Abstract
Specific cellulose hydrolysis rates (g of cellulose/g of cellulase per h) were shown to be substantially higher (2.7- to 4.7-fold) for growing cultures of Clostridium thermocellum as compared with purified cellulase preparations from this organism in controlled experiments involving both batch and continuous cultures. This “enzyme–microbe synergy” requires the presence of metabolically active cellulolytic microbes, is not explained by removal of hydrolysis products from the bulk fermentation broth, and appears due to surface phenomena involving adherent cellulolytic microorganisms. Results support the desirability of biotechnological processes featuring microbial conversion of cellulosic biomass to ethanol (or other products) in the absence of added saccharolytic enzymes.
Keywords: cellulase, cellulosome, consolidated bioprocessing, ethanol
Production of fuels from cellulosic biomass could offer large benefits in terms of sustainability, security, and rural economic development [refs. 1 and 2; Greene et al. (2004), www.nrdc.org/air/energy/biofuels/biofuels.pdf]. The cost of producing reactive intermediates from cellulosic biomass is the central technical obstacle to be overcome for these benefits to be realized (3). Accordingly, a substantial and ongoing effort has been devoted to describing the components and mode of action of multicomponent cellulase enzyme systems (4–8), and to improving their effectiveness (9–11). Synergism among cellulase components is an important and widely documented phenomenon in this context. Values for the degree of synergism (DS, the activity exhibited by mixtures of components divided by the sum of the activities exhibited by the components acting separately) >2 are common, and DS values >5 have been reported under some conditions (8).
Cellulose hydrolysis can be mediated by cellulase enzymes acting in the absence of cells, by cellulases acting in the presence of cells but with no cell–enzyme attachment, or by cellulases attached to cells. In the latter case, hydrolysis is mediated by ternary cellulose–enzyme–microbe (CEM) complexes rather than binary cellulose–enzyme (CE) complexes. For anaerobic cellulolytic bacteria, CEM complexes are commonly formed and are thought to be the major agent of cellulose hydrolysis (12). Potential benefits of CEM complexes for cellulolytic microorganisms have been suggested, including preferred access to hydrolysis products and local concentration of cellulases (12–16).
The possibility of enhanced effectiveness of cellulases present in CEM complexes has been mentioned (12) but has not been demonstrated or quantitatively evaluated. We investigate such “cell-enzyme synergy” here for Clostridium thermocellum, an anaerobic thermophilic bacterium that exhibits one of the highest rates of cellulose utilization among described microorganisms (12). C. thermocellum produces a cellulase complex, or “cellulosome,” a substantial fraction of which is bound to the cell surface under most culture conditions (17–20). Recently, Zhang and Lynd (21) showed that C. thermocellum assimilated cellodextrins with a mean degree of polymerization of ≈4 during growth on cellulose, and that these cellodextrins were subsequently cleaved by intracellular phosphorolytic enzymes. This organism is a starting point for developing anaerobic microbes capable of one-step processing of cellulosic biomass to ethanol or desired products in the absence of added saccharolytic enzymes (3, 12, 22).
Results
Hydrolysis of microcrystalline cellulose (Avicel, FMC, Philadelphia, PA) was compared in batch and continuous cultures for two systems.
(i) Growing cultures of C. thermocellum in the absence of added cellulase, “microbial hydrolysis,” in which hydrolysis is mediated by both CEM and CE complexes.
(ii) Enzymatic hydrolysis mediated by purified C. thermocellum cellulosome with fermentation of hydrolysis products by the noncellulolytic thermophilic anaerobe Thermoanaerobacterium thermosaccharolyticum. In such simultaneous saccharification and fermentation (SSF), hydrolysis is mediated by CE complexes only.
