Abstract
The changes that lead to activation of G protein-coupled receptors have not been elucidated at the structural level. In this work we report the crystal structures of both ground state and a photoactivated deprotonated intermediate of bovine rhodopsin at a resolution of 4.15 Å. In the photoactivated state, the Schiff base linking the chromophore and Lys-296 becomes deprotonated, reminiscent of the G protein-activating state, metarhodopsin II. The structures reveal that the changes that accompany photoactivation are smaller than previously predicted for the metarhodopsin II state and include changes on the cytoplasmic surface of rhodopsin that possibly enable the coupling to its cognate G protein, transducin. Furthermore, rhodopsin forms a potentially physiologically relevant dimer interface that involves helices I, II, and 8, and when taken with the prior work that implicates helices IV and V as the physiological dimer interface may account for one of the interfaces of the oligomeric structure of rhodopsin seen in the membrane by atomic force microscopy. The activation and oligomerization models likely extend to the majority of other G protein-coupled receptors.
Keywords: G protein-coupled receptor, G protein-coupled receptor activation, phototransduction, membrane protein structure
G protein-coupled receptors (GPCRs) comprise the largest family of transmembrane receptors in animals, accounting for ≈3% of the genome (1). GPCRs are involved in detecting a large variety of chemical and physical signals, and they are the targets of ≈50% of current therapeutics. Structural information on GPCRs has been limited because of difficulties with their expression, purification, intrinsic chemical heterogeneity, and instability. These biochemical problems were overcome by using rhodopsin as a model GPCR, as it is highly expressed in a homogeneous form in rod photoreceptors and stabilized in the ground state by its covalently bound chromophore, 11-cis-retinal (2).
The first crystal structure of rhodopsin revealed the arrangement of helices, the interhelical connections, the chromophore binding site, the extracellular “plug,” interactions involved in ligand binding in other GPCRs, and cytoplasmic helix 8 (3). Further improvements in the rhodopsin crystals yielded higher-resolution diffraction data that provided details about the effects of water molecules located close to the chromophore and more precise descriptions of the cytoplasmic loops. However, the improved crystals did not elucidate the mechanism of activation (4, 5). The arrangement of the seven transmembrane helices of rhodopsin differs from that in the more completely structurally studied bacterial retinoid-binding protein, bacteriorhodopsin (6).
Upon absorption of a single photon of light, rhodopsin's chromophore, 11-cis-retinal, isomerizes to form all-trans-retinal, a covalently bound, full agonist. Once all-trans-retinal is formed, the protein portion of rhodopsin progresses through a series of photostates, including bathorhodopsin, lumirhodopsin, and metarhodopsin I (Meta I), leading to the formation of the deprotonated state, metarhodopsin II (Meta II), its fully activated state. After attainment of the Meta II state, rhodopsin can progress to the metarhodopsin III state where the Schiff base becomes protonated once again. Eventually, the chromophore is hydrolyzed from metarhodopsin III rhodopsin or other protonated intermediates and dissociates from the binding pocket (2). Recently, the crystal structures of the bathorhodopsin and lumirhodopsin photointermediates were solved and confirmed that only small-scale, local changes occurred up to the formation of lumirhodopsin (7, 8). Furthermore, the structure of a predominantly Meta I intermediate (60%) was determined at low resolution by electron crystallography, revealing that the transition from ground state to Meta I does not involve large rigid-body movements of helices (9). Although most of the light energy is dispersed before rhodopsin reaches Meta I (10), some biophysical studies suggested large conformational changes of the tightly packed transmembrane helices during the transition from the inactive Meta I to the active Meta II state (11).
Changes in the extracellular domains have been proposed to be involved in the mechanism of activation of other GPCRs. The crystallographic structure of the extracellular ligand-binding region of the metabotropic glutamate receptor GluR1 demonstrated that the functional receptor is a disulphide-linked homodimer, whose “active” and “resting” conformations are modulated through the extracellular dimeric interface. Movements of the extracellular domains in the dimer can affect the separation of the transmembrane domains, activating the receptor without major conformational changes in the individual transmembrane domains (12, 13). A similar mechanism has been implicated from x-ray studies of the extracellular dimeric domains of the follicle-stimulating hormone receptor (14). However, up to this point, no crystallographic structure of a transmembrane domain of a GPCR in its activated state has been determined, and all hypotheses about large structural rearrangements upon activation have had to be estimated through EPR, Cys cross-linking, molecular modeling, and other indirect methods (2).
