Abstract
The heme-copper oxidases constitute a superfamily of terminal dioxygen-reducing enzymes located in the inner mitochondrial or in the bacterial cell membrane. The presence of a mechanistically important covalent bond between a histidine ligand of the copper ion (CuB) in the active site and a generally conserved tyrosine residue nearby has been shown to exist in the canonical cytochrome c oxidases. However, according to sequence alignment studies, this critical tyrosine is missing from the subfamily of cbb3-type oxidases found in certain bacteria. Recently, homology modeling has suggested that a tyrosine residue located in a different helix might fulfill this role in these enzymes. Here, we show directly by methods of protein chemistry and mass spectrometry that there is indeed a covalent link between this tyrosine and the copper-ligating histidine. The identity of the cross-linked tyrosine was determined by showing that the cross-link is not formed when this residue is replaced by phenylalanine, even though structural integrity is maintained. These results suggest a universal functional importance of the histidine-tyrosine cross-link in the mechanism of O2 reduction by all heme-copper oxidases.
Keywords: mass spectrometry
The heme-copper oxidases catalyze the final step of energy conservation in the respiratory chain. These enzymes couple the four-electron reduction of molecular oxygen to translocation of protons across the membrane, thereby contributing to the electrochemical potential that is used as a primary energy source for example in ATP synthesis (1–3). The major catalytic subunit I contains two hemes and a copper atom as redox centers, which are coordinated by fully conserved histidine residues in all of the heme-copper oxidases. The so-called binuclear center where the catalytic O2 reduction takes place is formed by a high-spin heme with a nearby copper ion (CuB). Apart from the metal ligands themselves, there are also a few amino acids close to this active site that are well conserved within the family. In helix VI of subunit I there is a valine residue located next to the high-spin heme and a tryptophan in the vicinity of the active site, both of which are characteristic of all heme-copper oxidases (4).
The bacterial cbb3-type cytochrome c oxidase is a distant member of the heme-copper oxidase family, characterized by its ability to maintain catalytic activity under low oxygen tension while retaining the capacity to translocate protons. These enzymes were first identified as a gene cluster fixNOQP (ccoNOQP) in the genome of rhizobial bacteria from root nodules, and since then they have been found and isolated from several other bacteria as well (5–11). Subunit CcoN, corresponding to subunit I of the canonical oxidases, contains two B-type hemes (protohemes) in addition to CuB. Instead of the copper center (CuA) located in subunit II of the aa3-type cytochrome c oxidases, the electron input pathway is composed of altogether three redox centers embedded in subunits CcoO and CcoP. These two other subunits, which do not have counterparts in the canonical oxidases, contain one and two C-type hemes, respectively. The fourth small subunit CcoQ does not seem to take part in catalysis (12). Even though several key structural features of the canonical oxidases are missing from the cbb3-type enzymes according to sequence alignment studies (4, 13, 14), the mechanism of four-electron reduction of oxygen to water is thought to be the same in all heme-copper oxidases.
In the canonical heme-copper oxidases there is a fully conserved tyrosine residue (tyrosine 244, Bos taurus) within the active site that forms a covalent bond to one of the three histidine ligands of CuB, which resides four amino acids apart in the same helix VI (histidine 240, B. taurus). This unusual posttranslational modification has been proposed to be of critical importance in the catalytic mechanism. The first evidence for its presence in the canonical heme-copper oxidases was obtained from crystallographic data (15, 16) and subsequently verified by protein-chemical analysis (17). According to sequence alignments this helix VI tyrosine is completely missing from the cbb3-type oxidases despite the presumed catalytic importance of the histidine-tyrosine dimer. However, recent homology modeling has suggested that another residue, tyrosine 311 (the numbering refers to subunit CcoN of Rhodobacter sphaeroides cytochrome cbb3 unless otherwise stated), which is fully conserved among the cbb3-type oxidases and is located in the nearby transmembrane helix VII, might structurally replace the missing tyrosine in the active site and thereby possibly fulfill the mechanistic requirement also in these distant members of the heme-copper oxidase family (Fig. 1) (13, 14). However, because there is no three-dimensional structure available for any cbb3-type oxidase, the existence of a cross-linked histidine-tyrosine in this group of enzymes has remained elusive. Here, we have applied methods of protein chemistry and mass spectrometry to directly address this question in the cbb3-type oxidase from R. sphaeroides.