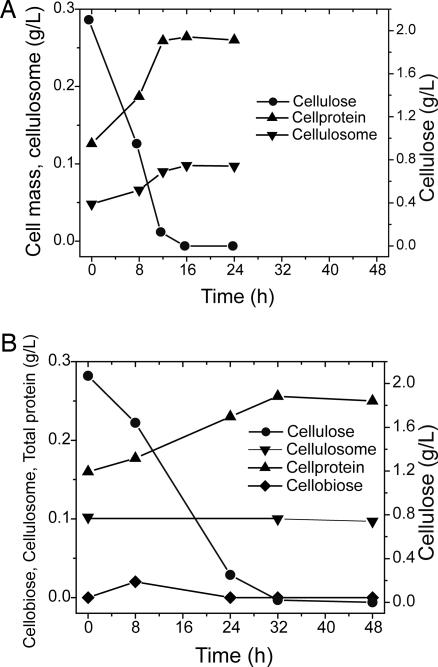
In batch culture under the conditions examined, microbial hydrolysis required 16 h for complete hydrolysis of 2 g of cellulose/liter, during which the cellulosome and cell protein concentrations roughly doubled from 48 to 98 and 125 to 264 mg/liter, respectively (Fig. 1A). SSF required 32 h for complete hydrolysis, during which the cellulosome concentration was 100 mg/liter, and the cell protein concentration increased from 160 to 260 mg/liter (Fig. 1B). The conditions under which microbial hydrolysis and SSF occurred were very similar (see Figs. 4 and 5, which are published as supporting information on the PNAS web site). The same growth medium and pH were used, and the concentrations of fermentation products were low. Hydrolysis products (total soluble glucans) in the growth medium were ≤0.02 g/liter at all times for both microbial hydrolysis and SSF, 2 orders of magnitude less than concentrations at which 50% inhibition of the C. thermocellum cellulase system is observed (23). The cellulase-specific activity was quite similar for enzymatic hydrolysis and microbial hydrolysis (measured with metabolically inactive cells) and remained nearly constant throughout the experiment (Table 2, which is published as supporting information on the PNAS web site).
Fig. 1.
Batch cellulose utilization. (A) Microbial cellulose utilization. The experiment was initiated by adding 2 g/liter cellulose at time 0 to a culture of C. thermocellum that had previously used 2 g/liter cellulose. (B) Enzymatic hydrolysis of cellulose (SSF). The experiment was initiated by adding 0.1 g/liter purified C. thermocellum cellulosome at time 0 to a culture of T. thermosaccharolyticum that had previously used 2 g/liter cellobiose in the presence of 2 g/liter cellulose. Hydrolysis products other than cellobiose were assayed but were below detection limits. pH and concentrations of fermentation products are presented in Figs. 4 and 5 for microbial and enzymatic hydrolysis, respectively.
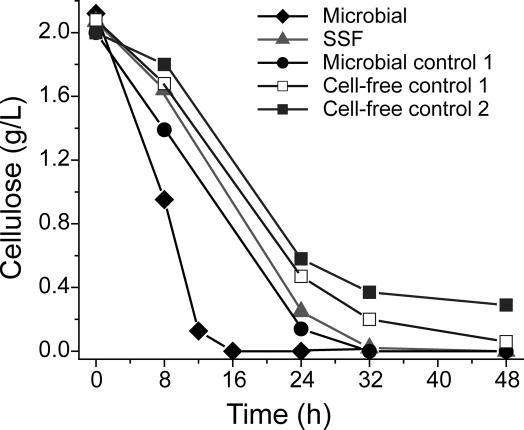
Cellulose concentration is plotted vs. time in Fig. 2 for microbial hydrolysis (from Fig. 1A), SSF (from Fig. 1B) and for controls as follows: cell-free control 1, 100 mg/liter purified cellulase with no fermenting organism; microbial control 1, a C. thermocellum culture (100 mg/liter cellulosome, 264 mg/liter cell protein) with 38.5 mM sodium azide, a fermentation inhibitor; cell-free control 2, as for cell-free control 1 with 38.5 mM sodium azide. Although concentrations of hydrolysis products were 1 order of magnitude higher for cell-free control 1 (Fig. 6, which is published as supporting information on the PNAS web site) than for SSF the hydrolysis rates were similar. Accumulation of hydrolysis products in the bulk fermentation broth (away from the surface of cellulose) thus is not a plausible explanation for the marked difference between microbial hydrolysis and SSF. The rate of hydrolysis was substantially higher for growing cells (microbial hydrolysis) than for metabolically inactive cells (microbial control 1), despite the fact that the cellulase concentration was lower for microbial hydrolysis through most of the experiment. The lower rates of hydrolysis by inhibited cells were not primarily due to inhibition of cellulase by azide, because the rates of cell-free hydrolysis observed in the presence and absence of azide were similar.
