433-500), pET32a/PRC(450-500), pET32a/PRC(467-500), pET32a/PRC(485-698), pET32a/PRC(1379-1450), pET32a/PRC(1379-1507), and pET32a/PRC(1379-1565) were prepared by cloning PCR products into BamHI/XhoI-digested pET32a.
PRC(1-1379)/pSV-Sport was constructed by combining a 4-kb XhoI/StuI fragment from FL-PRC/pBSII with the SalI/SnaBI fragment of pSV-Sport. The internal deletion in PRC(Δ433/467)/pSV-Sport was introduced by PCR. A PCR product containing the PRC internal HindIII site at the 5′ end and the deletion at the 3′ end was amplified from FL-PRC/pBSII as a template by using primers PRC/HindIIIS (AAAAAAAAAAAGCTTCCTAGCTGGAGACCC) and PRC/Del 433-467AS (ACAGGCTGCTGGCTCCCTGGGCTTCAATAAGC). Similarly, a product with the deletion at the 5′ end and the PRC internal EcoRI site at the 3′ end was amplified from the same template with primers PRC/Del 433-467S (AAGCCCAGGGAGCCAGCAGCCTGTGTGGAAGG) and PRC/EcoRIAS (AAAAAAAAAGAATTCTCCAAGGCAGCTGCC). Equal amounts of the two products were mixed and amplified using primers PRC/HindIIIS and PRC/EcoRIAS, and the resulting 850-bp fragment was digested with HindIII/EcoRI and subcloned into HindIII/EcoRI-digested FL-PRC/pBSII. PRC(Δ433-467)/pSV-Sport was generated by cloning the 5.2-kb XhoI/NotI fragment from PRC(Δ433-467)/pBSII into SalI/NotI-digested pSV-Sport.
pSG5/NRF-1 (34), pSG5/CREB (13), and pNCITE/HCF (35) have been described previously. Two vectors for the expression of CREB deletions, pSG5/CREB(1-174) and pSG5/CREB(1-280), were generated by PCR, using pSG5/CREB as a template. The 520-bp EcoRI/BamHI CREB fragment generated by using PCR primers pSG5MCS (GGGCAACGTGCTGGTTATTGTGCTGTCTCA) and CREB/BamHI174AS (TGGGATCCTGCTAAATTGGAGTTGGCACCG) was cloned into EcoRI/BamHI-digested pSG5 to create pSG5/CREB(1-174). pSG5/CREB(1-280) was constructed by substituting primer CREB/BamHI280AS (AAAAAAGGATCCTTATTCAGCAGGCTGTGTAGG) for CREB/BamHI174AS.
pSG5/CREB-HA was made by cloning the BglII/BamHI PCR product generated using CREB/BglIIS (CTGATGGACAGCAGATCTTAGTGCCCAGCA) and CREB/BamHI Stop HAAS (AAAGGATCCTTAAGCGTAATCGGGGACATCGTAAGGGTAATCTGATTTGTGGCAGTAAAG) into BglII-digested pSG5/CREB. pSG5/PRC(400-604)-HA was made using template FL-PRC/pBSII and primers PRC/BamHI/Sta400(S) (AAAAAAGGATCCCACCATGGCTGCTGTG) and PRC/BamHI/StpHA604(AS) (AAAAAAGGATCCTCAAGCGTAATCGGGGACATCGTAAGGGTAAGGGCCAGC) to generate a 650-bp fragment that was cloned into the BamHI site of pSG5. pSG5/PRC(1-221)-HA was prepared with the same template and the primer pair PRC/BamHI1(S) (AAAAAAGGATCCATGGCGGCGCGCCGG) and PRC/BamHI/StpHA221(AS) (AAAAAAGGATCCTCAAGCGTAATCGGGGACATCGTAAGGGTACTTGGGGGAAGAGGTCTC). The PCR product was digested with BamHI and cloned into pSG5.
Plasmid constructs for the production of glutathione fusion proteins used for antibody purification were made by cloning subfragments of PRC into pGEX-3X. A PCR product for the construction of pGEX-3X/PRC(400-467) was generated using primers PRC/BamHIxx400S (AAAAGGATCCCCGCTGCTGTGCCCAAGGTA) and PRC/EcoRI467AS (ACAGGCGAATTCCTGCTCCTTGCTCTTCTT), with FL-PRC/pBSII as a template. The amplification product was digested with BamHI/EcoRI and cloned into BamHI/EcoRI-digested pGEX-3X. pGEX-3X/PRC(1047-1379) was generated in a similar fashion. Sequence verification for all constructs was performed by the Northwestern Biotech Core Facility.
Cell culture and transfections.
293FT cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) with 10% fetal bovine serum (HyClone), 1% penicillin-streptomycin (Invitrogen), and 0.1 mM nonessential amino acids (Invitrogen). For transfection, proliferating cells were trypsinized, collected in phosphate-buffered saline (PBS), and counted with a hemocytometer. The cells were resuspended in Opti-MEM (Invitrogen) (2.3 × 106 cells per 400 μl) and mixed with plasmid DNA. Transfections were completed by electroporating the mixture with a Bio-Rad Gene Pulser Xcell in a 2-mm cuvette according to the manufacturer's setting for 293 cells. Following electroporation, the cells were diluted in 10 ml fresh medium and plated on 10-cm tissue culture dishes.
For serum starvation experiments, BALB/3T3 fibroblasts were plated at a density of 875,000 per 150-mm dish in DMEM containing 10% calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin and allowed to grow for 48 h. The cells were then washed twice with PBS and serum starved in DMEM containing 0.5% fetal bovine serum for 48 to 60 h. Following starvation, the cells were stimulated in DMEM containing 20% fetal bovine serum for 3 and 8 h. U2OS cells were grown in McCoy's 5a medium with 1.5 mM l-glutamine (Invitrogen) supplemented with 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (Invitrogen). Cells were starved in McCoy's 5a medium containing 0.1% fetal bovine serum for 18 h, followed by stimulation in medium containing 20% fetal bovine serum for the above-indicated times.
Transient transfection of BALB/3T3 cells was performed by calcium phosphate precipitation as described previously (1). BALB/3T3 cells used in transfections were maintained in DMEM (Invitrogen) supplemented with 10% calf serum (HyClone) and 1% penicillin-streptomycin (Invitrogen). Cells were plated at a density of 3,000 cells per cm2 in six-well plates and transfected with 100 ng of pGL3RC4/-326 reporter (13) and 4 ng of pRL-TK control vector (Promega). For trans-activation, 2 μg each of full-length PRC (PRC/pSV-Sport) or mutated derivatives [PRC(Δ433-467)/pSV-Sport or PRC(1-1379)/pSV-Sport] was cotransfected. For trans-inhibition, 2 μg of full-length PRC (PRC/pSV-Sport) was cotransfected with either 1 or 2 μg of pSG5/PRC(400-604)-HA or pSG5/PRC(1-221)-HA. After 5 h, cells were washed twice with Dulbecco's phosphate-buffered saline (Invitrogen) and grown for an additional 40 h in fresh media. Cell extracts were prepared, and luciferase assays were performed using the dual luciferase reporter assay system (Promega). Firefly luciferase activity from the cytochrome c promoter-luciferase reporter construct pGL3RC4/-326 (13) was normalized to Renilla luciferase luminescence from the cotransfected pRL-TK control vector.