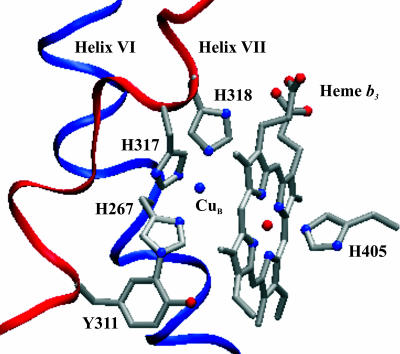
Fig. 1.
The active site of cytochrome cbb3 from R. sphaeroides together with residues Y267, H311, H317, H318, and H405. According to molecular modeling, the Nε2 of the histidine 311 imidazol and the Cε2 of the tyrosine 311 phenol ring may come close enough to enable the formation of a covalent bond even though the amino acids originate from different helices (13, 14). The figure was prepared by using the Visual Molecular Dynamics software (18).
Results
Cytochrome cbb3 from R. sphaeroides was produced in a Paracoccus denitrificans strain as described in ref. 14 and isolated as described in Materials and Methods, yielding a highly active and pure enzyme preparation (see Fig. 5, which is published as supporting information on the PNAS web site). Subunit CcoN of cytochrome cbb3 was isolated from heavily overloaded SDS-polyacrylamide gels by passive diffusion resulting in good recovery of the protein. To assess whether a covalent bond between the CuB-ligating histidine and the proposed tyrosine residue exists in the cbb3-type oxidases, a suitable set of enzymatic cleavages that would yield unique results was required. According to sequence alignment and modeling, the residues that may form the bond in the cbb3 oxidases originate from different helices (Fig. 1): the histidine (267) ligand of CuB is in helix VI, but the putative cross-linked tyrosine (311) is in helix VII. Hence, polypeptide fragments containing the potential covalent link were expected to be large and extremely hydrophobic.
We first performed a double digestion in two stages using endoproteinase Asp-N and trypsin (Fig. 2A). Endoproteinase Asp-N cleaves at the N-terminal side of aspartate residues and, on occasion, unspecifically at the N-terminal side of glutamate residues. An 11,715-Da polypeptide (corresponding to the D258 to W358 peptide, i.e., the entire sequence in Fig. 2A) that contains the putative link-forming residues should result from this cleavage {all polypeptides are referred to using mass to charge ratios (m/z) of singly charged ions [M+H]+; hence, individual masses of fragments do not add up}. If the link is present, the same fragment should result even if unspecific cleavage at E291 took place, as opposed to the case in the absence of the cross-link, where two separate polypeptides should be produced. As shown in Fig. 3 (black trace), a polypeptide fragment with a mass of 11,715 Da was systematically detected. No fragments corresponding to the unlinked protein were found (3,848 Da and 7,887 Da). The 11,715-Da fragment was also detected when the holoenzyme was cleaved (data not shown). Other peaks expected to arise from digestion fragments of CcoN were also identified (Fig. 3), and there was no evidence for unexpected cleavage patterns. Because no other unusual bonds between fragments were detected, the location of the covalent bond, if present, was presumed to be within the 11,715-Da fragment.
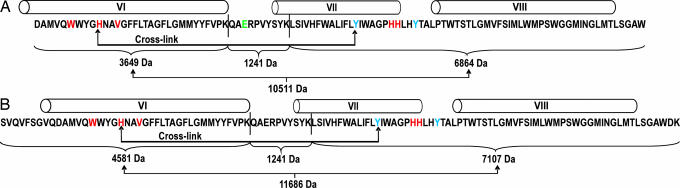
Fig. 2.