Fig. 2.
Batch cellulose hydrolysis. Microbial and SSF curves are from the experiment depicted in Fig. 1. Experimental conditions for controls are specified in the text.
Microbial hydrolysis and SSF were also compared in steady-state continuous cultures. Mean values for four or more steady-state data points are reported in Table 1, with cellulase-specific activities in Table 2 and standard deviations and carbon recovery in Table 3, which is published as supporting information on the PNAS web site. Microbial steady states 1 and 2 were obtained at residence times (τ = fermentor volume/feed flow rate) of 6.8 and 9.8 h. For microbial steady state 1, 65.3% of the feed cellulose was hydrolyzed in the presence of a total cellulosome concentration of 39 mg/liter, whereas 76.8% hydrolysis was achieved at 46 mg of cellulase/liter for microbial steady state 2. Steady-state continuous SSF mediated by purified C. thermocellum cellulosome in conjunction with fermentation by T. thermosaccharolyticum was carried out at conditions chosen to achieve conversion and total cellulase concentrations similar to those observed for microbial cellulose utilization. Time-course SSF data are presented in Fig. 3(SSF steady state 1) and Fig. 7, which is published as supporting information on the PNAS web site (SSF steady state 2), with steady-state data presented in Table 1. Cellulase-specific activity was similar for microbial and SSF steady states (Table 2). For SSF steady state 1, comparable to microbial steady state 1, cellulose hydrolysis of 67% was observed at τ = 24.4 h and added cellulosome at 52 mg/liter. For SSF steady state 2, cellulose hydrolysis of 75.3% was observed at τ = 19.2 h and 64 mg/liter cellulosome. The concentration of cellulose hydrolysis products was below detection limits (2.5 mg/liter) for both microbial and SSF steady states.
Table 1.
Continuous culture data and degree of synergy calculation
| Hydrolysis | Time | Cellulose, g/liter |
X | Cellulose, g/liter |
τ, hr | Specific rate, hr−1 |
Synergy |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Co | C | EP | ET | r CEt | r CEp | DSEMET | DSEMEP | ||||
| Batch* | |||||||||||
| Microbial | After 8 h | 2.12 | 0.95 | 0.55 | — | 0.057 | 8 | 2.52 | — | 4.69 | — |
| Final (16 h) | 2.12 | 0.0 | 1.0 | — | 0.073 | 16 | 1.80 | — | 2.81 | — | |
| SSF (enzymatic) | After 5 h | 2.07 | 1.64 | 0.21 | — | 0.102 | 8 | 0.54 | — | ||
| Final (32 h) | 2.07 | 0.0 | 1.0 | — | 0.101 | 32 | 0.65 | — | |||
| Continuous† | |||||||||||
| Microbial | Steady state 1 | 4.68 | 1.63 | 0.653 | 0.028 | 0.039 | 6.80 | 11.50 | 16.01 | 4.70 | 4.61 |
| Steady state 2 | 4.66 | 1.08 | 0.768 | 0.029 | 0.046 | 9.80 | 7.94 | 10.66 | 2.72 | 3.05 | |
| SSF (enzymatic) | Steady state 1 | 4.53 | 1.50 | 0.67 | 0.037 | 0.052 | 24.4 | 2.39 | 3.36 | ||
| Steady state 2 | 4.65 | 1.16 | 0.753 | 0.041 | 0.064 | 19.2 | 2.84 | 4.43 | |||
See text for definitions of Co, C, Ep, ET, τ, rC, and DSEM. X is equal to the fractional conversion (Co − C)/Co. All cellulose concentrations are reported in terms of glucose equivalent.