Because of the inherent instability of rhodopsin in its photoactivated deprotonated state, it was necessary to determine crystallization conditions for ground-state rhodopsin that yielded crystals that were stable upon exposure to light at room temperature. Because prior rhodopsin crystal forms lost diffraction when illuminated, we developed a purification protocol and crystallization conditions to obtain two additional crystal forms of rhodopsin that are capable of withstanding photoactivation (15). Using these procedures, crystals of ground-state rhodopsin were grown, and then the crystals were exposed to light to transform the rhodopsin into its photoactivated deprotonated intermediate. One is a rhombohedral structure that diffracts to 3.7-Å resolution but loses diffraction upon light exposure (space group R32, with one molecule in the asymmetric unit), whereas the other is trigonal (space group P3112, with three molecules in the asymmetric unit) (see Supporting Text, which is published as supporting information on the PNAS web site), which diffracts to 4.1–4.2 Å both before and after photoactivation.
Results
Characterization of the Photostate of Rhodopsin Crystals.
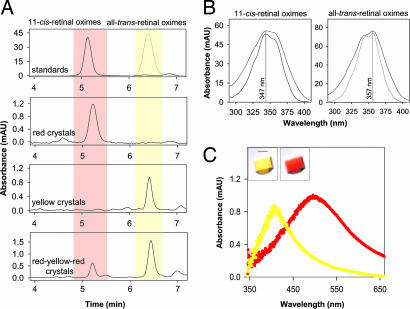
To characterize the photoactivation of these rhodopsin crystals, we first examined the retinoid composition of ground-state (red) and photoactivated (yellow) crystals. The red crystals exclusively contained 11-cis-retinal as determined by normal-phase HPLC (Fig. 1 A and B), indicating that they are composed solely of ground-state rhodopsin. Exposure of the red crystals to light led to the complete isomerization of 11-cis-retinylidene to its all-trans isomer within the protein crystals, as no 11-cis-retinal oximes were observed. Interestingly, photoactivated crystals left in the dark for several hours reverted to a light red color, and retinoid analysis of these crystals revealed the presence of ≈25% of the 11-cis chromophore (as measured by normal-phase HPLC) (Fig. 1A Bottom), indicating that chemical back-isomerization had occurred and the retinal had not been hydrolyzed. Freezing crystals in liquid nitrogen prohibits this reisomerization. In addition, red-reverted crystals exposed to light turned yellow, and when incubated in the dark, they turned reddish again (data not shown). The treatment of photoactivated rhodopsin crystals with hydroxylamine yields colorless crystals, indicating that the Schiff base linkage of the retinal is accessible to hydroxylamine. When rhodopsin in solution or membranes is treated with hydroxylamine, only rhodopsin that has progressed to or past the Meta II photointermediate reacts with hydroxylamine. Eventually, after prolonged exposure to light, the rhodopsin crystals also become colorless. Thus, two paths of relaxation can take place: a slow hydrolysis of the Schiff-base linkage or a slow thermal isomerization back to the 11-cis conformation. An important consequence of this observation is that the chromophore is not hydrolyzed (which would indicate that the photostate of the crystals has progressed beyond the Meta II intermediate in these crystals) and is therefore still attached to Lys-296 by its Schiff base linkage. This reisomerization could be promoted by the steric restraints imposed by contacts in the chromophore binding site.
Fig. 1.
Analysis of retinoid content and absorption spectra of single rhodopsin crystals. (A) Representative chromatograms showing detection and separation of the retinal oxime isomers found in red, yellow, and red-reverted rhodopsin crystals. Identification of the 11-cis and all-trans isomers of retinal oximes was based on comparisons of elution times and absorbance spectra to synthetic standards plotted in the top panel. The chromatograms shown are representative of 20 single crystal measurements. Similar results were obtained in 10 independent experiments. (B) Absorption spectra of the peak confirm the identity of isomerization state of the chromophore in red (Left) and yellow (Right) crystals. Black and gray lines correspond to the normalized spectra of the standards and samples, respectively. (C) Absorption spectra of ground-state rhodopsin (red) and photoactivated (yellow) crystals. Photoactivation of rhodopsin in the red crystals induces a shift of its maximum absorbance from 500 nm to ≈380 nm characteristic of the Meta II state of rhodopsin. (Scale bar: 0.1 mm.)
