Abstract
Crystallographic characterization of a ternary complex containing a monomeric gp120 core, parts of CD4, and a mAb, revealed a region that bridges the inner and outer domains of gp120. In a related genetic study, several residues conserved among primate lentiviruses were found to play important roles in CC-chemokine receptor 5 (CCR5) coreceptor utilization, and all but one were mapped to the bridging domain. To reconcile this finding with previous reports that the hypervariable region 3 (V3) of gp120 plays an important role in chemokine coreceptor utilization, elucidating the roles of various V3 residues in this critical part of the HIV type 1 (HIV-1) life cycle is essential. Alanine-scanning mutagenesis was carried out to identify V3 residues critical for CCR5 utilization. Our findings demonstrated that several residues in V3 were critical to CCR5 utilization. Furthermore, these residues included not only those conserved across HIV-1 subtypes, but also those that varied among HIV-1 subtypes. Although the highly conserved V3 residues may represent unique targets for antiviral designs, the involvement of variable residues raises the possibility that antigenic variation in the coreceptor binding domain could further complicate HIV-1 vaccine design.
The entry of HIV type 1 (HIV-1) into target cells generally requires the interaction of the exterior envelope glycoprotein, gp120, with the cellular receptor, CD4, and a chemokine coreceptor, such as CC-chemokine receptor 5 (CCR5) (1–5). Based on mutational analysis (6), it recently was reported that some highly conserved residues in a region that bridges outer and inner domains of gp120 play an important role in CCR5 coreceptor utilization (6, 7). These findings appear to be consistent with the structural characterization of a ternary complex consisting of a gp120 core, a two-domain CD4 molecule, and an antigen-binding fragment of a human mAb, 17b, which blocks interaction between gp120 and the CCR5 coreceptor (8).
Several lines of evidence also have indicated that the hypervariable region 3 (V3) loop of gp120, which was absent from the gp120 core analyzed, participates in chemokine coreceptor utilization. First, the deletion of most of the V3 residues from gp120 had no effect on CD4 receptor binding, but it did abolish CCR5 interaction (6). Second, several mAbs directed against V3 were shown to block the ability of gp120 to interfere with the interaction between CCR5 and its natural ligands (9–12). Furthermore, amino acid substitutions introduced to two V3 residues recently were found to affect CCR5 interactions (6, 13, 14). One of these was a highly conserved arginine residue located adjacent to the N-terminal base of V3 (13). The other was a hydrophobic residue located adjacent to the crest of V3 (6, 14).
The coreceptor binding step of the HIV-1 life cycle is a potential target for therapeutic and prophylactic interventions. To design intervention strategies targeting this critical step of HIV-1 entry requires knowing how V3 works in concert with those residues in the bridging sheet to interact with CCR5, the chemokine coreceptor most often used by the so-called R5 viruses of human and nonhuman primate lentiviruses (15, 16). To that end, it is essential to provide a full account of V3 residues involved in CCR5 utilization. In this study, we identified several V3 residues, both highly conserved and variable ones, that were shown to play a role in CCR5 utilization.
MATERIALS AND METHODS
Construction of Mutant Viruses.
The infectious molecular clone, ConB, which contains the consensus V3 sequence of HIV-1 subtype B and is known to use CCR5 as its entry coreceptor, has been described (13). Oligonucleotide-directed mutagenesis was performed on the 3.2-kb EcoRI–XhoI fragment of ConB, using the method of Kunkel (17). Mutants were identified by DNA sequencing as described (18). The 2.6-kb SalI–BamHI fragment containing the directed mutation was excised from the replicative form of each mutant and used to replace the 2.6-kb SalI–BamHI fragment of ConB. All mutations in the ConB construct were further verified by DNA sequencing.
Virus Infection in HOS-CD4.CCR5 Cells.