*Batch data are for the experiments presented in Figs. 1, 2, 4, 5, and 6. “Final” corresponds to 16 hours for microbial conversion and 32 hours for SSF.
†Continuous data are for the experiments presented in Figs. 3 and 7.
Fig. 3.
Continuous simultaneous saccharification and fermentation of Avicel by T. thermosaccharolyticum in the presence of 0.052 g/liter purified C. thermocellum cellulosome. Feed cellulose concentration = 4.53 g/liter, τ = 24.4 h. Data from this experiment are presented as “SSF steady state 1” in Table 1.
The cellulase-specific hydrolysis rate (rCE, g of cellulose·g of cellulosome−1·hr−1) may be calculated by using
where Co is the cellulose concentration in g/liter either initially (for batch) or in the feed (for steady state continuous), C is the fermentor cellulose concentration in g/liter after time t (for batch) or at steady state (for continuous), τ is the elapsed time (for batch) or the residence time (for continuous), and E is the average cellulosome concentration in g/liter over the elapsed time (for batch) or for multiple steady-state points (for continuous). The degree of enzyme–microbe synergism, DSEM, may be calculated from the cellulosome-specific hydrolysis rates observed for microbial hydrolysis and SSF by using
The degree of synergism on a total cellulosome basis, DSEMET, is found by using the total cellulosome concentration, ET, in Eq. 1. Alternatively, the degree of synergism on a pellet cellulosome basis, DSEMEP, is found if the pellet cellulase concentration (EP, potentially including both CE and CEM complexes) is used.
Values for DSEMET calculated from batch and continuous data are presented in Table 1. DSEMET based on batch data after 8 h is 4.69. If DSEMET is calculated after complete cellulose hydrolysis is achieved, a value of 2.81 is obtained. In continuous culture, a DSEMET value of 2.72 is obtained based on microbial and SSF steady states 1, for which ≈75% of the feed cellulose was hydrolyzed. For microbial and SSF steady states 2, for which ≈66% hydrolysis was achieved, DSEMET is equal to 4.70. Values for enzyme–microbe synergy on a pellet cellulase basis, DSEMEP, are quite similar to DSEMET values observed in continuous culture: 3.05 for microbial and SSF steady state 1, and 4.61 for microbial and SSF steady state 2. Decreasing synergy is seen with increasing extents of cellulose hydrolysis and with decreasing substrate-to-enzyme ratios for both batch and continuous culture. Variation of the degree of synergy as a function of experimental conditions is commonly observed in determination of synergy among cellulase components in cell-free experiments (9).
Discussion
The C. thermocellum cellulase complex is substantially more effective during microbial hydrolysis as compared with SSF under the conditions examined. Such enzyme–microbe synergy requires the presence of metabolically active cellulolytic microbes and is not explained by removal of hydrolysis products from the bulk fermentation broth. The key apparent difference between microbial hydrolysis and SSF as examined here is that CEM complexes are present during microbial hydrolysis, whereas this is not the case for SSF. Candidate mechanisms for the increased effectiveness of the cellulosome when presented on the surface of a cellulose-adhered cell as compared with when it acts without cell attachment involve events occurring at the site of cellulose hydrolysis, the cellulose surface.