RNA methods.
The requirement for protein synthesis on PRC mRNA induction was tested by subjecting BALB/3T3 fibroblasts to serum starvation as described above. The cells were stimulated with serum in the absence or presence of cycloheximide (10 μg/ml), and RNA was harvested at various times with TRIzol (Invitrogen). PRC mRNA levels relative to those for the 18S rRNA control were determined by real-time reverse transcription-PCR as described previously (9). Results are expressed as the induction levels (n-fold) for PRC mRNA in stimulated cells versus those for PRC mRNA in starved cells.
The half-life of PRC mRNA in serum-stimulated BALB/3T3 fibroblasts was determined by adding actinomycin D (5 μg/ml) to the medium at 8 h after serum stimulation. The half-life of PRC mRNA in starved BALB/3T3 fibroblasts was determined by adding actinomycin D (5 μg/ml) together with the starvation medium (DMEM plus 0.5% fetal bovine serum). The half-life of PRC mRNA in confluent BALB/3T3 fibroblasts was determined by adding actinomycin D (5 μg/ml) to confluent cultures. The effect of protein synthesis on PRC mRNA stability was determined by subjecting BALB/3T3 fibroblasts to serum starvation as described above. The cells were stimulated with serum for 1 h in the presence or absence of cycloheximide (10 μg/ml). After 1 h, the cells were washed twice with PBS and further stimulated for various times in the presence of 5 μg/ml actinomycin D. In each case, total RNA was harvested with TRIzol (Invitrogen) at various times following the addition of actinomycin D, and PRC mRNA levels relative to that for the 18S rRNA control were determined by real-time reverse transcription-PCR as described previously (9).
Antibodies.
The preparation of affinity-purified anti-PRC(95-533) has been described previously (1). Thioredoxin fusion proteins used for immunization were prepared from BL21 CodonPlus(DE3)-RIL cells (Stratagene) transformed with either pET32a/PRC(400-467) or pET32c/PRC(1047-1379). Transformants were grown to “log” phase (optical density at 600 nm, approximately 0.5) at 37°C in LB with 0.1 mg/ml ampicillin and 34 μg/ml chloramphenicol. Induction was carried out at room temperature in the same media supplemented with IPTG (isopropyl-β-d-thiogalactopyranoside) to 1 mM and grown for 2.5 h. The cells were resuspended in a 1/20 culture volume of binding wash buffer (BWB) (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride), lysed on ice for 30 min with lysozyme at 1 mg/ml, and frozen overnight at −80°C. The lysate was thawed on ice, sonicated briefly in 10-s bursts, and centrifuged at 20,000 × g for 20 min. The thioredoxin fusion proteins trdx-PRC(400-467) and trdx-PRC(1047-1379) were purified from the soluble fraction of the lysates with 1 ml Ni-nitrilotriacetic acid Superflow (QIAGEN) resin per 10 ml lysate. The slurry was mixed on a rocking table for 1 h at 4°C. The resin was washed with 50 resin volumes of 300 mM NaCl, 50 mM NaH2PO4, pH 7.5, and 20 mM imidazole. The thioredoxin fusion proteins were eluted with 300 mM NaCl, 50 mM NaH2PO4, pH 7.5, and 250 mM imidazole and dialyzed overnight in PBS at 4°C. Rabbit anti-PRC(400-467) and anti-PRC(1047-1379) sera were prepared commercially using the purified thioredoxin fusion proteins as antigens (Harlan Bioproducts for Science). Antibodies were affinity purified on columns prepared by coupling glutathione S-transferase (GST) fusion proteins GST-PRC(400-467) and GST-PRC(1047-1379), produced from pGEX-3X/PRC(400-467) and pGEX-3X/PRC(1047-1379), respectively, to CNBr-activated Sepharose 4B (Amersham) according to the manufacturer's instructions. The GST fusion proteins were purified from soluble bacterial lysate by adding 1 ml glutathione-Sepharose 4B (Amersham) resin per 10 ml lysate and rocking the slurry for 1 h at 4°C. The resin was washed with 50 resin volumes of 10 mM Tris-HCl, pH 8, and 500 mM NaCl. The fusion proteins were eluted with 100 mM glycine, pH 2, and collected in 0.1 elution volume of 2.5 M Tris-HCl, pH 8. The eluted GST fusion proteins were dialyzed in coupling buffer (0.1 M NaHCO3, pH 8.5, 0.5 M NaCl) prior to coupling. The binding and elution of the antibodies from the affinity columns have been described (1). The affinity-purified antibodies were concentrated by ammonium sulfate precipitation and dialyzed against PBS.
Preparation and affinity purification of goat anti-NRF-1 serum have been described previously (1, 33). Mouse antihemagglutinin (anti-HA) monoclonal antibody was purchased from Covance Research Products (MMS-101R). The antibodies were used in coimmunoprecipitation experiments to detect NRF-1 and hemagglutinin-tagged CREB (CREB-HA), respectively.
S-tag pulldown assay.
Pulldown assays were performed by a minor modification of a previously described method (1). The expression of thioredoxin fusion proteins and preparation of soluble bacterial lysates were carried out as described above for the production of fusion proteins for antibody production. The thioredoxin fusion proteins were purified with S-protein agarose resin (Novagen) by adding 100 μl of 50% S-protein agarose slurry to 1 ml bacterial lysate. The slurry was rocked at 4°C for 1 h, and the resin-bound protein was removed by centrifugation for 1 min at 270 × g. Following aspiration of the supernatant, the resin was washed five times with 1 ml BWB and suspended in 150 μl BWB. The amount of a given fusion protein bound to the resin was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of an aliquot followed by Coomassie blue staining.
NRF-1, CREB (and its deletions), and HCF were 35S labeled by in vitro translation (TNT reticulocyte lysate system; Promega) according to the manufacturer's instructions. A 5-μl aliquot of the translation reaction mixture was mixed with approximately 1 μg thioredoxin fusion protein (in S-protein slurry) in 500 μl binding buffer (32) and rocked at 4°C for 1 h. The resin was washed five times with 500 μl binding buffer at 4°C, suspended in 20 μl sample buffer (with β-mercaptoethanol), and boiled for 5 min. The samples were separated by SDS-PAGE and visualized by autoradiography.
Coimmunoprecipitation and immunoblotting.