Previously described procedures were followed in this analysis (19–21). HOS-CD4.CCR5, HOS-CD4.pBABE-puro, and HOS-CD4.CXCR4 are HOS-CD4 cells stably expressing CCR5, pBABE-puro, and CXCR4, respectively. They were obtained from the AIDS Research and Reference Reagent Program (National Institutes of Health, Bethesda, MD) and propagated at 37°C in DMEM supplemented with 10% heat-inactivated FBS, 1% penicillin-streptomycin, and puromycin at 1 mg/ml as described (19). COS-7 cells were propagated in DMEM supplemented with 10% heat-inactivated FBS and 1% penicillin-streptomycin. Four micrograms of wild-type or mutant DNA were transfected to 3–5 × 106 COS-7 cells by the DEAE-dextran method (22). Cell-free supernatants were collected 72 hr after transfection, filtered through 0.45-mm filters, and assayed by p24 ELISA (DuPont). Equal amounts of wild-type and mutant viruses, as measured by p24, were used to infect 3 × 104 HOS-CD4.CCR5, HOS-CD4.pBABE-puro, or HOS-CD4.CXCR4 cells in a 24-well plate with 1.5 ml of medium per well. A half-milliliter of the culture medium was collected from each culture every 3 days for p24 analysis. The culture was monitored for 7 days.
Western Blot Analysis of Envelope Expression in Transfectants.
Four micrograms of wild-type or mutant DNA were transfected to 3–5 × 106 COS-7 cells by the DEAE-dextran method (22). Mock-transfected, wild-type, and mutant-transfected COS-7 cells were collected 72 hr posttransfection, washed with PBS, and subjected to centrifugation at 2,500 rpm. Cell pellets were resuspended with 100 μl RIPA lysis buffer (0.15 M NaCl/0.05 M Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) and spun at 28,000 rpm (F-13 rotor, Sorvall) at 4°C for 40 min. Ten microliters of each cell lysate was electrophoresed in a 10% SDS-polyacrylamide gel. A reference HIV-1 positive serum (1:200 dilution) was used for Western blot analysis as described (18).
Envelope Incorporation into Virion.
Cell-free supernatants were collected from wild-type and mutant transfectants and filtered through 0.45-mm filters (Nalge). The supernatants were centrifuged at 20,000 rpm (Beckman, SW28 rotor) for 2 hr through a 20% (wt/vol) sucrose cushion. Virus pellets were dissolved in RIPA lysis buffer and electrophoresed on a 12% SDS-polyacrylamide gel. A reference HIV-1 positive serum was used for Western blot analysis as described above.
CD4 Binding Assay.
CV-1 cells were infected with recombinant vaccinia virus vCB5, a soluble CD4-expression vector, for 3 hr in cysteine-free medium and metabolically labeled with S35-cysteine overnight. Equal amounts of supernatants (1.5 ml) were incubated with cell lysates derived from wild-type or mutant transfectants at 4°C for 3 hr, and the mixtures subsequently were incubated with reference serum preabsorbed to protein A-Sepharose beads (23). After washing, the coprecipitated gp120/CD4 complex was eluted in a sample buffer and electrophoresed on 10% SDS-polyacrylamide gel.
RESULTS
Substitution of Highly Conserved Alanine328 of V3.
The first V3 residue studied in this report was the alanine at position 328 (alanine328). A closer examination of various V3 sequences reveals that this V3 residue is highly conserved among HIV-1, HIV-2, and simian immunodeficiency viruses known to use CCR5 as a coreceptor (Figs. 1 and 2) (16). One possibility considered was that this highly conserved alanine residue, like the previously identified arginine at position 298 of V3 (13), may have contributed to functional convergence in CCR5 utilization by this large family of genetically divergent human and nonhuman primate lentiviruses.
Figure 1.
Highly conserved alanine residue at position 328 of V3. The alanine at position 328 is marked by ∗. The two conserved cysteine residues are numbered on top (296 and 330). † indicates consensus V3 sequences of different subtypes of HIV-1, HIV-2, and simian immunodeficiency (16).
Figure 2.
V3 sequences of R5 viruses of different HIV-1 subtypes. Residues identified to be important for CCR5 utilization (∗) are numbered and marked. The two conserved cysteine residues are numbered on top (296 and 330). †, Parentheses indicate the genetic subtypes of HIV-1. The information was derived from refs. 6 and 13–16. For brevity, R5X4 viruses, which use both CCR5 and CXCR 4, have not been included.
