Abstract
The Gα subunits of heterotrimeric G proteins (Gαβγ) mediate signal transduction via activation by receptors and subsequent interaction with downstream effectors. Crystal structures indicate that conformational changes in “switch” sequences of Gα, controlled by the identity of the bound nucleotide (GDP and GTP), modulate binding affinities to the Gβγ subunits, receptor, and effector proteins. To investigate the solution structure and dynamics of Gαi1 through the G protein cycle, nitroxide side chains (R1) were introduced at sites in switch II and at a site in helix α4, a putative effector binding region. In the inactive Gαi1(GDP) state, the EPR spectra are compatible with conformational polymorphism in switch II. Upon complex formation with Gβγ, motions of R1 are highly constrained, reflecting direct contact interactions at the Gαi1–Gβ interface; remarkably, the presence of R1 at the sites investigated does not substantially affect the binding affinity. Complex formation between the heterotrimer and activated rhodopsin leads to a dramatic change in R1 motion at residue 217 in the receptor-binding α2/β4 loop and smaller allosteric changes at the Gαi1–Gβγ interface distant from the receptor binding surface. Upon addition of GTPγS, the activated Gαi1(GTP) subunit dissociates from the complex, and switch II is transformed to a unique conformation similar to that in crystal structures but with a flexible backbone. A previously unreported activation-dependent change in α4, distant from the interaction surface, supports a role for this helix in effector binding.
Keywords: G protein-coupled receptor, signal transduction, site-directed spin labeling, switch II, transducin
Heterotrimeric G proteins (Gαβγ) mediate signal transduction and amplification in a variety of cell signaling pathways (1–3). A constellation of homologous but distinct Gα subunits confers specificity for a particular pathway. In the inactive state, the Gα subunit of the heterotrimer contains a bound GDP [Gα(GDP)] and has a high affinity for Gβγ. When activated by an appropriate signal, membrane-bound G protein-coupled receptors bind the heterotrimer in a quaternary complex and catalyze exchange of the bound GDP for GTP in Gα. The affinity of Gα(GTP) for Gβγ is dramatically reduced relative to Gα(GDP), resulting in functional dissociation of active Gα(GTP) from the membrane-bound complex. The active Gα(GTP) subsequently binds downstream effector proteins to trigger a variety of regulatory events, depending on the particular system. Intrinsic GTPase activity of Gα returns it to the inactive Gα(GDP) form that again binds Gβγ, completing the G protein cycle.
The structural changes in Gα that accompany nucleotide exchange and the means by which the activated receptor catalyzes the nucleotide exchange are central to understanding the mechanism of signal transduction. Crystal structures for α subunits of the homologous G proteins Gt (transducin) and Gi1 have been determined for both the GDP and GTPγS bound forms (4–7) and reveal that the nucleotide modulates the structure of three sequences, designated switches I–III. Two of these sequences (switch I and II) are located at the Gα–Gβ interface in the heterotrimer (8) and at the interface with effector proteins (9), and both make direct contact with the bound nucleotide. In Gαt(GTPγS) a short helix (α2) in switch II is displaced ≈8 Å relative to Gαt(GDP) (Fig. 1A); in Gαi1(GDP), switch II is not resolved because of disorder in the lattice, but in Gαi1(GTPγS) it has a similar conformation to that in Gαt(GTPγS). The difference between switch II conformation in Gαt(GDP) and Gαi1(GDP) may be due to crystal lattice contacts at switch II in the former, which are absent in Gαi1(GDP). If this is the case, switch II might be expected to be flexible in the Gα(GDP) states in solution, a view supported by NMR studies of corresponding regions of the small G proteins ras and Cdc42Hs (10–13). Crystal lattice forces are sufficiently strong to distort flexible structures; indeed, crystal structures of GDP bound ras from different space groups have different structures in switch II (14). This raises the possibility that the structures of the switches in the active Gα(GTPγS) states in solution may differ from that in crystal as well, because they are located at lattice contacts in the crystals of Gtα(GTPγS) and Gαi1(GTPγS) (5, 7). Moreover, it has recently been shown that the unusual solvent conditions required for crystallization can strongly affect protein conformation of flexible sequences (15). Thus, it is of interest to compare the status of the switch regions of Gα(GDP) and Gα(GTPγS) in solution with those in the crystal. Crystal structures are available for the heterotrimeric Gtαβγ(GDP) (8) and Gi1αβγ(GDP) (16) (Fig. 1B), in which switch II adopts a yet another conformation, but little structural information is available on the important complex between any heterotrimeric G protein and the activated receptor.