The presence of a cellulose-adherent cellulolytic microbe may increase hydrolysis rates by lowering the local concentration of inhibitory hydrolysis products. Cellobiose and to a lesser extent glucose are known to inhibit the C. thermocellum cellulosome (23); other hydrolysis products could also be inhibitory. Because synergy is observed with hydrolysis products in the fermentation broth at concentrations far below those at which inhibition has been documented (23), a large concentration gradient would have to exist between the cellulose surface and the bulk solution for inhibition by locally high concentrations of hydrolysis products to be important. The occurrence of such gradients is consistent with a highly structured water layer at the surface of cellulose, which has been hypothesized to limit the rate of enzymatic hydrolysis of cellulose by impeding either the escape of hydrolysis products and/or the approach of cellulase enzymes (24). The surface of a cellulose-adherent cell, including peptidoglycan, other cell wall components, and perhaps a glycocalyx, very likely modifies the physical chemistry of the gap between cellulose and an adhered cellulolytic microorganism. It also seems likely that modifications to the chemical environment in this gap that are of benefit to the adherent microorganism would be rewarded from an evolutionary perspective, because organisms with improved substrate access (higher concentration of substrate at the cell surface and/or less loss of substrate to the bulk medium) would presumably grow faster and thus have a selective advantage (11, 25). Microbial adhesion to cellulose may facilitate substrate capture (26), and assimilation of cellodextrins in lieu of cellobiose and glucose (21) has been implicated in molecular evolution (12) and invites consideration of cellulose hydrolysis as a reacting biofilm (26).
From an applied perspective, the 2.7- to 4.7-fold synergistic effect reported here is significant in the context of the search for strategies to decrease the cost of enzymatic hydrolysis, a focus of considerable effort since the late 1990s (2, 11, 27, 28). Further work is warranted to establish the generality of enzyme–microbe synergy, e.g., with respect to a diversity of organisms, substrates/feedstocks, and conditions.
The development of microbes capable of fermenting cellulose to ethanol or other desired products in an industrial setting is a challenging proposition for biotechnology. However, there are increasing indications that such development is both desirable and achievable. Results presented herein suggest that cellulose hydrolysis may be more rapid when mediated by adherent cellulolytic microbes in a consolidated bioprocessing (CBP) configuration as compared with cellulase enzymes acting independently of cellulolytic microbes. The feasibility of CBP is supported by our recent finding that C. thermocellum realizes bioenergetic benefits specific to growth on cellulose that exceed the bioenergetic cost of cellulase synthesis (21). Process design studies indicate that CBP represents a potential breakthrough for low-cost processing of cellulosic biomass (3).
If development of a CBP-enabling microorganism is to be based on naturally occurring cellulolytic microorganisms such as C. thermocellum, it will be necessary to perform metabolic engineering to improve product yield and titer relative to performance obtained to date with available strains. Substantial advances have recently been made in developing requisite genetic tools for this approach (29, 30). Alternatively, organism development could be based on a microbe that already produces high product yields and titers (e.g., yeast) but is not cellulolytic. In this case, it will likely be desirable to understand, incorporate, and perhaps enhance physiological features that underlie enzyme–microbe synergy. Progress and prospects for both CBP organism development strategies are reviewed in detail elsewhere (3, 12).
Materials and Methods
Microbial Cultures and Chemicals.
C. thermocellum ATCC 27405 was provided by Arnold Demain (Massachusetts Institute of Technology, Cambridge, MA) and has been maintained in our laboratory, as described (31). T. thermosaccharolyticum ATCC 31960 was purchased from American Type Culture Collection (Manassas, VA). Chemically defined MTC medium was prepared as described (18) with changes as noted below. All chemicals were reagent grade and were obtained from Sigma (St. Louis, MO), unless indicated otherwise.
Batch Culture of C. thermocellum and Relevant Controls.