The amino acid sequence and putative transmembrane helices of the R. sphaeroides subunit CcoN of cytochrome cbb3 in the region of the cross-link. (For the full sequence of subunit CcoN, GenBank entry U58092 with six histidine-tag added to the C terminus.) (A) The sequence of the 11,715-Da (D258 to W358) polypeptide resulting from endoproteinase Asp-N cleavage is presented with glutamate 291 highlighted in green and the alternative fragments after double digestion with endoproteinase Asp-N and trypsin are shown. (B) The sizes of the alternative fragments resulting from endoproteinase Lys-C cleavage in the area of S249 to K360 with or without cross-link are shown. The fully conserved amino acids in heme-copper oxidases are highlighted in red (W263, H267, V270, H317, and H318), and the tyrosine residues 311 and 321 are marked with blue. The putative cross-link is indicated between histidine 267 and tyrosine 311 and the cleavage sites of trypsin and endoproteinase Lys-C within the presented sequences are indicated with vertical bars between K288/Q289 and K298/L299.
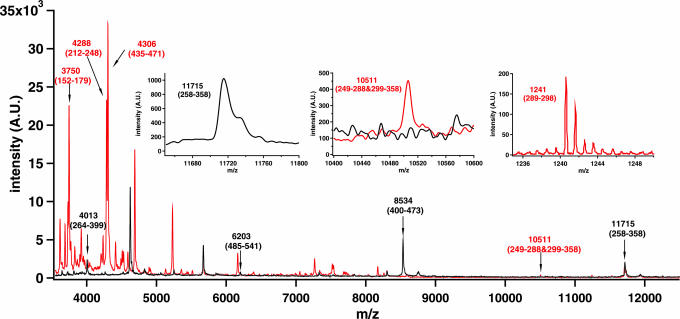
Fig. 3.
The MALDI-TOF mass spectra of the fragments from the subunit CcoN of cytochrome cbb3 after cleavage with endoproteinase Asp-N (black trace) and after double digestion with both endoproteinase Asp-N and trypsin (red trace). The assignments of the subunit CcoN fragments as average (>3,300 Da) or monoisotopic (<3,300 Da) masses are indicated in the spectrum and the respective amino acid numbering are shown in brackets. The most important parts of the spectra are shown as Insets: the 11,715-Da fragment produced by endoproteinase Asp-N cleavage (Left), the 10,511-Da region before and after the trypsin digestion (Center), and the 1,241-Da region after the double digestion (Right).
After the endoproteinase Asp-N treatment, subunit CcoN was further cleaved with trypsin, which cuts at the C terminus of lysine and arginine residues, except when the C-terminal residue is a proline. Trypsin was expected to release a 1,241-Da fragment (Q289 to K298) from the 11,715-Da polypeptide, resulting in the case of an unlinked protein in two fragments, 3,649 Da (D258 to K288) and 6,864 Da (L299 to W358) in size. In contrast, a single 10,511-Da polypeptide having the two previous fragments linked together was expected if the cross-link is present (Fig. 2A). Although the trypsin cleavage was not complete, resulting in some remnant uncleaved 11,715-Da fragment, only the 10,511-Da polypeptide was detected, which proves the existence of the cross-link between the 3,649- and 6,864-Da protein fragments, because the latter were not observed individually (Fig. 3, red trace). Also, the expected 1,241-Da fragment released by trypsin from the middle of the 11,715-Da polypeptide (Fig. 2A) could be detected in the spectrum (Fig. 3 Insets), proving that the 11,715-Da fragment had indeed been cleaved. Separate digestions of both holoenzyme and subunit CcoN with trypsin alone yielded no contradictory evidence (data not shown). Altogether, these results clearly show that a covalent bond exists between the described polypeptide fragments. No other combination of peptides, including the possibility of incomplete digestions, could result in a protein fragment of this size. However, these data do not identify the residues forming the link.