Approximately 13.5 × 106 293FT cells were transfected by electroporation with pSG5/CREB-HA and plated on three 15-cm tissue culture dishes as described above. An additional 13.5 × 106 nontransfected 293FT cells were plated on three 15-cm tissue culture dishes. The cells were maintained at 37°C for approximately 48 h and then lysed in NP-40 lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 2.5 mM Na2VO4, 5 mM NaF, 0.1 mM phenylmethylsulfonyl fluoride, 0.1 mM EDTA, and mini-complete protease inhibitor cocktail [Roche]) as described previously (1). The protein concentration of each whole-cell extract was determined by the Bradford assay (Bio-Rad), using known concentrations of bovine serum albumin as standards. Purified rabbit immunoglobulin G (IgG; Sigma) control, affinity-purified anti-PRC(95-533), or affinity-purified anti-PRC(1047-1379) (2.5 or 7.5 μg of each) was added to 500 μg whole-cell extract (pSG5/CREB-HA transfected or nontransfected) in a total volume of 250 μl NP-40 lysis buffer. After 1 h of incubation on ice, 15 μl protein A-agarose (Roche) was added, and the incubation continued for an additional 1 h on a rocking table at 4°C. The immunoprecipitate was centrifuged for 1 min at 270 × g and washed four times with 500 μl NP-40 lysis buffer. The rabbit IgG control and anti-PRC immunoprecipitates were resuspended in 20 μl sample buffer containing β-mercaptoethanol, subjected to 10% SDS-PAGE, and transferred to a nitrocellulose membrane (Schleicher and Schuell) by using a Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad) with Towbin transfer buffer (31). Immunoblots from pSG5/CREB-HA-transfected cells were probed with mouse monoclonal anti-HA antibody (Covance), whereas those from nontransfected cells were probed with affinity-purified goat anti-NRF-1 antibody.
For detection of PRC protein, whole-cell extracts were prepared as described above, subjected to 7.5% SDS-PAGE, and transferred to nitrocellulose membranes (Schleicher & Schuell) with high-molecular-weight transfer buffer (50 mM Tris, 380 mM glycine, 0.1% SDS, and 20% methanol) by using a Mini Trans-Blot cell tank transfer system (Bio-Rad). Immunoblots were probed with anti-PRC(95-533) (1), anti-PRC(400-467), or anti-PRC(1047-1379). Proteins were visualized using SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology).
ChIP.
Starved and 8-h-serum-stimulated BALB/3T3 fibroblasts were fixed in 1% formaldehyde for 10 min at 25°C. Chromatin immunoprecipitations (ChIP) were performed as described previously (9, 30) with control IgG antibodies (Sigma) and anti-NRF-1 (12), anti-PRC(1047-1379) (this work), anti-CREB, and anti-phospho-CREB antibodies (Santa Cruz Biotechnology). Salmon sperm DNA/protein G agarose (Roche Diagnostics) was used instead of salmon sperm DNA/protein A agarose (Roche Diagnostics) for the precipitation with goat anti-phosho-CREB because protein A cannot efficiently bind goat IgG. Immunoprecipitated promoter fragments were quantitated by real-time PCR on an ABI PRISM 7900HT sequence detection system with SYBR green PCR Mastermix (Applied Biosystems). The primers used for real-time PCR were specific for the mouse cytochrome c promoter (forward, GTTACCTGAGCCGAGCCACAC; reverse, TGACGTAACCGCACCTCATTGG) and were used to amplify a promoter fragment that includes the NRF-1 (−156/−145 relative to the transcription initiation site) and CREB (−262/−255 and −110/−103) recognition sites. The ΔΔCT method (18) was used to calculate the relative quantities of immunoprecipitated cytochrome c promoter DNA from serum-starved or -stimulated cells. The ΔCT value was calculated by subtracting the average cycle threshold (CT) value of the input DNA from the average CT value of the immunoprecipitated DNA. ΔΔCT was then calculated by subtracting the ΔCT value of the ChIP with control IgG from the ΔCT value of the ChIP with specific antibody. The results were expressed as the relative levels of promoter occupancy by the various factors compared with levels for control IgG for quiescent and serum-stimulated cells.
Lentivirus methods and cell growth determinations.
Stable BALB/3T3 cell lines constitutively expressing PRC(400-604) or the tetR gene encoding the Tet repressor (TR) (as a control) were generated using a ViraPower T-REx lentiviral expression system according to the manufacturer's protocol (Invitrogen). Briefly, lentivirus was produced by transfecting the 293FT producer cell line with the expression construct pLenti4/TO/V5-DEST-PRC(400-604) or pLenti6/TR. BALB/3T3 cells were infected with the viral supernatants, and stably transduced cells were selected using the appropriate antibiotic. Growth rates for both cell lines were determined by growing freshly thawed cells to approximately 75% confluence and then plating 20,000 cells in 6-cm dishes containing DMEM supplemented with 10% calf serum, 1% penicillin-streptomycin, 1 mM sodium pyruvate, and either 5 mM glucose or 5 mM galactose. Cells were fed daily and counted from days 3 through 6 by using a hemocytometer.
Statistical analysis.
Statistical comparisons of the data were made by Student's t test. The level of significance was set at P values of <0.05 in all cases.
RESULTS
Immediate early expression of PRC.
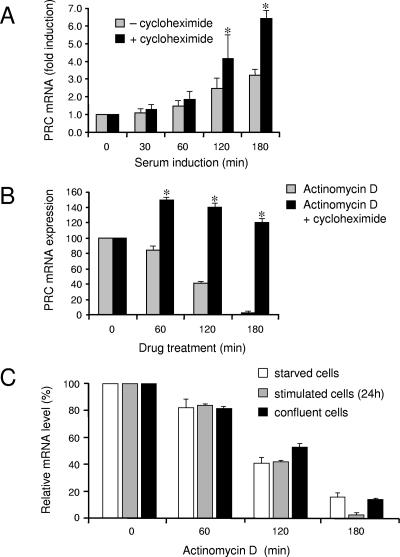
PRC mRNA is induced upon serum stimulation of quiescent BALB/3T3 fibroblasts and is downregulated upon serum withdrawal or contact inhibition (1). The rapid induction of PRC under these conditions is reminiscent of the class of immediate early genes, which are defined by their induction in the absence of de novo protein synthesis (21). To test whether PRC belongs to this class, the levels of serum induction of PRC were compared in the presence and absence of the cytosolic protein synthesis inhibitor cycloheximide. As shown in Fig. 1A, PRC transcripts are rapidly induced with similar kinetics in both the absence and the presence of cycloheximide. This indicates that PRC mRNA synthesis and/or stabilization occurs through the use of preexisting proteins and does not require de novo protein synthesis. In addition, like other immediate early mRNAs, PRC transcripts are induced to a much higher level in the presence of cycloheximide (superinduced), suggesting that PRC mRNA is stabilized in the absence of protein synthesis. This possibility was tested by measuring the half-life of PRC mRNA by treating cells with actinomycin D in both the absence and the presence of cycloheximide. As shown in Fig. 1B, PRC mRNA has a half-life of about 2 h and the PRC transcript is markedly stabilized in the presence of cycloheximide. It is possible that the regulated stabilization of PRC mRNA may account for its rapid induction in response to serum. This appears not to be the case because the half-lives of PRC mRNA are similar in serum-starved, serum-stimulated, and confluent cells (Fig. 1C). This indicates that PRC mRNA stabilization cannot account for PRC mRNA induction upon entry to the cell cycle and suggests that PRC upregulation in proliferating cells occurs at the level of increased transcription initiation. This was confirmed by a nuclear run-on experiment showing increased levels of PRC nascent transcripts in response to serum stimulation (not shown).