Five milliliters of a C. thermocellum stock culture was inoculated by syringe into 100 ml of MTC medium containing 2 g/liter Avicel PH105 and 10 g/liter 4-morpholinepropanesulfonic acid buffer (initial pH 7.6) in triplicate 200-ml sealed serum vials (Bellco Biotechnology, Vineland, NJ) under an N2 atmosphere. Cultures were incubated at 60°C in a water bath with rotary shaking at 200 rpm. Once 2 g/liter Avicel was consumed, as determined by visual inspection, supplemental Avicel was added by syringe as a 40 g/liter sterile suspension to a concentration of 2 g/liter, the pH was adjusted to 7.6 by addition of 4 M NaOH, and the gas phase was replaced by flushing with filter-sterilized N2. The microbial cellulose utilization data presented in Figs. 1 and 2 and Table 1, as well as Fig. 8, which is published as supporting information on the PNAS web site, are taken with the initial (time 0) data point just after supplemental Avicel addition. Microbial control 1 was carried out as above except that a sterilized 1 M sodium azide solution was added to a final concentration of 38.5 mM in conjunction with supplemental Avicel addition. Addition of azide as specified above resulted in cessation of fermentation as indicated by constant concentrations of fermentation products over time measured by HPLC.
Cellulase Preparation and Purification.
Cellulase used for batch SSF experiments was obtained from batch cultures of C. thermocellum grown in MTC medium in a 200-ml serum vial with Avicel as the growth substrate at an initial concentration of 4 g/liter. Cellulase for continuous SSF experiments was obtained from steady-state continuous cultures of C. thermocellum grown in MTC medium at a dilution rate (flow rate/fermentor working volume) of 0.052 h−1 and feed cellulose concentration of 4 g/liter. Purification of cellulase from culture supernatants was carried out by affinity digestion as described (18, 32). Purified cellulase preparations used for batch and continuous SSF experiments contained ≈1.2 g/liter cellulase with a specific activity of 2.8 units/mg of cellulase in Tris buffer (50 mM, with 10 mM CaCl2, pH 6.8). The concentration of soluble hydrolysis products in the purified cellulase preparation was verified by HPLC to be sufficiently small (<0.002 g/liter) to not complicate the interpretation of SSF experiments. The cellulase enzymes used for SSF experiments may not be exactly the same as those present in growing C. thermocellum cultures. However, several factors suggest that any differences between the cellulases present under the conditions examined are not large and do not confound the interpretation of synergy as presented herein (see Supporting Text, which is published as supporting information on the PNAS web site).
Batch SSF and Relevant Enzyme Controls.
Five milliliters of a T. thermosaccharolyticum stock culture was inoculated into 100 ml of MTC medium containing 2 g/liter Avicel PH105 and 2 g/liter cellobiose in triplicate 200-ml serum vials under an N2 atmosphere. Cultures were incubated at 60°C in a water bath with rotary shaking at 200 rpm. Once cellobiose was exhausted, as determined by HPLC, the pH was adjusted back to 7.6, and a purified cellulosome preparation (above) was sterilized by using a filter (Millex-GV, 0.22 μm pore size, Millipore, Billerica, MA) and added to the culture by syringe to a final concentration of 100 mg/liter. The SSF data presented in Figs. 1 and 2 and Table 1, as well as Fig. 5, are taken with the initial (time 0) data point just after cellulase addition. Cell-free control 1 was carried out in the presence of 2 g/liter Avicel and 100 mg/liter purified cellulase as above, except that no fermenting organism was present. Cell-free control 2 was carried out as for cell-free control 1, except that a sterilized 1 M sodium azide solution was added to a final concentration of 38.5 mM.
Continuous Culture.
A modified 1-liter fermentor (Applikon Dependable Instruments, Foster City, CA, modified by NDS) with an overflow sidearm (i.d. 0.38 in.) and 0.5-liter working volume was used both for microbial fermentation by C. thermocellum and for SSF carried out in continuous mode. pH was controlled at 6.8 by a Delta V process control system (New England Controls, Mansfield, MA) with addition of 4 M NaOH, the fermentor was stirred at 250 rpm, and temperature was controlled at 60°C by circulating hot water through the fermentor jacket (19). MTC medium containing 4 g/liter Avicel PH was fed by a peristaltic pump as described elsewhere (31) to achieve residence times as specified in the text. For SSF experiments, an additional peristaltic pump was used to deliver purified cellulase in 50 mM Tris buffer (pH 6.8) and stored at 4°C. A diagram of the SSF system is presented in Fig. 8. The composition of MTC medium and the concentration of Avicel used for SSF experiments were adjusted to provide the same concentrations as those used in C. thermocellum fermentation experiments (e.g., final concentration of 4 g/liter Avicel). SSF experiments were initiated by inoculating 50 ml of a late-exponential phase culture of T. thermosaccharolyticum into MTC medium containing 4 g/liter Avicel supplemented with 2 g/liter cellobiose. Once growth was evident, cellulase addition was commenced. Samples used to calculate steady-state values for continuous fermentations were taken at intervals of at least one residence time. Results are reported as the average of at least four steady-state samples.