From sequence alignments (4, 13, 14, 19), we know that there is a limited number of completely conserved amino acids within the family of cytochrome cbb3-type oxidases that could form the cross-link. We can safely assume that histidine 267 in helix VI (marked in red in Fig. 2A) is a CuB ligand because it is completely conserved throughout the heme-copper oxidases and is crucial for the cross-link in the canonical enzymes. The fact that the middle of helix VI close the active site is among the best conserved areas in the heme-copper oxidases makes it very likely that histidine 267 is a partner in the cross-link. Moreover, it is the only histidine residue within the 3,649-Da fragment shown to be a component of the cross-linked peptide (Fig. 2A). However, the other partner is less obvious. There are two well conserved tyrosines (Y311 and Y321) within the 6,864-Da fragment (marked in blue in Fig. 2A), either of which could be filling the role of second partner, and we cannot exclude the possibility that some other residue may take this role.
We used site-directed mutagenesis to determine which amino acid forms the link with the CuB ligand histidine 267. The proposed partner, tyrosine 311, was mutated to phenylalanine, which resulted in an inactive but correctly assembled enzyme (13, 14). Then, both WT and Y311F mutant enzymes were digested by using endoproteinase Lys-C, which cleaves at the C-terminal side of lysine residues. As presented for WT enzyme in Fig. 2B, fragments of 4,581 Da (S249 to K288) and 7,107 Da (L299 to K360) should be observed in the case of unlinked protein, but with the cross-link they should be connected resulting in an 11,686-Da polypeptide. As shown in Fig. 4 (black trace), no fragments corresponding to the unlinked protein were observed in the WT enzyme, but the 11,686-Da polypeptide was clearly present. After the same endoproteinase Lys-C cleavage with the mutated enzyme, the mass spectra (Fig. 4, red trace) now show only the two peaks corresponding to the unlinked protein fragments (4,581 Da and 7,091 Da), whereas the peak of the cross-linked 11,670-Da structure was absent. Other peaks identified as CcoN fragments were identical to those in the spectrum of the WT enzyme. Hence, it is evident that site-specific mutation of tyrosine 311 to phenylalanine specifically abolishes the histidine-tyrosine cross-link otherwise observed, identifying this tyrosine as the second partner of the cross-link.
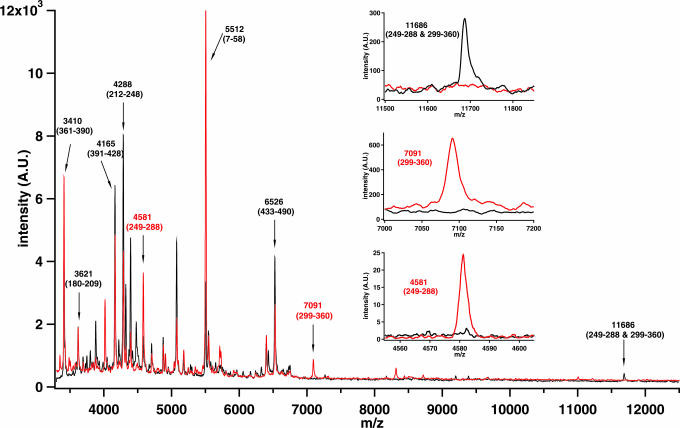
Fig. 4.
The MALDI-TOF mass spectra of the fragments from the subunits CcoN of WT (black trace) and Y311F mutant (red trace) after endoproteinase Lys-C digestion. The assignments of the subunit CcoN fragments as average masses together with respective amino acid numbering in brackets are presented. In the Insets the key regions are shown in detail: the 11,686-Da peak (Top) composed of two fragments held together by a covalent bond is clearly visible only in the WT mass spectrum, whereas the separate 7,091-Da (Middle) and 4,581-Da (Bottom) fragments are observed solely in the Y311F mutant spectrum.