FIG. 1.
Effect of cycloheximide on PRC mRNA induction and stabilization. (A) Serum-starved, quiescent BALB/3T3 fibroblasts were serum stimulated for the indicated times in the absence or presence of 10 μg/ml cycloheximide. An asterisk denotes a P value of <0.05 compared to corresponding values for untreated cells. (B) Quiescent BALB/3T3 cells were serum stimulated for 1 h in either the absence or the presence of 10 μg/ml of cycloheximide, followed by treatment with 5 μg/ml actinomycin D. An asterisk denotes a P value of <0.05 compared to values for cells treated with actinomycin D alone. (C) The PRC mRNA half-life was determined following treatment of starved cells, serum-stimulated cells, or confluent cells with 5 μg/ml actinomycin D. For all three panels, total RNA was isolated at the indicated times and the relative amounts of PRC mRNA were determined by quantitative real-time PCR, using 18S rRNA as an internal standard. Results are expressed as the means ± standard errors of the means for three independent determinations.
Antibodies to PRC were raised to measure PRC protein levels. These antibodies were initially tested with human 293FT cells because they were raised against human PRC subfragments. These cells also have a high transfection efficiency, which facilitates the detection of protein expressed from transfected plasmids. The initial antibody, directed against a PRC subfragment encompassing amino acids 95 to 533, revealed multiple bands in extracts from cells transfected with empty control vector pSV-Sport. Only one of these bands corresponded to the 177-kDa molecular mass predicted based on the PRC amino acid sequence (Fig. 2A, lane 1). However, this band was much diminished in an immunoblot using a second antibody raised against amino acids 1047 to 1379 (Fig. 2A, lane 3) and was absent using a third antibody directed against amino acids 400 to 467 (Fig. 2A, lane 5). A major band migrating at 250 kDa was detected by all three antibodies, suggesting that PRC migrates upon gel electrophoresis to a position greater than its predicted mass. To confirm that the 250-kDa band was PRC, extracts were prepared from cells transfected with an expression vector (PRC/pSV-Sport) containing the entire PRC open reading frame. Immunoblots of these extracts using each of the three antibodies revealed enhanced expression of the 250-kDa band (Fig. 2A, lanes 2, 4, and 6) compared to what was found for extracts from the empty-vector controls (Fig. 2A, lanes 1, 3, and 5). These results unambiguously identify the 250-kDa protein as PRC.
FIG. 2.
Identification and quantitation of PRC protein levels. (A) PRC protein was identified in 293FT cell extracts by using antibodies raised to three different subregions [anti-PRC(95-533), anti-PRC(1047-1379), and anti-PRC(400-467)] of the molecule. Extracts were prepared from cells transfected with either empty vector (pSV-Sport) or vector expressing the full PRC open reading frame (PRC/pSV-Sport). (B) Total BALB/3T3 or U2OS cell extracts were prepared from proliferating cells, serum-starved cells, or starved cells stimulated with serum for either 3 or 8 h. In both panels, proteins were detected following denaturing gel electrophoresis and immunoblotting with either the indicated antibodies (A) or anti-PRC(1047-1379) (B).
Mouse (BALB/3T3) and human (U2OS) serum-responsive cell lines were used to determine whether the 250-kDa PRC protein is induced in the transition between G0 and G1. Extracts were prepared from proliferating cells, serum-starved cells, and starved cells that were serum stimulated for 3 or 8 h. Immunoblots revealed that steady-state PRC protein levels were diminished upon serum starvation and induced upon serum stimulation of both mouse and human lines (Fig. 2B). In both cases, PRC was induced to levels equivalent to those found in proliferating cells within 3 to 8 h of serum addition. This is consistent with the serum induction of PRC mRNA, which begins after 1 h of serum addition (Fig. 1A) and demonstrates that the upregulation of the PRC mRNA leads to increased steady-state levels of PRC protein.
In vitro and in vivo interaction of PRC with CREB.
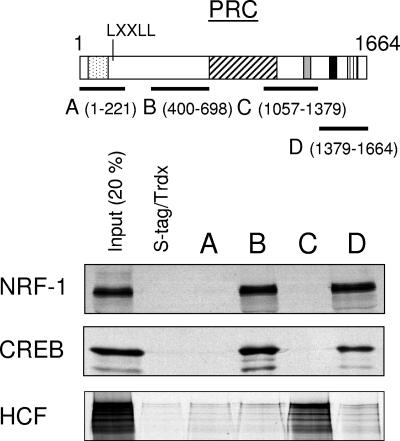
We demonstrated previously that cytochrome c is induced upon serum stimulation of quiescent BALB/3T3 cells and that the induced expression of this respiratory protein coincides with enhanced mitochondrial respiration (13). The cytochrome c promoter is a target for both NRF-1 and CREB transcription factors, and promoter recognition sites for both of these factors were necessary for maximal serum-dependent promoter activation. Because PRC can trans-activate NRF-1 target genes through a specific interaction with the NRF-1 DNA binding domain (1), it was of interest to determine whether CREB is also a target for this coactivator. To this end, levels of NRF-1 and CREB binding to PRC were compared using an S-tag pulldown assay (1) (Fig. 3). The results demonstrate that both NRF-1 and CREB bind to PRC subfragments B (amino acids 400 to 698) and D (amino acids 1379 to 1664) but not to LXXLL-containing subfragment A (amino acids 1 to 221) or to subfragment C (amino acids 1057 to 1379). As a control for binding specificity, host cell factor, which binds to a DHDY motif, binds only subfragment C containing this motif but not any of the other PRC subfragments, including those binding both NRF-1 and CREB (Fig. 3). These results demonstrate that PRC can interact specifically with both NRF-1 and CREB, suggesting that it may be involved in cytochrome c promoter activation by both factors.
FIG. 3.
Comparison of the in vitro interactions between PRC and NRF-1, CREB, or HCF. A schematic representation of PRC is shown at top, with the various functional domains indicated (stippled, activation domain; cross-hatched, proline-rich region; gray, consensus recognition site [DHDY] for host cell factor [HCF]; black, R/S domain; vertical hatched, RNA recognition motif). Subfragments of PRC designated A, B, C, or D, with their amino acid coordinates shown in parentheses, were used in S-tag pulldown assays with 35S-labeled transcription factors NRF-1, CREB, or HCF. Binding of the various subfragments to each 35S-radiolabeled transcription factor was compared to that of S-tagged thioredoxin as a negative control. Bound proteins were eluted from the S-protein agarose and visualized by autoradiography.