To further characterize the photoactivated rhodopsin crystals, we analyzed the absorption spectra of both red and yellow crystals by single-crystal microspectrophotometry. All red crystals that were examined had the characteristic absorption maximum at ≈500 nm of ground-state bovine rhodopsin in solution (Fig. 1C). Upon exposure to light, all examined crystals (n = 20) showed the same shift of absorbance maxima to 390–400 nm. Two spectroscopically observed photointermediate states of rhodopsin exhibit this characteristic spectra: Meta I-380 (also called Meta IIa) and Meta II (Meta IIb) rhodopsin in solution both exhibit λmax close to this value (380 nm) and likely only differ in the protonation state of an amino acid side chain (not the Schiff base Lys-296). This maximum is attributed to the protonation state of the Schiff base linkage of the chromophore to Lys-296, a step both necessary for the attainment of the active state and indicative of the attainment of the activated state.
Importantly, photoactivated rhodopsin crystals strongly activate G protein when dissolved in detergent as determined by fluorescence assays (Fig. 5, which is published as supporting information on the PNAS web site). Meta II binds heterotrimeric transducin and induces the exchange of the bound GDP molecule for a GTP by the Gαt subunit, which causes its dissociation from the Gβγt subunits. During this dissociation, the increase of Trp fluorescence of Gαt can be monitored, which indicates that nucleotide exchange has occurred within Gαt, yielding activated subunits. In the direct intrinsic fluorescence assay (16), rhodopsin from yellow crystals activated Gαt with an initial activation rate of k0 ≈1.4 × 10−3 s−1 (Fig. 5A). Rhodopsin from red crystals, illuminated in the reaction solution, activated Gαt with a similar rate of k0 ≈1.9 × 10−3 s−1 (Fig. 5B). In a control experiment, the activation of Gαt by rhodopsin from red crystals in the dark was also detected, but the activation rate (k0 ≈2.8 × 10−8 s−1) was much slower (Fig. 5C) and could be caused by photoactivation by the 300-nm excitation beam of the instrument. These results indicate that in photoactivated crystals the ability of rhodopsin to activate Gαt remains unaffected. Thus, the photoactivated rhodopsin in the crystals exclusively contains the all-trans-chromophore and exhibits the anticipated bathochromic shift in the absorption maxima to ≈380, indicating the formation of a deprotonated Schiff-base photointermediate of rhodopsin. When dissolved, it can activate Gαt, demonstrating that the rhodopsin in the yellow crystals has been photoactivated.
Dimeric Structure of Rhodopsin.
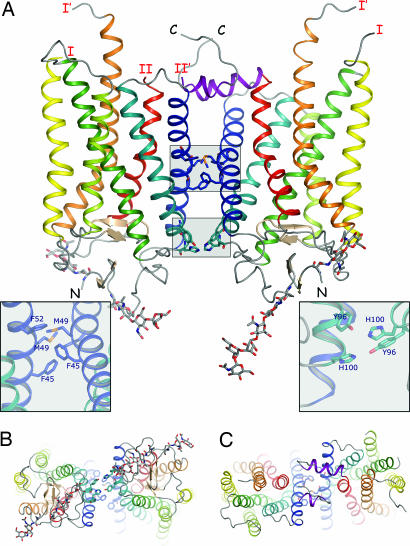
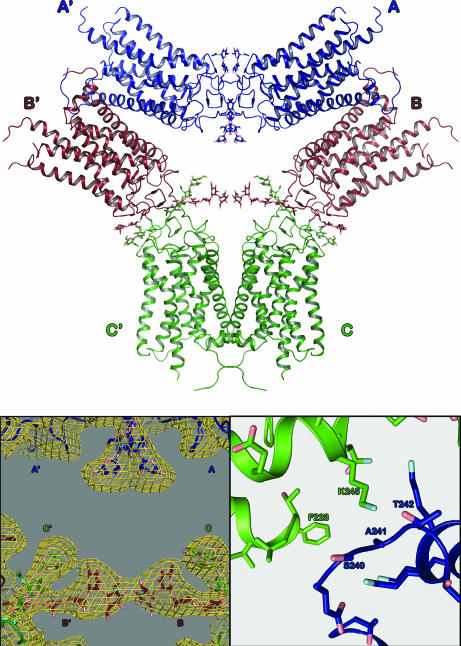
The trigonal crystal form contains three molecules per asymmetric unit, with the A and B molecules arranged as a dimer (Fig. 2) and the C subunit making a similar dimer with a crystallographically related C subunit (Fig. 3Upper). The dimer is stabilized through three symmetric contact patches consisting of a hydrophobic interaction at the top of helix I centered at Met-49 (Fig. 2A Left Inset), or 1.34 in nomenclature for GPCRs (17), polar interactions between Tyr-96 (2.63) and His-100 (2.67) (Fig. 2A Right Inset), and hydrophobic interactions at the C terminus that may involve the palmitoylate groups on C322 and C323 (data not shown). In the ground-state structure, no density is observed for the chromophore in any of the three copies of rhodopsin. For a complete description of the extent of various models, see Table 1, which is published as supporting information on the PNAS web site. The total buried surface area in the interface between rhodopsin molecules in the dimer is ∼800 Å2. The arrangement of the pairs of dimers within the structure forms a triangle (Fig. 3 Upper) with the primary interaction between dimers consisting of interactions at the extracellular face of the protein (Fig. 3). In the rhombohedral crystal form, a similar dimer interaction is also observed, which is imposed by the crystallographic space group (unpublished results).