Discussion
Our data show unequivocally that a histidine-tyrosine cross-link is present also in the active site of the cbb3-type oxidase from R. sphaeroides. However, in this case the cross-linked amino acids originate from different helices as had indeed been proposed by molecular modeling (13, 14). We used systematic proteolytic digestions to study the putative covalent bonding between the fragments close to the active site, and we found that there is a very stable cross-link keeping the resulting fragments together in the entire protein population even after denaturation, as also verified by using several independent enzyme preparations along with detailed comparison of WT and mutant enzymes. This is in harmony with the result reported for the other types of oxidases. The chemical structure of the cross-link was shown to be a covalent bond in the Thermus thermophilus ba3-type oxidase; sequencing was used to provide evidence for the position of the link also in other oxidases (17). This method was not applicable in our case because of the large size of the fragments. Thus, we took two different approaches to test the existence of the link and used site-directed mutagenesis to pinpoint its location.
One unifying property of the family of heme-copper oxidases is that the histidine ligands coordinating the cofactors in the catalytic subunit are fully conserved. Although there is no three-dimensional structure available for the cbb3-type oxidases, it is likely that these histidines have the same roles also in these enzymes (20). The three histidine ligands of CuB are all located in helices VI and VII (Fig. 2). The region of the CuB ligand within helix VI is one of the best conserved domains and therefore it is predicted to be structurally very similar among all heme-copper oxidases. Although helix VII, in which the tyrosine partner of the cross-link resides in the cbb3 oxidases, has less sequence similarities the two other histidine ligands of CuB in that helix set restrictions to its structure. The fully conserved histidine in helix VI takes part in the formation of the histidine-tyrosine cross-link in the canonical oxidases. Thus far, amino acids shown to be capable of forming cross-linked dimers in biological systems always have tyrosine as one partner. In addition to histidines, also for instance cysteines have been shown to have this ability (21). By excluding other options, our results show that histidine 267 is one of the partners in the cross-link. According to mutation data, substitution of this histidine by alanine leads to fully assembled although inactive enzyme in Bradyrhizobium japonicum cytochrome cbb3. The CuB center is also present despite the mutation, implying that the lack of activity is not due to lack of cofactor (20).
A priori it was impossible to exclude that some other residue than tyrosine 311 would be the second partner of the cross-link. Indeed, there is a second well conserved tyrosine residue at position 321, which is included in the same cleavage product as tyrosine 311. We used site-directed mutagenesis to replace the most likely candidate, tyrosine 311, with phenylalanine to minimize structural disturbance of the active site. This mutation leads to an inactive phenotype (13), but the assembly of the mutant enzyme is correct based on detection of all subunits on SDS/PAGE gels, and a virtually unchanged optical spectrum, as we also confirmed (data not shown). Our results showed that the cross-link failed to form when phenylalanine replaced tyrosine 311, yielding an entirely different digestion pattern. We conclude that the cross-link between the histidine ligand of CuB and a tyrosine is another unifying feature of the heme-copper oxidases, even though the origin of the tyrosine varies in the primary structure.
It has been proposed that the histidine-tyrosine bond forms during the very first turnovers of the enzyme and that it is stable during the catalytic cycle, as supported by our data. The absence of the cross-link in the tyrosine-phenylalanine mutant may give a clue about the mechanism of its formation. The only difference is the lack of the OH group in the aromatic ring while the atoms forming the link are still present. Because the metal centers of the active site can provide only three electrons out of the four needed in the full reduction of molecular oxygen to water, tyrosine has been suggested to have a key role in the O2 reduction chemistry; the hydroxyl group of the tyrosine may donate both an electron and a proton to break the O–O bond, thereby forming a neutral tyrosine radical in the PM state of the catalytic cycle (22–24). This may avoid production of reactive oxygen species and might initialize the formation of the cross-link itself. Consequently, the absence of the hydroxyl group might be the simple reason for the absence of the cross-link in the mutant enzyme.
Several studies of histidine-tyrosine model compounds have been reported (25–32) supporting stabilization of a neutral tyrosine radical. However, the ligated copper, not included in these models, also is likely to affect the properties of the histidine-tyrosine dimer, which is the actual ligand of CuB (33), and might increase its acidity and stabilize the neutral radical beyond that shown for the model compounds. However, we stress that our data cannot identify the cross-linked atoms in the histidine-tyrosine dimer of cytochrome cbb3, although the modeled cytochrome cbb3 structures suggest identity with the canonical oxidases in this respect (13, 14).