If the interactions revealed by the in vitro binding assays are significant physiologically, it should be possible to detect a complex between PRC and CREB in cell extracts by coimmunoprecipitation. A vector designed to express CREB-HA was introduced into 293FT cells by electroporation, and cell extracts were immunoprecipitated either with control IgG or with purified anti-PRC(95-533) or anti-PRC(1047-1379). These cells were used for immunoprecipitation experiments because of their high transfection efficiency. Following gel electrophoresis, immunoblots were probed with mouse anti-HA monoclonal antibody. Both PRC antibodies at 2.5 μg resulted in the specific coprecipitation of a protein coinciding with the expected molecular mass of CREB (Fig. 4A, lanes 3 and 5) and comigrating with the CREB protein expressed in the cell extracts (Fig. 4A, lane 7). Notably, increasing the concentration of anti-PRC(1047-1379) to 7.5 μg markedly enhanced the CREB signal (Fig. 4A, lane 6), whereas the same increase in anti-PRC(95-533) reduced the signal (Fig. 4A, lane 4). This difference in the antibodies can be explained by the fact that a strong CREB interaction domain is localized to PRC amino acids 400 to 698, making it likely that the anti-PRC(95-533) antibody competes for CREB binding to PRC at the higher concentration. No such CREB interaction domain is localized to PRC amino acids 1047 to 1379, and thus the antibody directed against this region does not compete for CREB binding. No specific CREB band was detected in the IgG control at either concentration (Fig. 4A, lanes 1 and 2). The results obtained for CREB were nearly identical to those obtained using goat anti-NRF-1 to probe an immunoblot following immunoprecipitation with anti-PRC(1047-1379) (Fig. 4B). NRF-1 was precipitated by anti-PRC(1047-1379) in a concentration-dependent manner, confirming the specific interaction between PRC and NRF-1 observed previously (1) and demonstrating the validity of the assay for detecting PRC transcription factor interactions. These results confirm that CREB can enter into a complex with PRC in a manner that is indistinguishable from that observed for NRF-1.
FIG. 4.
In vivo interaction between PRC and CREB. (A) HA-tagged CREB was expressed in 293FT cells following electroporation with pSG5/CREB-HA. Cell extracts were immunoprecipitated with either 2.5 or 7.5 μg, respectively, of rabbit IgG as a negative control (lanes 1 and 2), anti-PRC(95-533) (lanes 3 and 4), or anti-PRC(1047-1379) (lanes 5 and 6). Immune complexes were brought down with protein A-agarose, washed, and run on an SDS-10% PAGE gel. For comparison, 20 μg of cell extract was run (lane 7). After transfer, the immunoblot was probed with mouse anti-HA monoclonal antibody. (B) 293FT cell extracts were immunoprecipitated with either 2.5 or 7.5 μg rabbit IgG as a negative control (lanes 1 and 2) or anti-PRC(1047-1379) (lanes 3 and 4). Immune complexes were precipitated and electroblotted as described for panel A. For comparison, 2 ng of recombinant NRF-1 was run (lane 5). After transfer, the immunoblot was probed with goat anti-NRF-1. Molecular mass standards in kilodaltons are indicated at the left in each panel.
Molecular determinants of CREB binding to PRC.
Both CREB and NRF-1 bind to PRC subfragments B and D, encompassing amino acids 400 to 698 and 1379 to 1664, respectively (Fig. 3). Deletion fine mapping within these regions was performed to assess whether the transcription factors bind to distinct sites within these PRC domains. A series of deletions within each domain was subjected to S-tag pulldown assays by using in vitro-translated NRF-1 and CREB. As shown in Fig. 5, all of the fragments that bind CREB also bind NRF-1 and those that fail to bind CREB also fail to bind NRF-1. The 3′ deletion breakpoint for the upstream domain was localized between amino acids 450 and 467, while the 5′ deletion breakpoint was between amino acids 433 and 485. The smallest fragment binding both transcription factors was bounded by amino acids 433 to 467. Similarly, the downstream domain was defined as amino acids 1379 to 1450, a region coinciding with the R/S domain. A comparison of the amino acid sequences for these binding sites does not reveal obvious sequence similarities, although both contain clusters of basic amino acid residues. Thus, within the limits of resolution of this analysis, CREB and NRF-1 share the same binding sites within PRC.
FIG. 5.
Deletion fine mapping of the NRF-1/CREB interaction domains in PRC. A schematic representation of PRC is shown at top, with various functional domains indicated as described in the legend to Fig. 3. A series of deletions of the PRC(400-698) and the PRC(1379-1664) subfragments, shown to bind NRF-1 and CREB, was constructed, and their levels of in vitro binding to the two 35S-labeled transcription factors were compared using the S-tag pulldown assay. Binding of the various subfragments was compared to that of S-tagged thioredoxin as a negative control. Bound proteins were eluted from the washed beads and visualized by autoradiography. A schematic of the relative length of each deletion, with a number at its right indicating its amino acid coordinates, is shown. The ability to bind (+) or not bind (−) each transcription factor in the in vitro pulldown assay is indicated at the right of each subfragment. The amino acid sequence of the smallest subfragment to display significant binding is shown at bottom.
Previous domain-mapping experiments demonstrated that PRC interacts with the NRF-1 DNA binding domain (1). A similar requirement for the CREB DNA binding domain was investigated by assaying a series of carboxy-terminal CREB deletions for the ability to bind both upstream and downstream CREB binding sites within PRC. Deletion of the CREB b-Zip DNA binding domain between amino acids 240 and 341 eliminated CREB binding to PRC sites encompassing amino acids 400 to 604 (Fig. 6, left panel) and 1379 to 1664 (Fig. 6, right panel). Thus, the CREB DNA binding domain is required for the observed activator-coactivator interaction.
FIG. 6.
Deletion mapping the region of CREB required for binding PRC. A schematic representation of CREB is shown at top, with various functional domains indicated. Full-length CREB and C-terminal deletions, shown as solid lines below the diagram, were 35S labeled and subjected to S-tag pulldown assays using either the upstream CREB binding domain in PRC (amino acids 400 to 604) (left panel) or the downstream CREB binding domain (amino acids 1379 to 1664) (right panel). Binding of the CREB subfragments to each PRC domain was compared to that of S-tagged thioredoxin as a negative control. Bound proteins were eluted from the washed S-protein agarose and visualized by autoradiography.
PRC trans activation and dominant negative inhibition of respiratory growth.
Both NRF-1 and CREB recognition sites are required for maximal trans activation of the cytochrome c promoter by PRC (1). The finding of two distinct transcription factor interaction domains on PRC raises the question of whether they are functionally equivalent for promoter activation. Experiments with PGC-1α revealed that the carboxy-terminal R/S domain is dispensable for transcription initiation but serves as a protein-protein interaction interface that recruits splicing factors and promotes the coupling of transcription and RNA processing (19). As shown in Fig. 7, the full-length PRC [PRC(1-1664)] activates the cytochrome c promoter at severalfold-higher levels than the control in a transient-cotransfection assay. However, most of this activity is eliminated by deleting the CREB/NRF-1 interaction domain localized to amino acids 433 to 467 within the context of the full-length PRC [PRC(Δ433-467)]. To test further the importance of the CREB/NRF-1 binding domain within this region, a fragment containing this domain [PRC(400-604)] was expressed in trans in a PRC cotransfection assay. Under conditions of increasing PRC(400-604), PRC trans activation of the promoter was inhibited to a level identical to that achieved upon deletion of amino acids 433 to 467. Introduction of the same amounts of vector expressing an adjacent control fragment [PRC(1-221)], which lacks a CREB/NRF-1 binding site, showed little or no inhibition. These results are consistent with the conclusion that the transcription factor interactions within this region are important for the transcriptional activity of PRC. Since the carboxy-terminal CREB/NRF-1 interaction domain maps to the R/S domain of PRC, one might predict, based on the results with PGC-1α, that this domain would not be required for trans activation of the cytochrome c promoter. Interestingly, deletion of the PRC carboxy terminus, containing both the R/S domain and the RNA recognition motif [PRC(1-1379)], did not significantly reduce PRC trans activation of the promoter, suggesting that the CREB/NRF-1 recognition site within the R/S domain is not required for transcription initiation. The results obtained are unlikely to result from differences in expression because the full-length PRC [PRC(1-1664)] and PRC(Δ433-467) were expressed at similar levels, whereas PRC(1-1379) was expressed at severalfold-higher levels (not shown).