To examine this possibility, site-directed mutagenesis was carried out first to replace alanine328 with lysine, a positively charged residue, in a previously described molecular infectious clone, ConB (13). This molecular clone contains the consensus V3 sequence of HIV-1 subtype B (Fig. 3A) and is known to use CCR5 as its entry coreceptor (13, 24). Several macrophage-tropic nonsyncytion-inducing viruses, including HIV-1JRFL and HIV-1ADA, have a V3 sequence identical to that of ConB (24). The resulting mutant was designated 328AK.
Figure 3.
(A) Schematic presentation showing the envelope region and V3 sequences of ConB and alanine328 mutants. ConB contains the consensus V3 sequence of HIV-1 subtype B in the backbone of an infectious molecular clone, HXB2RU3 (6). (B) Effect of amino acid substitutions to alanine328 on CCR5 utilization as measured by log reduction of p24 in virus-infected cultures. The log reduction of p24 between each alanine328 mutant and the wild type also is summarized in A.
Characterization of Alanine328 Mutant.
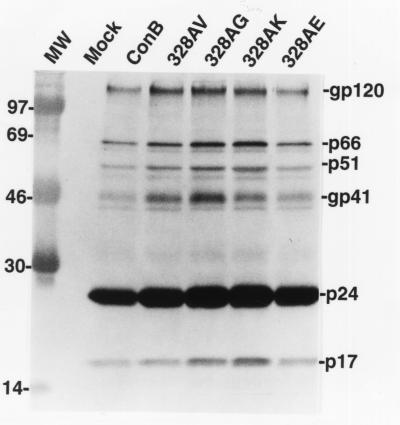
Western blot analysis of envelope proteins derived from the 328AK transfectants readily detected gp160 and gp120 in cell lysates, indicating that this single amino acid substitution did not have a significant effect on the expression and processing of the mutant envelope protein (data not shown). Consistent with the interpretation that this substitution did not result in drastic changes of gp120 conformation, the formation of the gp120/CD4 complex was readily detectable for 328AK by using a standard coprecipitation assay (13, 23, 25, 26) (data not shown). Further analyses of viral lysates derived from culture supernatants of 328AK transfectants also readily detected gp120 and gp41, indicating again that this single amino acid substitution did not cause drastic conformational changes of gp120 (Fig. 4). These findings are in agreement with previous observations that V3 is likely to assume a structural domain relatively independent of the rest of gp120 (27, 28).
Figure 4.
Western blot analysis of envelope proteins incorporated into virions of ConB and alanine328 mutants. Viral lysates prepared from ConB, 328AV, 328AG, 328AK, and 328AE viruses were reacted with a HIV-1-positive human serum. Molecular mass markers, 97 kDa, 69 kDa, 46 kDa, 30 kDa, and 14 kDa are shown in lane MW.
The Effect of Substitution on CCR5 Utilization.
The ability of mutant 328AK to interact with the CCR5 coreceptor was addressed by using a previously described assay in which replication of the mutant virus in three cell lines was compared with that of the wild type (19–21). The three cell lines used were HOS-CD4.CCR5, which stably expresses CD4 and CCR5, HOS-CD4.CXCR4, which stably expresses CD4 and CXCR4, and HOS-CD4.pBABE-puro, which stably expresses CD4, but not CCR5 or CXCR4 (19–21).
As shown in Fig. 3B, when compared with the wild-type virus, mutant 328AK had a more than 1,000 fold reduction of p24 in the culture supernatants of HOS-CD4.CCR5, suggesting that the conserved alanine328 is likely involved in CCR5 coreceptor utilization. In the two control cell lines, HOS-CD4.CXCR4 and HOS-CD4.pBABE-puro, only background levels of p24 were registered for the wild-type or mutant 328AK (data not shown).
Further Characterization of Alanine328 Mutants.