Fig. 2.
Structure of photoactivated rhodopsin. (A) The A and B subunits form a dimer along helices I and II. This dimer contact depends upon three small areas of contact. The main interaction involves hydrophobic contacts at the level of Phe-45 (1.30), Met-49 (1.34), and Phe-52 (1.37) residues on both subunits (Left Inset). This interaction is further enhanced by two polar interactions between Tyr-96 (2.63) and His-100 (2.67) on adjacent molecules (Right Inset) and a hydrophobic contact between the palmitoylate group covalently attached to Cys-322 in one subunit and the protein backbone of residues 308 and 309 in the opposite subunit (data not shown). Formation of this dimer buries only ≈800 Å of surface area. Right Inset has been rotated ≈90° about the horizontal axis for clarity. The C subunit forms a similar crystallographic dimer with one of its symmetry-related copies with similar buried surface area and contacts. Helices are denoted by color: H-I, blue; H-II, teal; H-III, green; H-IV, yellow-green; H-V, yellow; H-VI, orange; H-VII, red; H-8, purple. Loops C-II and C-III are partially disordered in the photoactivated structure, and the last and first observed residues in the structure are labeled I/I′ and II/II′, respectively. N and C denote the respective termini of the protein. (B) The cytoplasmic face of the dimer. (C) The extracellular face of the dimer. All molecular graphics representations were generated with PYMOL (34).
Fig. 3.
Superassembly of rhodopsin dimers found in the unit cell. (Upper) The trimeric assembly of rhodopsin dimers is held together by interactions between their extracellular regions. These interactions are further strengthened by interactions between the polysaccharide chains of each molecule. This interface, which buries ≈2,400 Å2, is the most extensive in the crystallographic unit cell. The hexameric superassembly consists of two asymmetric units (ASU). A similar superassembly is seen in the rhombohedral (R32) crystal form. Chains in one ASU are denoted with the letters A, B, and C, and in the other ASU they are denoted A′, B′, and C′. Monomers A, B, and C are colored blue, red, and green, respectively, and pertinent oxygen and nitrogen atoms are denoted in pink and cyan, respectively, for clarity. (Lower Left) A 3-fold averaged, density modified map calculated by using DM, contoured at 1σ, clearly indicates that the polysaccharide chains wrap around one another to strengthen this interface. Additionally, σA-weighted 2 Fo − Fc maps also are consistent with these structures for the polysaccharides. (Lower Right) The interface between adjacent superassemblies (not shown in Upper), only buries ≈500 Å2 of surface area and consists of both charged and hydrophobic interactions. The 2 Fo − Fc density for this region in the photoactivated crystals is discontinuous, and no Fo − Fc density is observed in this region. Because this interface is minimal, the rhodopsin is largely unconstrained, and the crystal is capable of accommodating the small conformational changes associated with photoactivation. The small extent of the intermonomer contacts could also account for the limited resolution.