The cross-link has been proposed to maintain the structure of the active site (34, 35). Studies with R. sphaeroides cytochrome aa3 showed that in the tyrosine to phenylalanine mutant, one of the histidine ligands of CuB might coordinate heme a3, CuB is largely lost, and the optical spectrum is changed (34). This is in contrast to the observations with cbb3 oxidase, where the structure of the active site appears less affected (see above). As pointed out earlier (34), the cross-link may have an important role in holding the tyrosine residue in the correct orientation. In cytochrome aa3 there is a hydrogen bonding interaction between the hydroxyl group of the tyrosine and the hydroxyl group of the hydroxyethyl farnesyl side chain of the high spin heme. The predicted position of the tyrosine in the structure of cytochrome cbb3 puts it in a very similar position as in the canonical oxidases (13), but in the cbb3 oxidases the heme of the binuclear site is of type B (protoheme), which lacks the hydroxyethyl farnesyl side chain of hemes A and O in the canonical oxidases. Therefore, the orientation of the cross-linked tyrosine in the cbb3 oxidases cannot be “steered” by hydrogen-bonding to the heme hydroxyethyl group. Switching the critical tyrosine residue to a helix closer to the histidine with which the cross-link is formed may therefore be coupled to the employment of heme A or O in the active site.
It is evident that the histidine-tyrosine cross-link is a completely conserved feature of the heme-copper oxidases because it persists even in the cbb3-type enzymes although the tyrosine stems from a different helix. Such structural substitution of a functionally important residue has been observed also for other amino acids in heme-copper oxidases, for instance glutamate 278 in P. denitrificans cytochrome aa3 (36, 37).
Materials and Methods
Protein Purification and Subunit Isolation.
R. sphaeroides cytochrome cbb3 WT enzyme and the Y311F mutant produced as described in ref. 14 were first isolated by using chromatographic steps essentially as described in ref. 20 at pH 7.5. The samples were further purified by using a Source 15Q column (Amersham Biosciences) and concentrated to 10 mg/ml in 20 mM Tris·Cl (pH 7.5)/0.05% n-dodecyl β-d-maltoside (DM) (Anatrace, Maumee, OH). Subunit CcoN was isolated from a 12% SDS-polyacrylamide minigel (Bio-Rad) supplemented with 20% glycerol by using 100 μg of protein denatured in Laemmli sample buffer (38) at room temperature. The band position of the subunit CcoN was detected by Coomassie staining of a small slice of the gel (see Fig. 5) containing also a molecular weight standard (LMW standard; Amersham Biosciences). The subunit CcoN to be continued with was excised from the unstained gel, sliced into very small pieces, and placed into an Eppendorf tube, and 20 mM Tris·Cl (pH 7.5)/0.05% DM was added to cover the gel pieces. The passive elution of subunit CcoN to the liquid phase was performed overnight at 37°C with shaking at 200 rpm. The supernatant was collected and concentrated by using Microcon concentrators (Amicon; cut-off of 50 kDa), and the buffer was exchanged to 50 mM sodium phosphate (pH 8.0)/0.05% DM. The purity and quantity of the sample were estimated on an SDS/PAGE gel (see Fig. 5).
Enzymatic Cleavages.
Isolated subunit CcoN was exposed to proteolytic degradation by incubating it with endoproteinase Lys-C (sequencing grade; Waco), endoproteinase Asp-N (sequencing grade; Roche), or trypsin (sequencing grade; Sigma-Aldrich, St. Louis, MO). All digestions were performed overnight at 37°C with 200-rpm agitation (protein to enzyme ratio of 10:1). The endoproteinase Asp-N (in 10 mM Tris·Cl, pH 7.5/0.05% DM) and trypsin (in H2O supplemented with 0.05% DM) were used both for separate and sequential digestions of isolated subunit CcoN as well as for holoenzyme control digestions. The endoproteinase Lys-C (in 25 mM Tris·Cl, pH 8.8/0.05% DM) was used for cleavages of both WT and Y311F mutant subunit CcoN. All of the protein digests were analyzed by mass spectrometry.