Further analysis was carried out to determine whether substitution by other classes of amino acid residues had a similar negative effect on CCR5 utilization. Three more mutants that had the conserved alanine328 replaced by the negatively charged glutamic acid (328AE), and one of two hydrophobic residues varying in side-chain length, valine (328AV) and glycine (328AG), were constructed for this purpose. None of these substitutions drastically altered the overall conformation of gp120, as was the case for mutant 328AK (Fig. 4 and data not shown).
As shown in Fig. 3, mutants 328AE, 328AG, and 328AV, all had impairment in CCR5 utilization, though the effect of the valine substitution was less prominent. The finding that neither charged residues, nor hydrophobic residues were well tolerated at this position is compatible with the interpretation that the highly conserved alanine328 is likely to play an important role in CCR5 utilization.
The Role of Other V3 Residues in CCR5 Utilization.
To gain a more complete understanding of the extent of the involvement of V3 in CCR5 utilization, alanine-scanning mutagenesis subsequently was carried out on those V3 residues whose possible involvement had not been investigated. Alanine was chosen as a substituent because its small nonpolar methylene side chain is less likely to impose severe constraints on gp120. Among the 33 V3 residues of the virus we studied, 24 had yet to be analyzed. The nine that had been studied included arginine298 and its seven adjacent residues (13), as well as alanine328 examined in this study.
The alanine scanning approach adopted in this study addressed the role of V3 residues from lysine at position 305 to arginine at position 327, and the histidine at position 329 in CCR5 utilization. In the case of the alanine residue at position 314, substitution by a lysine residue was carried out (Fig. 5). The resulting mutant viruses, which were designated according to the position of each alanine substitution, are shown in Fig. 5. As was the case with alanine328 mutants, there was no evidence of global perturbation of gp120 conformation for these additional mutants (data not shown).
Figure 5.
Effect of amino acid substitutions to V3 residues on CCR5 utilization as measured by log reduction of p24 in virus-infected cultures. ∗ indicates less than 1 log difference in p24 between V3 mutant and the wild type.
Among this series of 24 mutants, only substitutions introduced to lysine305, isoleucine307, arginine313, and phenylalanine315 resulted in significant reduction of p24 in HOS-CD4.CCR5 cells when compared with the wild type (Fig. 5). The difference between the other 20 mutants and the wild-type virus was less than 10-fold, a cut-off used in this study for comparative purposes. None of the mutants analyzed infected control cell lines, HOS-CD4.CXCR4 and HOS-CD4.pBABE-puro, as only background levels of p24 were scored (data not shown). These results indicated that the majority of V3 residues were not absolutely required for interaction with the CCR5 coreceptor. However, four residues flanking the relatively conserved “GPG” residues in the crest of V3 appeared to be involved in CCR5 utilization.
DISCUSSION
A rather unusual feature revealed by the crystal structure of a ternary complex of gp120, CD4 and 17b, a presumed surrogate of the CCR5 coreceptor, is that only the heavy chain of 17b was in contact with gp120 (8). The presence of a large gap between the gp120 core and the light chain of 17b raises the question of how 17b binds to native gp120. Based on the position of two V3 residues retained in the complex, it is believed that the base of V3 is in close proximity to the bridging sheet, where other residues critical for CCR5 utilization were identified (6–8). This interpretation allows for the possibility that the light chain of 17b could have interacted with V3, if most of the V3 residues were antigenically compatible and had not been deleted from the gp120 core (Fig. 6). Our finding that several V3 residues have a role in CCR5 utilization is in agreement with the inference that both the bridging sheet and the V3 of gp120 are functionally related domains with regard to HIV-1 chemokine coreceptor utilization.
Figure 6.
Schematic drawing of the ternary complex consisting of gp120 core, two-domain CD4, and Fab fragment of 17b, as revealed by crystallographic study (8). A large gap was present between the base of V3 and light chain of 17b, a surrogate molecule of CCR5. The V3 loop is depicted as occupying this gap if there was an antigenic match. V3 residues identified to be critical for CCR5 interaction in this study are circled. BRS, bridging sheet; H.C., heavy chain; L.C., light chain.