Differences in the Crystal Lattice of the Ground-State Structure and Previously Published Structures.
The cytoplasmic loop III (C-III) follows the trace of the Schertler group's model (Protein Data Bank ID code 1GZM) (4), including the extension of helix V (Fig. 6, which is published as supporting information on the PNAS web site). This result is not surprising, because those crystals were also presumably delipidated, unlike the tetragonal crystals used for the initial structure determination and subsequent improvements (Protein Data Bank ID codes 1F88 and 1U19) (5). In the crystal structures described herein, portions of the C terminus appear to take a decidedly different path than they do in either models Protein Data Bank ID codes 1GZM or 1U19 (data not shown), although this difference could be caused by intermonomer contacts close to helix 8. In both red and yellow crystals, the electron density for the region between residues 121 (3.36) and 127 (3.42) in the middle of helix III is discontinuous, more so in the yellow than the red structure, indicating that this region is mobile within the rhodopsin molecules. In lumirhodopsin, this region is also disordered (8). It is interesting to note that in the first rhodopsin structures that had rhodopsin arranged in dimers in a head-to-tail orientation contacts along helix I were crucial to support that interface; however, their formation buried >2,400 Å2 of surface area (3). Some of the residues used to form the dimer interface are located between rhodopsin monomers in both our structure and the head-to-tail dimer.
Dimer Interfaces within the Structures Compared with Other Observed and Proposed Dimer Interfaces.
When the dimer observed in these crystals is superimposed onto the electron density maps that were generated from 2D crystals of ground-state or Meta I rhodopsin, it becomes clear that although both use helix I to form the interface, the contacts and angles that the two monomers use to form the dimer are distinct. These differences can account for the formation of crystals that progress to the deprotonated intermediate, whereas rhodopsin in the 2D crystals formed a mixture of photointermediates that were predominately Meta I. It should be noted that the contacts between adjacent dimers in the 2D crystals are significantly different from the contacts seen in our crystals, which could also account for the inability of the 2D crystals to fully attain the Meta II state. Furthermore, the lipid content of the 2D crystals is likely different from the largely delipidated, purified rhodopsin from which our crystals are grown.
Differences Between Ground-State and Photoactivated Rhodopsin.
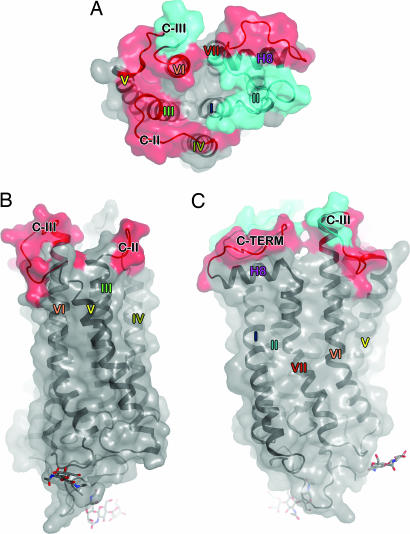
The photoactivated structure found in the yellow crystals is similar to that of the ground-state structure with several significant exceptions. Although both structures were refined at comparable resolution limits, portions of the cytoplasmic loops C-II and C-III are found to be disordered in photoactivated rhodopsin (Figs. 2–4 and 6). Furthermore, C-III does not follow the path that it does in the ground-state crystals as there is no Fo − Fc density observed for it, even at very low contour levels. Interestingly, in the B subunit, density that may correspond to the β-ionone ring is seen between H-V and H-VI (chromophore is modeled into this density in Fig. 7, which is published as supporting information on the PNAS web site), close to Phe-212 (5.43), Trp-265 (6.48), Leu-266 (6.49), and Tyr-268 (6.51), residues that have been implicated as having direct interactions with the β-ionone ring (3). Most importantly, our placement of the β-ionone ring in the activated structure is supported by solid-state NMR studies of changes within the chromophore and surrounding residues upon activation of rhodopsin to Meta II (18). It has been suggested (18) that upon formation of Meta II, a change in the conformation of Tyr-268 (6.51) (induced by the chromophore) could stabilize the Glu-181 anion and modify its water-modulated interaction with Glu-113 (4.70), the residue that interacts with the protonated Schiff base nitrogen (19). The reorganization of loop C-III has been inferred recently from monoclonal antibody studies (20). Residues 175 and 176 are also different between the two structures, possibly because of the flexibility of the adjacent Gly-174 residue. These models are consistent with the photoactivated structure and position of the chromophore. It is important to note that because of the low resolution of both of these structures, the locations of the main-chain atoms are not precisely determined, and side-chain information is largely absent. However, large displacements of individual helices would be evident at this resolution.