Mass Spectrometry.
MALDI-TOF spectrometry of protein digests was carried out at the Protein Chemistry Unit of Biomedicum Helsinki by using a Bruker Daltonics (Bremen, Germany) Autoflex mass spectrometer operated in the linear positive ion mode (large proteins) or the reflector mode (smaller peptides). The instrument was calibrated with commercially available protein/peptide standards. Samples were prepared in the same buffers as used in each digestion. Sinapic acid was used as the matrix for proteins and α-cyano-4-hydroxycinnamic acid as the matrix for peptides (prepared as saturated solutions in 50% acetonitrile/0.1% TFA in water). One- to 2-μl aliquots of mixed matrix and sample were deposited on the target plate and allowed to dry in air. Spectra were acquired as a sum of signals recorded during 50–200 laser shots. The spectra were analyzed by using FlexAnalysis software (Bruker Daltonics) and visualized by using IGOR Pro (WaveMetrics).
Supplementary Material
Acknowledgments
This work was supported by the National Graduate School in Informational and Structural Biology (V.R.), the Sigrid Jusélius Foundation, Biocentrum Helsinki, and the Academy of Finland.
Abbreviation
- DM
n-dodecyl β-d-maltoside.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Wikström MKF. Nature. 1977;266:271–273. doi: 10.1038/266271a0. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson-Miller S, Babcock GT. Chem Rev. 1996;96:2889–2907. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 3.Wikström M. Biochim Biophys Acta. 2004;1655:241–247. doi: 10.1016/j.bbabio.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Pereira MM, Santana M, Teixeira M. Biochim Biophys Acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 5.Preisig O, Anthamatten D, Hennecke H. Proc Natl Acad Sci USA. 1993;90:3309–3313. doi: 10.1073/pnas.90.8.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandon K, Kaminski PA, Elmerich C. J Bacteriol. 1994;176:2560–2568. doi: 10.1128/jb.176.9.2560-2568.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Horsman JA, Berry E, Shapleigh JP, Alben JO, Gennis RB. Biochemistry. 1994;33:3113–3119. doi: 10.1021/bi00176a046. [DOI] [PubMed] [Google Scholar]
- 8.Gray KA, Grooms M, Myllykallio H, Moomaw C, Slaughter C, Daldal F. Biochemistry. 1994;33:3120–3127. doi: 10.1021/bi00176a047. [DOI] [PubMed] [Google Scholar]
- 9.Preisig O, Zufferey R, Thöny-Meyer L, Appleby CA, Hennecke H. J Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 11.Urbani A, Gemeinhardt S, Warne A, Saraste M. FEBS Lett. 2001;508:29–35. doi: 10.1016/s0014-5793(01)03006-x. [DOI] [PubMed] [Google Scholar]
- 12.Pitcher RS, Watmough NJ. Biochim Biophys Acta. 2004;1655:388–399. doi: 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 13.Hemp J, Christian C, Barquera B, Gennis RB, Martinez TJ. Biochemistry. 2005;44:10766–10775. doi: 10.1021/bi050464f. [DOI] [PubMed] [Google Scholar]
- 14.Sharma V, Puustinen A, Wikström M, Laakkonen L. Biochemistry. 2006;45:5754–5765. doi: 10.1021/bi060169a. [DOI] [PubMed] [Google Scholar]