Taken together, six V3 residues have been identified to be important for CCR5 utilization by the R5 viruses studied (6, 13, 14). Of those residues, two (arginine298 and alanine328) are located in the base of V3 and are highly conserved by R5 viruses of all HIV-1 subtypes (Fig. 1) (16). With the emergence of resistant mutants to highly active antiretroviral therapy (29), these highly invariable residues as well as some of those identified in the bridging sheet (6) may represent new targets for antiviral designs aimed at blocking the coreceptor entry step of HIV-1 replication.
The other four residues identified to be critical for CCR5 utilization (lysine305, isoleucine307, arginine313, and phenylalanine315) are believed to be located in two β-strands flanking the crest of V3 (24). The symmetry these four residues displayed in relation to the highly conserved glycine310 at the crest of V3 raises the interesting question of whether these residues help preserve some as-yet-to-be-defined local conformation critical for CCR5 utilization. Although such a possibility cannot be completely ruled out, a more direct role for these residues is also possible. It is known that the interaction between CCR5 and its natural ligands can be inhibited by gp120 (9–12). Anti-V3 mAbs targeting residues at and around the crest of V3 have been shown to be able to block such inhibition. Furthermore, it should be noted that mutational studies of CCR5 have reported several acidic and aromatic residues in the extracellular domains of CCR5 as critical for CCR5 utilization (30–32). Our findings that critical V3 residues included both basic and hydrophobic residues are compatible with the interpretation that the interactions between V3 and CCR5 could involve electrostatic and hydrophobic interactions. Whatever the mechanism, our finding clearly indicates that residues adjacent to the crest of V3 can have a role in CCR5 utilization.
A closer examination of the V3 sequences of various R5 viruses reveals that among the four V3 residues found to be important for CCR5 utilization, only isoleucine307 is conserved by all known R5 viruses of HIV-1 subtype B (Fig. 2). For R5 viruses outside of the HIV-1 subtype B, isoleucine307 and arginine313 are not conserved by HIV-1 subtypes A, C, and E (16). Similarly, lysine305 and arginine313 are not conserved by R5 viruses of HIV-1 subtypes B, C, and E (16). Although phenylalanine315 is conserved among R5 viruses of HIV-1 and the more distantly related HIV-2 and simian immunodeficiency, this residue is not conserved by R5 viruses of HIV-1 subtypes F and G (16). More studies are required to determine which V3 residues contribute to the functional convergence in CCR5 utilization by these genetically divergent R5 viruses. One of the possibilities is that V3 residues at corresponding positions to those identified in this study are also critical for CCR5 utilization by other R5 viruses. Alternatively, V3 residues mapped to positions different from those identified here may be involved. For instance, a V3 residue corresponding to isoleucine325 of the virus we studied was previously reported to play a role in CCR5 utilization (33). Either of the two scenarios would indicate that functional convergence in CCR5 utilization by different R5 viruses is not just mediated by conserved residues.
Virus entry is an important target for HIV-1 vaccine development. Two functionally important steps of HIV-1 entry that have been extensively studied are CD4 binding and chemokine coreceptor interaction. Structural analysis recently has revealed that the CD4 binding domain of gp120 involves not only conserved, but also variable residues (7, 8). The interaction between gp120 and CD4 appears to be mediated by backbone carbons, rather than the characteristic sidechains of those variable gp120 residues (7, 8). It is believed that HIV-1 may have evolved to rely on antigenic variation in this functionally important domain of gp120 to evade immune surveillance (7, 34). Our finding that CCR5 coreceptor utilization by HIV-1 involves not only conserved, but also variable residues raises the possibility that antigenic variation in the coreceptor-binding domain of gp120 poses another challenge to HIV-1 vaccines targeting the entry steps of the HIV-1 life cycle.
Acknowledgments
We thank V. Philippon, H. G. Brumblay, and S. Y. Chang for helpful discussions; M. F. McLane, E. J. Tsai, Y. J. Zhao, and M. Chang for technical assistance; R. Rawat for editorial assistance, and Dr. B. Moss for the soluble CD4 expression vector. The following reagents were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: HOS-CD4.CCR5 and HOS-CD4.pBABE-puro, and HOS-CD4.CXCR4 cells were from Dr. Nathaniel Landau, Aaron Diamond AIDS Research Center. This work was supported in part by Public Health Service Grants CA-39805 from the National Institutes of Health.