Fig. 4.
Differences between ground-state and photoactivated structures. Regions found to be disordered and not observed in all three subunits in the photoactivated structure are depicted in cyan. Regions that are found in different orientations in the photoactivated structure when compared with their equivalent subunit in the low-resolution ground-state structure are denoted in red on a molecular surface representation of the most complete rhodopsin structure to date (Protein Data Bank ID code 1U19). The regions of the cytoplasmic surface that are mobile are important for activation of transducin [cytoplasmic loop II (C-II), cytoplasmic loop III (C-III), and the C terminus]. Because of the low resolution and completeness of the trigonal data sets, estimated coordinate errors were calculated with the program ESCET, indicating coordinate precisions of 1.4 and 1.1 Å for the photoactivated and ground-state data sets, respectively. Differences of less than these amounts are within experimental error at this resolution (see Supporting Text). (A) View of the intracellular side of rhodopsin. (B and C) Side views of the monomer. C depicts the monomer after a 90° rotation about the vertical axis.
Discussion
In this work we developed procedures for the purification and crystallization of highly delipidated rhodopsin. These crystals diffract to 4.15-Å resolution, lower than the previously reported tetragonal and trigonal crystals (4, 5). However, in contrast to previous rhodopsin crystals, our crystals remain ordered when exposed to light. The photoactivated deprotonated Schiff base intermediate state of rhodopsin in these crystals absorbed similarly to Meta II in biological membranes, activated Gαt when dissolved in detergent, and contained all-trans-retinylidene in the binding pocket.
Although earlier work that relied upon indirect methods such as EPR, Cys cross-linking, and fluorescence predicted that there are large rigid body movements of the transmembrane helices upon attainment of the Meta II state (11, 17, 21) the scale of movements we predict based on the photoactivated crystal structure is much smaller. This finding is not altogether surprising as analysis of the structural changes upon activation of the phototaxic archeal protein bacteriorhodopsin were predicted to be much larger when measured via indirect biochemical methods than what was seen in the activated crystal structure (22). Furthermore, these biochemical methods are limited in their resolution and give only pairwise distances that can be explained by rigid body movements, rotations, displacement of single residues, or a combination of all these factors. Our crystal structure indicates that the extent of the structural changes that were predicted by these indirect biophysical methods is most likely overestimated. It is important to add that restraints imposed by the crystal lattice may lead to an underestimation of the scale of the changes upon activation as well, but the paucity of intermonomeric contacts within the crystals suggests otherwise. In summary, the transformation from the ground state to the photoactivated deprotonated intermediate state involves minor changes in the structure, in contrast to earlier predictions of large structural rearrangements (11). Additional work is needed to determine the relationship of this activated form of rhodopsin in the crystals to Meta II.
Based on our crystallographic models, we hypothesize that upon photoactivation rhodopsin relaxes and loses its counter ion (23). Its cytoplasmic loops become more flexible, allowing the induced fit of heterotrimeric transducin (Gt). The flexibility of the structures of both the receptor and Gt (24) allows the adaptation of the proteins to form a productive complex. The dimer found in these crystals fits well with the atomic force microscopy-derived model (25–27) (Fig. 8, which is published as supporting information on the PNAS web site). The observed dimer contains both rhodopsin monomers in the same orientation unlike the dimers found in previous rhodopsin crystal structures (3, 4). The dimer is stabilized through a series of intermonomer contacts not observed previously in 3D crystals, allowing rhodopsin to undergo changes within the crystal lattice without disruption caused by the small contact surfaces in these crystals. The crystallographic dimer further validates contacts involved in the proposed higher-order organization of rhodopsin dimers (25–27) (Fig. 8).