Measurement of Residual Cellulose and Fermentation Products.
Residual cellulose was determined by quantitative saccharification (18, 21, 31). Concentrations of sugars (cellobiose and glucose) and fermentation products (lactic acid, acetic acid, and ethanol) were analyzed by HPLC as described (19, 21). Oligomer sugars were analyzed according to the modified NREL posthydrolysis procedure (33). Carbon recovery was calculated as described in Supporting Text.
Measurement of Protein, Cellulase Concentration, and Cellulase Activity.
The protein content in supernatant samples was determined with BSA as the standard by the Bradford protein assay (34). Protein content in the pellet was measured by using the pellet protein assay as described (18, 19, 21). Supernatant and pellet cellulase concentrations were determined by an ELISA method developed previously (18). Avicelase activity in supernatant and pellet samples was measured at 60°C as described (18), based on soluble sugar production as determined by the phenol-sulfuric acid method (35). Results are expressed in terms of International Units (IU) = 1 μmol glucose equivalent/liter per min.
Supplementary Material
Acknowledgments
We thank P. J. Weimer, G. Wolfaardt, and J. Brady for useful discussions. We are grateful for support from the Department of Energy (Grant DE-FG02-02ER15350) and the National Institute of Standards and Technology (Grant 60NANB1D0064).
Abbreviations
- CEM
cellulose–enzyme–microbe
- CE
cellulose–enzyme
- SSF
simultaneous saccharification and fermentation
- CBP
consolidated bioprocessing.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Farrell AE, Plevin RJ, Turner BT, Jones AD, O'Hare M, Kammen DM. Science. 2006;311:506–508. doi: 10.1126/science.1121416. [DOI] [PubMed] [Google Scholar]
- 2.Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ, Jr, Hallett JP, Leak DJ, Liotta CL, et al. Science. 2006;311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 3.Lynd LR, van Zyl WH, McBride JE, Laser M. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Bayer EA, Belaich JP, Shoham Y, Lamed R. Annu Rev Microbiol. 2004;58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 5.Doi RH, Kosugi A. Nat Rev Microbiol. 2004;2:541–551. doi: 10.1038/nrmicro925. [DOI] [PubMed] [Google Scholar]
- 6.Fierobe HP, Mingardon F, Mechaly A, Belaich A, Rincon M, Pages S, Lamed RL, Tardif C, Belaich JP, Bayer EA. J Biol Chem. 2005;280:16325–16334. doi: 10.1074/jbc.M414449200. [DOI] [PubMed] [Google Scholar]
- 7.Davies GJ, Gloster TM, Henrissat B. Curr Opin Struct Biol. 2005;15:637–645. doi: 10.1016/j.sbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y.-H. P., Lynd LR. Biotechnol Bioeng. 2004;88:797–824. doi: 10.1002/bit.20282. [DOI] [PubMed] [Google Scholar]
- 9.Murashima K, Kosugi A, Doi RH. J Bacteriol. 2005;187:7146–7149. doi: 10.1128/JB.187.20.7146-7149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker JO, McCarley JR, Lovett R, Yu CH, Adney WS, Rignall TR, Vinzant TB, Decker SR, Sakon J, Himmel ME. Appl Biochem Biotechnol. 2005:121–124. 129–148. [PubMed] [Google Scholar]