FIG. 7.
Determinants of PRC trans activation of the cytochrome c promoter. A cytochrome c promoter-luciferase reporter construct was trans activated by full-length PRC [PRC(1-1664)] or its mutated derivatives [PRC(Δ433-467) and PRC(1-1379)] in a transient-cotransfection assay. Relative luciferase activity was measured in the absence of PRC subfragments or in the presence of 1 or 2 μg of plasmid expressing PRC(400-604) or PRC(1-221). Values were normalized for transfection efficiency by using a Renilla luciferase control in a dual luciferase reporter system and represent the averages ± standard errors of the means for six separate determinations. A single asterisk indicates a P value of <0.05 compared with values for the uninduced control. Double asterisks indicate P values of <0.05 compared to values obtained from full-length PRC(1-1664) trans activations. wt, wild type.
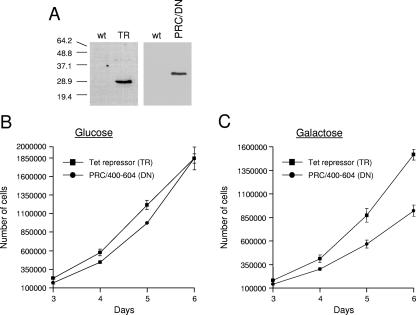
The dominant negative inhibitory effect of the PRC(400-604) subfragment in a transient-transfection assay raised the question of whether this domain could inhibit biological function in growing cells. This possibility was tested by constructing a lentivirus vector designed to express this subfragment constitutively. Stable integrants were obtained following infection of BALB/3T3 cells with virus derived from the PRC(400-604) subfragment-expressing construct and a control virus expressing bacterial TR. TR was selected as the control because it is similar in size to the PRC subfragment and it has no known biological activity in mammalian cells. As shown in Fig. 8A, these proteins were expressed in their respective cell lines but not in the wild-type controls. When plated on glucose growth medium, the PRC(400-604)-expressing cells displayed a small but reproducible growth lag relative to the TR control but showed no significant difference in growth rate (Fig. 8B). However, when plated on galactose growth medium, which requires mitochondrial respiration for the production of ATP, the PRC(400-604)-expressing cells exhibited both a growth lag and a significant reduction in growth rate (approximately twofold) relative to the control (Fig. 8C). This result is consistent with the transfection data and supports an in vivo function for PRC in regulating respiratory growth.
FIG. 8.
Growth comparison of PRC(400-604)-expressing cells to TR-expressing controls on media containing either glucose or galactose as the primary carbon source. (A) Immunoblotting of whole-cell extracts from TR- or PRC/DN-expressing cells (DN, dominant negative) compared to that of the wild type (wt). TR was detected using rabbit anti-TR antibody (Imgenex), whereas PRC/DN was expressed from the lentivirus with a carboxy-terminal V5 tag and detected using anti-V5 antibody (Invitrogen). (B) Growth of TR- and PRC/DN-expressing cells on glucose media. (C) Growth of TR- and PRC/DN-expressing cells on galactose media. Growth curves in panels B and C represent the averages ± standard errors of the means for three separate determinations.
Serum-induced promoter occupancy in vivo.
If PRC targets the cytochrome c promoter through NRF-1 and CREB, one might expect that the upregulation of PRC in the serum-induced transition from G0 to G1 would coincide with increased occupancy of the promoter by PRC in vivo. This was investigated by devising a quantitative ChIP assay using antibodies directed against NRF-1, CREB, phospho-CREB, and PRC to assay the in vivo occupancy of the promoter by these factors in serum-starved and serum-stimulated cells. CREB and phospho-CREB serve as ideal internal controls because serum stimulation leads to the increased phosphorylation of CREB with little or no increase in CREB protein expression (13). The results for three independent experiments show significant occupancy of the cytochrome c promoter by PRC under conditions where the DNA-bound transcription factors NRF-1 and CREB are also present (Table 1). This supports the conclusion that all three factors associate with the promoter in vivo since they are all cross-linked to the same immunoprecipitated chromatin fragment. In addition, there is a significant increase in the occupancy of the promoter by both PRC and NRF-1 at 8 h following serum stimulation of quiescent BALB/3T3 cells (Table 1). This is the time frame where both PRC and cytochrome c expression are induced. CREB occupancy was increased only 1.4-fold, whereas phospho-CREB was enhanced 3-fold, to a level similar to that observed for PRC. These results provide in vivo evidence that PRC occupies the cytochrome c promoter and plays a regulatory role in transcriptional expression in the G0-to-G1 transition.
TABLE 1.
Chromatin immunoprecipitation (ChIP) analysis of cytochrome c promoter occupancy by PRC, NRF-1, and CREB upon serum stimulation of quiescent fibroblasts
| Precipitating antibody | Promoter occupancy valuea for indicated fibroblast group
|
Fold increase (stimulated/starved)b | |
|---|---|---|---|
| Starved (60 h) | Stimulated (8 h) | ||
| Rabbit IgG | 1.0 | 1.0 | 1.0 |
| Anti-NRF-1 | 38.9 ± 17.1 | 98.5 ± 9.1 | 2.5 |
| Anti-PRC(1047-1379) | 3.8 ± 1.1 | 11.9 ± 1.5 | 3.1 |
| Anti-CREB | 8.6 ± 2.3 | 12.3 ± 1.8 | 1.4 |
| Anti-phospho-CREB | 15.3 ± 3.3 | 45.7 ± 3.4 | 3.0 |
Values for relative promoter occupancy represent the averages ± standard errors of the means for three separate determinations.
Increases in levels of relative promoter occupancy for stimulated fibroblasts compared to corresponding levels for starved fibroblasts.
DISCUSSION
Investigations of the bigenomic expression of the mitochondrial respiratory chain have contributed significant insights to the understanding of nucleomitochondrial interactions (15, 28). The characterization of mammalian cytochrome c and cytochrome oxidase genes has led to the identification of nuclear respiratory factors NRF-1 and NRF-2 (GA binding protein). In addition to their role in respiratory chain expression, these transcription factors have been associated with the expression of a variety of genes controlling diverse aspects of mitochondrial biogenesis, including the transcription and replication of mtDNA. Important insights into the means by which transcription factors, such as NRF-1 and -2, can be linked to extracellular signals came with the discovery of the PGC-1 family of regulated coactivators (24). Although the three members of this family, PGC-1α, PGC-1β, and PRC, exhibit clear differences in regulation and transcription factor specificities, they all share the ability to bind NRF-1 and to trans activate NRF-1 target genes (17). PGC-1α is the best-characterized family member, and both gain- and loss-of-function experiments have substantiated its role in mitochondrial biogenesis (37).