Our crystallographic data may have significant implications for understanding how other GPCRs are organized as oligomers and activated. The rhodopsin-Gt induced-fit model implies that activation may simply involve the relaxation of the somewhat more rigid structure of the inactive state. This process can occur within GPCR monomers or could also be accommodated by an activating rearrangement of an oligomeric structure, or both. This hypothesis is consistent with a model for metabotropic Glu receptor 1α for which ligand binding does not change the structure of each subunit, but does change the dimeric location of the cytoplasmic regions (13). In addition, our structural studies further confirm that rhodopsin is a good template for homology models of other GPCRs (28) used in docking calculations of both agonists and antagonists, because ground-state and photoactivated rhodopsin are structurally similar. Thus, our study is a major step in understanding the molecular basis of GPCR activation by crystallography.
Methods
Single-Crystal Spectrometry.
Absorbance spectra of 20 single crystals were recorded by using a microspectrophotometer model XSPECTRA (4DX System AB, Uppsala, Sweden) equipped with a halogen lamp and diode array detector. Crystals placed in the nylon loop were transferred onto a goniometer head and aligned. The light beam was delivered through a 50-μm fiber optic cable. All measurements were performed at 100 K in a nitrogen cold stream (X-Stream 2000, Rigaku, Woodlands, TX) for both red and yellow crystals.
Retinoid Analysis.
Single crystals were dissolved in 200 μl of buffer containing 25 mM bis-Tris-propane (pH 7.5), 50% (vol/vol) methanol, and 40 mM hydroxylamine. Hydroxylamine was added to cleave the Schiff base, which covalently attaches the chromophore to Lys-296, forming a retinal oxime that can be extracted, and its isomerization state was determined by normal-phase HPLC. After 30 min of incubation at room temperature, retinoids were extracted with 300 μl of hexane, evaporated to dryness, redissolved in 100 μl of hexane, and injected onto a normal-phase HPLC column (Ultrasphere-Si, Beckman-Coulter, Fullerton, CA; 5 μm; 4.5 × 250 mm), which had been equilibrated with 10% ethyl acetate in hexane and eluted in the same mobile phase at a flow rate of 2 ml/min. Retinoids were monitored at 360 nm.
Data Collection/Processing/Structure Determination.
Diffraction data for several ground-state and photoactivated data sets were collected (Table 2, which is published as supporting information on the PNAS web site). The diffraction patterns for the ground-state crystals extend to 4.1-Å resolution. A molecular replacement solution for the ground-state structure was determined by using MOLREP (29, 30) with one 1-HZX monomer as a search model. In general, diffraction for individual photoactivated crystals was poor; after optimization of crystallization, photoactivation, and cryoprotection conditions, most crystals diffracted to no better than 6 Å. However, a 4.15-Å resolution data set for a photoactivated crystal was collected at the microfocus beamline at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland, which was used to determine the photoactivated structure by molecular replacement using portions of the trigonal ground-state asymmetric unit as search models.
Model Building and Structure Refinement.
Initial models derived from molecular replacement solutions were built into threefold symmetry-averaged, density-modified maps and/or σA-weighted 2 Fo − Fc and Fo − Fc maps. Structures were refined in REFMAC by using strong noncrystallographic symmetry (NCS) restraints and strong stereochemical restraints (30). B factors were fixed at the Wilson B factor during refinement. During model building, stereochemistry was monitored with PROCHECK (31) and enforced during model building in O and XFIT (32, 33). Regions with poor density were excised, maps were recalculated, and the regions were rebuilt by hand using the prior high-resolution structures as a guide. For the photoactivated structure, omit maps for both H-VI and H-III suggest that there are no major movements of either helix. Furthermore, Fobs(red) − Fobs(yellow) maps fail to reveal large peaks, which would be present if there were large differences between the ground-state and photoactivated crystals. The solvent region in both structures contains features that are not modeled. NCS symmetry-averaged maps contain less unmodeled density. This unmodeled density could be caused by components in the crystallization conditions (nonyl-glucoside, the TETSQVAPA peptide used to elute the rhodopsin from the immunoaffinity column or Merpol HCS) but the resolution of our structures limits our ability to assign identities to these peaks.