- 15.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 16.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 17.Buse G, Soulimane T, Dewor M, Meyer HE, Blüggel M. Protein Sci. 1999;8:985–990. doi: 10.1110/ps.8.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 19.Toledo-Cuevas M, Barquera B, Gennis RB, Wikström M, Garcia-Horsman JA. Biochim Biophys Acta. 1998;1365:421–434. doi: 10.1016/s0005-2728(98)00095-4. [DOI] [PubMed] [Google Scholar]
- 20.Zufferey R, Arslan E, Thöny-Meyer L, Hennecke H. J Biol Chem. 1998;273:6452–6459. doi: 10.1074/jbc.273.11.6452. [DOI] [PubMed] [Google Scholar]
- 21.Ito N, Phillips SEV, Yadav KDS, Knowles PF. J Mol Biol. 1994;238:794–814. doi: 10.1006/jmbi.1994.1335. [DOI] [PubMed] [Google Scholar]
- 22.MacMillan F, Kannt A, Behr J, Prisner T, Michel H. Biochemistry. 1999;38:9179–9184. doi: 10.1021/bi9911987. [DOI] [PubMed] [Google Scholar]
- 23.Proshlyakov DA, Pressler MA, DeMaso C, Leykam JF, Dewitt DL, Babcock GT. Science. 2000;290:1588–1591. doi: 10.1126/science.290.5496.1588. [DOI] [PubMed] [Google Scholar]
- 24.Tomson F, Bailey JA, Gennis RB, Unkefer CJ, Li ZH, Silks LA, Martinez RA, Donohoe RJ, Dyer RB, Woodruff WH. Biochemistry. 2002;41:14383–14390. doi: 10.1021/bi026370c. [DOI] [PubMed] [Google Scholar]
- 25.McCauley KM, Vrtis JM, Dupont J, van der Donk WA. J Am Chem Soc. 2000;122:2403–2404. [Google Scholar]
- 26.Naruta Y, Sasaki T, Tani F, Tachi Y, Kawato N, Nakamura N. J Inorg Biochem. 2001;83:239–246. doi: 10.1016/s0162-0134(00)00170-7. [DOI] [PubMed] [Google Scholar]
- 27.Aki M, Ogura T, Naruta Y, Le TH, Sato T, Kitagawa T. J Phys Chem A. 2002;106:3436–3444. [Google Scholar]
- 28.Cappuccio JA, Ayala I, Elliott GI, Szundi I, Lewis J, Konopelski JP, Barry BA, Einarsdottir O. J Am Chem Soc. 2002;124:1750–1760. doi: 10.1021/ja011852h. [DOI] [PubMed] [Google Scholar]
- 29.Kim SH, Aznar C, Brynda M, Silks LAP, Michalczyk R, Unkefer CJ, Woodruff WH, Britt RD. J Am Chem Soc. 2004;126:2328–2338. doi: 10.1021/ja0303743. [DOI] [PubMed] [Google Scholar]
- 30.Bu YX, Cukier RI. J Phys Chem B. 2005;109:22013–22026. doi: 10.1021/jp053046t. [DOI] [PubMed] [Google Scholar]
- 31.Kim E, Kamaraj K, Galliker B, Rubie ND, Moenne-Loccoz P, Kaderli S, Zuberbuhler AD, Karlin KD. Inorg Chem. 2005;44:1238–1247. doi: 10.1021/ic048907b. [DOI] [PubMed] [Google Scholar]
- 32.Pratt DA, Pesavento RP, van der Donk WA. Org Lett. 2005;7:2735–2738. doi: 10.1021/ol050916g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas JW, Calhoun MW, Lemieux LJ, Puustinen A, Wikström M, Alben JO, Gennis RB. Biochemistry. 1994;33:13013–13021. doi: 10.1021/bi00248a010. [DOI] [PubMed] [Google Scholar]
- 34.Das TK, Pecoraro C, Tomson FL, Gennis RB, Rousseau DL. Biochemistry. 1998;37:14471–14476. doi: 10.1021/bi981500w. [DOI] [PubMed] [Google Scholar]
- 35.Pinakoulaki E, Pfitzner U, Ludwig B, Varotsis C. J Biol Chem. 2002;277:13563–13568. doi: 10.1074/jbc.M112200200. [DOI] [PubMed] [Google Scholar]
- 36.Backgren C, Hummer G, Wikström M, Puustinen A. Biochemistry. 2000;39:7863–7867. doi: 10.1021/bi000806b. [DOI] [PubMed] [Google Scholar]
- 37.Pereira MM, Teixeira M. Biochim Biophys Acta. 2004;1655:340–346. doi: 10.1016/j.bbabio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.