Here, we establish that PRC is a growth-regulated member of the PGC-1 coactivator family that has characteristics of an immediate early gene product. It is induced rapidly at both the mRNA and the protein level in response to serum stimulation of quiescent fibroblasts, and its mRNA is superinduced upon the inhibition of protein synthesis by cycloheximide. The similarity of PRC mRNA half-lives in serum-starved, serum-stimulated, and confluent cells, coupled with the serum-induced increase in PRC nascent transcripts (not shown), indicates that transcriptional mechanisms regulate PRC expression. The rapid induction through preexisting factors, coupled with a relatively short mRNA half-life, is typical of immediate early gene products, which include chemokines, growth factors, proto-oncogenes, serine-threonine kinases, and key enzymes involved in nucleic acid metabolism, among others (36). Although transcription factors are also well represented among immediate early gene products, to our knowledge, serum-inducible transcriptional coactivators are not widely known. Coactivators that have been associated with immediate early gene expression, such as p62TCF and MKL1, are activated by phosphorylation and bind serum response factor, a potent activator of the growth factor response (3).
In addition to NRF-1, the cytochrome c promoter contains canonical CREB/ATF recognition sites and these sites have been linked to cytochrome c expression in response to both cAMP (11) and serum stimulation of quiescent fibroblasts (13). In the latter case, induction of cytochrome c mRNA was correlated with elevated levels of heme-containing holo-cytochrome c and enhanced mitochondrial respiration, presumably to help meet energy demands associated with cell division. It is of interest in this context that PRC utilizes the same NRF-1 and CREB/ATF recognition sites for maximal trans activation of the cytochrome c promoter (1). The fact that PRC is rapidly induced upon serum stimulation, can bind NRF-1, and can trans activate NRF-1 target genes suggested that it may function through a specific interaction with CREB as well. This possibility is intriguing in light of the fact that CREB, in addition to mediating a cAMP response to hormones, plays a role in growth factor signaling. This mitogenic pathway involves phosphorylation of CREB by the pp90rsk family of protein kinases and the mitogen- and stress-activated kinases (4, 29).
Here, we find that CREB and NRF-1 bind the same sites on PRC in vitro and that both factors exist in a complex with PRC in vivo. In addition, deletion of the CREB/NRF-1 binding site at amino acids 433 to 467 on PRC inhibits trans activation of the cytochrome c promoter, and a fragment containing this site [PRC(400-604)], when expressed in trans, can inhibit cytochrome c activation by full-length PRC. The same fragment specifically inhibits growth on galactose when stably expressed from an integrated lentivirus. In contrast to glucose growth, where the bulk of the ATP produced is derived from glycolysis, growth on galactose requires mitochondrial respiration for ATP production because of the restricted metabolism of galactose through glucose 6-phosphate (26). Thus, specific dominant negative inhibition of respiratory growth on galactose by PRC(400-604) is consistent with a biological role for PRC in directing cell growth under conditions where energy is predominantly derived through oxidative phosphorylation. On this basis, one might predict that PRC deficiency would affect early postnatal development, when respiratory growth is enhanced, as well as tissues that rely heavily on oxidative energy. It will be of interest to determine both the stage of the cell cycle and the battery of genes affected by dominant negative inhibition of PRC. We note that prolonged growth on galactose abrogates the growth rate differential between the TR control cells and PRC(400-604)-expressing cells. This suggests that compensatory mechanisms may contribute to the maintenance of respiratory growth when PRC function is impaired.
These findings, together with the observation that both CREB and NRF-1 recognition sites are required for maximal PRC activation of the promoter (1), provide compelling evidence that PRC works through the interaction with both CREB and NRF-1. As previously observed for NRF-1 (1), CREB binding to PRC requires its DNA binding domain. NRF-1 has a unique DNA binding domain (33), while CREB binds DNA through a basic leucine zipper (b-Zip) domain (10, 20). A similar requirement for the NRF-1 and PPARγ DNA binding domains, which are also structurally diverse, was found for the interaction of these transcription factors with PGC-1α (37). It remains an open question as to how the same regions on these coactivators recognize disparate DNA binding motifs in their cognate transcription factors. A second CREB/NRF-1 binding site within the PRC R/S domain appears not to function in transcription initiation. The PGC-1α R/S domain is also dispensable for transcription initiation but is required for coupling transcription to RNA processing (19). A requirement for splicing would not be detected in the assay system reported here because the cytochrome c promoter construct is devoid of introns, and thus the expression of the luciferase reporter is independent of RNA splicing.
As shown here by quantitative ChIP assay, PRC occupies the cytochrome c promoter in vivo, and the PRC occupancy of this promoter is enhanced upon serum stimulation of quiescent cells. We also observe PRC occupancy of the human cytochrome c promoter in 293FT cells (not shown). The increase in PRC at the promoter in serum-stimulated BALB/3T3 cells parallels the increase in phospho-CREB detected using anti-phospho-CREB antibody, in keeping with the fact that CREB phosphorylation occurs under these conditions. Anti-CREB antibody is an ideal negative control because steady-state levels of CREB protein show little or no change upon serum stimulation (13), and likewise, the level of occupancy of the promoter by CREB is only modestly elevated. Several members of the CREB/ATF family of factors recognize the canonical CREB binding site. The ChIP results for both anti-CREB and anti-phospho-CREB antibodies provide strong evidence that CREB is actually present at the cytochrome c promoter in vivo. The methodology does not allow absolute comparisons between antibodies because of differences in antibody affinity, accessibility of the various factors, and efficiency of cross-linking. Nevertheless, the results support the conclusion that PRC induction and association with growth-regulated promoters represent a novel pathway for mediating the cellular response to proliferative signals.
The results in this work provide an alternative to the current model of CREB activation. It is well established that phosphorylation of CREB on Ser 133 promotes CREB binding to two structurally related coactivators, CBP and p300 (14). Binding of these coactivators occurs through the CREB P-box, which contains Ser 133 (25). In contrast to PRC and other PGC-1 family members, both CBP and p300 have intrinsic histone acetyltransferase activity, which facilitates transcriptional activation by promoting the remodeling of chromatin (16). PRC has a potent amino-terminal transcriptional activation domain that is highly similar to that found in PGC-1α (1). In PGC-1α, this domain associates with several coactivators that have intrinsic histone remodeling activities, including CBP/p300 and SRC-1 (23). It is likely that targeting of CREB and NRF-1 by PRC would allow recruitment of these same coactivators to the transcription complex via the conserved PRC activation domain. It is possible that phosphorylation of either or both transcription factors stabilizes their interaction with PRC. Thus, PRC induction and interaction with promoter-bound CREB and NRF-1 may represent an alternative mechanism for promoting chromatin remodeling in response to mitogenic stimulation.