Supplementary Material
Acknowledgments
We thank Dr. Ning Li and Dr. Li Zhang for their help during the course of the project; Dr. David Teller for his continuous contribution to this project; and beamline personnel at Advanced Light Source 8.3.1, 5.0.2, and 4.2.2, Stanford Synchrotron Radiation Laboratory 9.2, and Swiss Light Source X06SA and X10SA for assistance during the many data collection trips. This research was supported in part by U.S. Public Health Service Grant EY08061 from the National Eye Institute and National Institute of General Medical Sciences Grant GM63020. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences; at the Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program, which is supported by the Department of Energy, Office of Biological and Environmental Research and the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences; at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Department of Energy under Contract DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory; and at the Swiss Light Source, beamlines X06SA and X10SA.
Abbreviations
- GPCR
G protein-coupled receptor
- Meta I
metarhodopsin I
- Meta II
metarhodopsin II.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2I35, 2I36, and 2I37).
References
- 1.Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin JP, Davenport AP, Spedding M, Harmar AJ. Pharmacol Rev. 2005;57:279–288. doi: 10.1124/pr.57.2.5. [DOI] [PubMed] [Google Scholar]
- 2.Palczewski K. Annu Rev Biochem. 2006;75:743–767. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 5.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 6.Schertler GF, Villa C, Henderson R. Nature. 1993;362:770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- 7.Nakamichi H, Okada T. Angew Chem Int Ed Engl. 2006;45:4270–4273. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- 8.Nakamichi H, Okada T. Proc Natl Acad Sci USA. 2006;103:12729–12734. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruprecht JJ, Mielke T, Vogel R, Villa C, Schertler GF. EMBO J. 2004;23:3609–3620. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okada T, Ernst OP, Palczewski K, Hofmann KP. Trends Biochem Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- 11.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 12.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 13.Tateyama M, Abe H, Nakata H, Saito O, Kubo Y. Nat Struct Mol Biol. 2004;11:637–642. doi: 10.1038/nsmb770. [DOI] [PubMed] [Google Scholar]
- 14.Fan QR, Hendrickson WA. Nature. 2005;433:269–277. doi: 10.1038/nature03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salom D, Le Trong I, Pohl E, Ballesteros JA, Stenkamp RE, Palczewski K, Lodowski DT. J Struct Biol. 2006 doi: 10.1016/j.jsb.2006.05.003. 10.1016/j.jsb 2006.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Farrens DL, Khorana HG. J Biol Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 17.Visiers I, Ballesteros JA, Weinstein H. Methods Enzymol. 2002;343:329–371. doi: 10.1016/s0076-6879(02)43145-x. [DOI] [PubMed] [Google Scholar]
- 18.Spooner PJ, Sharples JM, Goodall SC, Bovee-Geurts PH, Verhoeven MA, Lugtenburg J, Pistorius AM, Degrip WJ, Watts A. J Mol Biol. 2004;343:719–730. doi: 10.1016/j.jmb.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 19.Yan EC, Kazmi MA, De S, Chang BS, Seibert C, Marin EP, Mathies RA, Sakmar TP. Biochemistry. 2002;41:3620–3627. doi: 10.1021/bi0160011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piscitelli CL, Angel TE, Bailey BW, Hargrave P, Dratz EA, Lawrence CM. J Biol Chem. 2006;281:6813–6825. doi: 10.1074/jbc.M510175200. [DOI] [PubMed] [Google Scholar]
- 21.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 22.Lanyi JK. Annu Rev Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 23.Zhukovsky EA, Oprian DD. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 24.Mazzoni MR, Hamm HH. Methods Enzymol. 2000;315:363–376. doi: 10.1016/s0076-6879(00)15854-9. [DOI] [PubMed] [Google Scholar]
- 25.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. FEBS Lett. 2004;564:281–288. doi: 10.1016/S0014-5793(04)00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 27.Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. J Biol Chem. 2003;278:21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filipek S, Teller DC, Palczewski K, Stenkamp R. Annu Rev Biophys Biomol Struct. 2003;32:375–397. doi: 10.1146/annurev.biophys.32.110601.142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruickshank D. Acta Crystallogr D. 1999;55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 31.Laskowski RA, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 32.McRee DE. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.DeLano W. PYMOL. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.