ABBREVIATIONS
- HIV-1
HIV type 1
- V3
hypervariable region 3
- CCR5
CC-chemokine receptor 5
References
- 1.Berger E A. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 2.Moore J P, Trkola A, Dragic T. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 3.Doms R W, Peiper S C. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 4.Doranz B J, Berson J F, Rucker J, Doms R W. Immunol Res. 1997;16:15–28. doi: 10.1007/BF02786321. [DOI] [PubMed] [Google Scholar]
- 5.Broder C C, Dimitrov D S. Pathobiology. 1996;64:171–179. doi: 10.1159/000164032. [DOI] [PubMed] [Google Scholar]
- 6.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 7.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. Nature (London) 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 8.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Nature (London) 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, et al. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 10.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 11.Verrier F C, Charneau P, Altmeyer R, Laurent S, Borman A M, Girard M. Proc Natl Acad Sci USA. 1997;94:9326–9331. doi: 10.1073/pnas.94.17.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W K, Dudek T, Zhao Y J, Brumblay H G, Essex M, Lee T H. Proc Natl Acad Sci USA. 1998;95:5740–5745. doi: 10.1073/pnas.95.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S, Caron M G, Cullen B R. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger E A, Doms R W, Fenyo E M, Korber B T, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. Nature (London) 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 16.Korber B, Hahn B, Foley B, Mellors J W, Leitner T, Myers G, McCutchan F, Kuiken K. Human Retroviruses and AIDS 1997: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences. Los Alamos National Laboratory, Los Alamos, NM: Theoretical Biology and Biophysics; 1997. [Google Scholar]
- 17.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee W R, Syu W J, Bin D, Matsuda M, Tan S, Wolf A, Essex M, Lee T H. Proc Natl Acad Sci USA. 1992;89:2213–2217. doi: 10.1073/pnas.89.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Huang Y, He T, Cao Y, Ho D D. Nature (London) 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- 21.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W K, Essex M, Lee T H. J Virol. 1996;70:607–611. doi: 10.1128/jvi.70.1.607-611.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C-N, Robinson J, Mazzara G, Cheng Y-L, Essex M, Lee T-H. AIDS Res Hum Retroviruses. 1995;11:777–780. doi: 10.1089/aid.1995.11.777. [DOI] [PubMed] [Google Scholar]
- 24.Myers G, Korber B, Foley B, Jeang K-T, Mellors J W, Wain-Hobson S. Human Retroviruses and AIDS 1996: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences. Los Alamos National Laboratory, Los Alamos, NM: Theoretical Biology and Biophysics; 1996. [Google Scholar]
- 25.Thali M, Olshevsky U, Furman C, Gabuzda D, Li J, Sodroski J. J Virol. 1991;65:5007–5012. doi: 10.1128/jvi.65.9.5007-5012.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olshevsky U, Helseth E, Furman C, Li J, Haseltine W, Sodroski J. J Virol. 1990;64:5701–5707. doi: 10.1128/jvi.64.12.5701-5707.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollard S R, Rosa M D, Rosa J J, Wiley D C. EMBO J. 1992;11:585–591. doi: 10.1002/j.1460-2075.1992.tb05090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen J. Science. 1997;277:32–33. doi: 10.1126/science.277.5322.32. [DOI] [PubMed] [Google Scholar]
- 30.Doranz B J, Lu Z-H, Rucker J, Zhang T-Y, Sharron M, Cen Y-H, Wang Z-X, Guo H-H, Du J-G, Accavitti M A, et al. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dragic T, Trkola A, Lin S W, Nagashima K A, Kajumo F, Zhao L, Olson W C, Wu L, MacKay C R, Allaway G P, et al. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, et al. J Virol. 1998;72:1160–1164. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyatt R, Sodroski J. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]