Acknowledgments
We thank Winship Herr of Cold Spring Harbor Laboratories for the gift of pNCITE/HCF.
This work was supported by United States Public Health Service grant GM32525-23.
Footnotes
Published ahead of print on 14 August 2006.
REFERENCES
- 1.Andersson, U., and R. C. Scarpulla. 2001. PGC-l-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell. Biol. 21:3738-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cam, H., E. Balciunaite, A. Blias, A. Spektor, R. C. Scarpulla, R. Young, Y. Kluger, and B. D. Dynlacht. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 16:399-411. [DOI] [PubMed] [Google Scholar]
- 3.Cen, B., A. Selvaraj, and R. Prywes. 2004. Myocardin/MKL family of SRF coactivators: key regulators of immediate early and muscle specific gene expression. J. Cell. Biochem. 93:74-82. [DOI] [PubMed] [Google Scholar]
- 4.De, C. D., G. M. Fimia, and P. Sassone-Corsi. 1999. Signaling routes to CREM and CREB: plasticity in transcriptional activation. Trends Biochem. Sci. 24:281-285. [DOI] [PubMed] [Google Scholar]
- 5.Evans, M. J., and R. C. Scarpulla. 1989. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF and intron Sp1 recognition sites. J. Biol. Chem. 264:14361-14368. [PubMed] [Google Scholar]
- 6.Evans, M. J., and R. C. Scarpulla. 1990. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 4:1023-1034. [DOI] [PubMed] [Google Scholar]
- 7.Garesse, R., and C. G. Vallejo. 2001. Animal mitochondrial biogenesis and function: a regulatory cross-talk between two genomes. Gene 263:1-16. [DOI] [PubMed] [Google Scholar]
- 8.Gaspari, M., N. G. Larsson, and C. M. Gustafsson. 2004. The transcription machinery in mammalian mitochondria. Biochim. Biophys. Acta 1659:148-152. [DOI] [PubMed] [Google Scholar]
- 9.Gleyzer, N., K. Vercauteren, and R. C. Scarpulla. 2005. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 25:1354-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez, G. A., P. Menzel, J. Leonard, W. H. Fischer, and M. R. Montminy. 1991. Characterization of motifs which are critical for activity of the cyclic AMP-responsive transcription factor CREB. Mol. Cell. Biol. 11:1306-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gopalakrishnan, L., and R. C. Scarpulla. 1994. Differential regulation of respiratory chain subunits by a CREB-dependent signal transduction pathway. Role of cyclic AMP in cytochrome c and COXIV gene expression. J. Biol. Chem. 269:105-113. [PubMed] [Google Scholar]
- 12.Gugneja, S., and R. C. Scarpulla. 1997. Serine phosphorylation within a concise amino-terminal domain in nuclear respiratory factor 1 enhances DNA binding. J. Biol. Chem. 272:18732-18739. [DOI] [PubMed] [Google Scholar]
- 13.Herzig, R. P., S. Scacco, and R. C. Scarpulla. 2000. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J. Biol. Chem. 275:13134-13141. [DOI] [PubMed] [Google Scholar]
- 14.Johannessen, M., M. P. Delghandi, and U. Moens. 2004. What turns CREB on? Cell. Signal. 16:1211-1227. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, D. P., and R. C. Scarpulla. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 18:357-368. [DOI] [PubMed] [Google Scholar]
- 16.Kouzarides, T. 1999. Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev. 9:40-48. [DOI] [PubMed] [Google Scholar]
- 17.Lin, J., C. Handschin, and B. M. Spiegelman. 2005. Metabolic control through the PGC-1 family of transcriptional coactivators. Cell Metab. 1:361-370. [DOI] [PubMed] [Google Scholar]
- 18.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 19.Monsalve, M., Z. Wu, G. Adelmant, P. Puigserver, M. Fan, and B. M. Spiegelman. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6:307-316. [DOI] [PubMed] [Google Scholar]
- 20.Montminy, M. 1997. Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 66:807-822. [DOI] [PubMed] [Google Scholar]
- 21.Nathans, D., L. F. Lau, B. Christy, S. Hartzell, Y. Nakabeppu, and K. Ryder. 1988. Genomic response to growth factors. Cold Spring Harbor Symp. Quant. Biol. 53:893-900. [DOI] [PubMed] [Google Scholar]
- 22.Puigserver, P. 2005. Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-α. Int. J. Obesity 29:55-59. [DOI] [PubMed] [Google Scholar]
- 23.Puigserver, P., C. Adelmant, Z. D. Wu, M. Fan, J. M. Xu, B. O'Malley, and B. M. Spiegelman. 1999. Activation of PPARgamma coactivator-1 through transcription factor docking. Science 286:1368-1371. [DOI] [PubMed] [Google Scholar]
- 24.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829-839. [DOI] [PubMed] [Google Scholar]
- 25.Radhakrishnan, I., G. C. Pérez-Alvarado, D. Parker, H. J. Dyson, M. R. Montminy, and P. E. Wright. 1997. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell 91:741-752. [DOI] [PubMed] [Google Scholar]
- 26.Robinson, B. H. 1996. Use of fibroblast and lymphoblast cultures for detection of respiratory chain defects. Methods Enzymol. 264:454-464. [DOI] [PubMed] [Google Scholar]
- 27.Scarpulla, R. C. 1999. Nuclear transcription factors in cytochrome c and cytochrome oxidase expression, p. 553-591. In S. Papa, F. Guerrieri, and J. M. Tager (ed.), Frontiers of cellular bioenergetics: molecular biology, biochemistry, and physiopathology. Plenum Publishing Company, London, United Kingdom.
- 28.Scarpulla, R. C. 2006. Nuclear control of respiratory gene expression in mammalian cells. J. Cell. Biochem. 97:673-683. [DOI] [PubMed] [Google Scholar]
- 29.Shaywitz, A. J., and M. E. Greenberg. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68:821-861. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 31.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vega, R. B., J. M. Huss, and D. P. Kelly. 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 20:1868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virbasius, C. A., J. V. Virbasius, and R. C. Scarpulla. 1993. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 7:2431-2445. [DOI] [PubMed] [Google Scholar]
- 34.Virbasius, J. V., C. A. Virbasius, and R. C. Scarpulla. 1993. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev. 7:380-392. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, A. C., R. N. Freiman, H. Goto, T. Nishimoto, and W. Herr. 1997. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol. Cell. Biol. 17:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkles, J. A. 1998. Serum- and polypeptide growth factor-inducible gene expression in mouse fibroblasts. Prog. Nucleic Acid Res. Mol. Biol. 58:41-78. [DOI] [PubMed] [Google Scholar]
- 37.Wu, Z., P. Puigserver, U. Andersson, C. Zhang, G. Adelmant, V. Mootha, A. Troy, S. Cinti, B. Lowell, R. C. Scarpulla, and B. M. Spiegelman. 1999. Mechanisms controlling mitochondrial biogenesis and function through the thermogenic coactivator PGC-1. Cell 98:115-124. [DOI] [PubMed] [Google Scholar]