- 11.Zhang Y-HP, Himmel M, Mielenz JR. Biotechnol Adv. 2006;22:452–481. doi: 10.1016/j.biotechadv.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams JJ, Pal G, Jia Z, Smith SP. Proc Natl Acad Sci USA. 2006;103:305–310. doi: 10.1073/pnas.0507109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miron J, Ben-Ghedalia D, Morrison M. J Dairy Sci. 2001;84:1294–1309. doi: 10.3168/jds.S0022-0302(01)70159-2. [DOI] [PubMed] [Google Scholar]
- 15.Schwarz WH. Appl Microbiol Biotechnol. 2001;56:634–649. doi: 10.1007/s002530100710. [DOI] [PubMed] [Google Scholar]
- 16.Shoham Y, Lamed R, Bayer EA. Trends Microbiol. 1999;7:275–281. doi: 10.1016/s0966-842x(99)01533-4. [DOI] [PubMed] [Google Scholar]
- 17.Mayer F, Coughlan MP, Mori Y, Ljungdahl LG. Appl Environ Microbiol. 1987;53:2785–2792. doi: 10.1128/aem.53.12.2785-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y-HP, Lynd LR. Anal Chem. 2003;75:219–227. doi: 10.1021/ac020271n. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y-HP, Lynd LR. J Bacteriol. 2005;187:99–106. doi: 10.1128/JB.187.1.99-106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayer EA, Kenig R, Lamed RL. J Bacteriol. 1983;156:818–827. doi: 10.1128/jb.156.2.818-827.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y-HP, Lynd LR. Proc Natl Acad Sci USA. 2005;102:7321–7325. doi: 10.1073/pnas.0408734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demain AL, Newcomb M, Wu JHD. Microbiol Mol Biol Rev. 2005;69:124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson EA, Reese ET, Demain AL. J Appl Biochem. 1982;4:64–71. [Google Scholar]
- 24.Matthews JF, Skopec CE, Mason PE, Zuccato P, Torget RW, Sugiyama J, Himmel ME, Brady JW. Carbohydr Res. 2006;341:138–152. doi: 10.1016/j.carres.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 25.Fan Z, McBride JE, van Zyl WH, Lynd LR. Biotechnol Bioeng. 2005;92:35–44. doi: 10.1002/bit.20576. [DOI] [PubMed] [Google Scholar]
- 26.Lynd LR, Weimer PJ, Wolfaardt G, Zhang Y-HP. In: Cellulosome. Kataeva IA, editor. Hauppauge, NY: Nova Science; 2006. in press. [Google Scholar]
- 27.Knauf M, Moniruzzaman M. Int Sugar J. 2004;106:147–150. [Google Scholar]
- 28.Sheehan J, Himmel M. Biotechnol Prog. 1999;15:817–827. doi: 10.1021/bp990110d. [DOI] [PubMed] [Google Scholar]
- 29.Tyurin MV, Desai SG, Lynd LR. Appl Environ Microbiol. 2004;70:883–890. doi: 10.1128/AEM.70.2.883-890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tyurin MV, Sullivan CR, Lynd LR. Appl Environ Microbiol. 2005;71:8069–8076. doi: 10.1128/AEM.71.12.8069-8076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynd LR, Grethlein HE, Wolkin RH. Appl Environ Microbiol. 1989;55:3131–3139. doi: 10.1128/aem.55.12.3131-3139.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morag E, Bayer EA, Lamed RL. Enzyme Microb Technol. 1992;14:289–292. [Google Scholar]
- 33.Sluiter A, Hames B, Ruiz R, Scarlata C, Sluiter J, Templeton D, Crocker D. Laboratory Analytic Procedure LAP-002. 2006 http://www1.eere.energy.gov/biomass/analytical_procedures.html. [Google Scholar]
- 34.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y-HP, Lynd LR. Biomacromolecules. 2005;6:1510–1515. doi: 10.1021/bm049235j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.