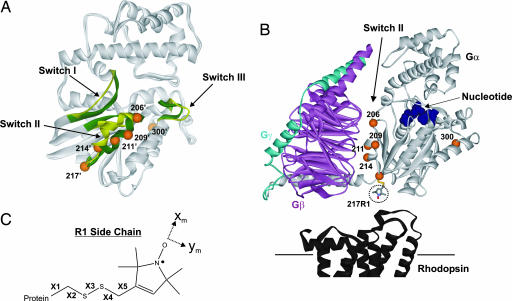
Fig. 1.
Ribbon diagrams of Gα and Gαβγ structures showing spin-labeled sites. (A) Overlay of Gαt(GDP) (Protein Data Bank ID code 1TAG) and Gαt(GTPγS) (Protein Data Bank ID code 1TND) structures highlighting the Switch regions (green in 1TND and yellow in 1TAG) and showing sites where R1 was introduced as spheres (1TND only); numbering is according to the corresponding position in Gαi1. (B) Structure of Gαi1(GDP)βγ (Protein Data Bank ID code 1GP2) showing positions in Gαi1 (light gray) where R1 was introduced in Switch II and α4 (300). The Gβ and Gγ subunits are magenta and cyan, respectively, and the cytoplasmic surface of rhodopsin (dark gray, Protein Data Bank ID code 1GZM) is positioned according to current models of the empty complex. (C) The nitroxide side chain, R1, with definitions of the side chain dihedral angles (X1–X5) and the x and y axes of the magnetic tensor frame.
In the present study, site-directed spin labeling (SDSL) is used to investigate the structure and dynamics of switch II and another putative switch region in helix α4 (17) along the activation pathway in the visual signal transduction system. Rhodopsin serves as a model G protein-coupled receptor, and Gαi1 was used as a fully functional mimic of the visual Gαt because it can be expressed in bacterial systems and is homologous to the transducin α subunit (18). In SDSL, a nitroxide side chain (R1) is introduced (Fig. 1C), and the motion of the nitroxide on the nanosecond time scale is extracted from the EPR spectra by using qualitative evaluation (19, 20) or spectral simulations (21). In helices and loops at solvent exposed sites where the nitroxide does not contact other residues, the internal motion of R1 is dominated by torsional oscillations of dihedrals X4 and X5 (Fig. 1C) (22, 23), giving rise to a characteristic z axis anisotropic motion of the nitroxide (23). This inherent motion is modulated by backbone motion on the nanosecond time scale, and variations in motion of R1 at different sites in helices reflect variations in local backbone motions (24, 25). The internal motion of R1 is also modulated by weak interactions of the nitroxide, a situation readily recognized in the lineshape by the appearance of features corresponding to reduced amplitudes and/or rates of motion. This property confers spectral sensitivity to the local structure of the protein (26, 27). Thus, the EPR spectra of R1 at a selected set of sites can be used to gain information on both local backbone dynamics and conformation.
To investigate Gαi1 with SDSL, R1 residues were introduced, one at a time, at sites along the switch II sequence and one within helix α4. For each of the spin-labeled mutants in solution, EPR spectra were recorded in four states: (i) Gαi1(GDP) alone; (ii) Gαi1(GDP)Gβγ; (iii) the heterotrimer in complex with activated rhodopsin (R*) in native membranes in the absence of added nucleotide; and (iv) the activated Gαi1(GTPγS), formed by addition of GTPγS to the complex. As described below, the EPR spectra report salient structural and dynamical features of switch II in each state, providing a basis for comparison with crystal structure where available, and providing the first information on the status of the Gα–Gβγ interface in the receptor complex at the membrane surface.
Results
Fig. 1 shows the sites of introduction of R1 in the structure of the Gαi1βγ heterotrimer (16) (Fig. 1B) and the corresponding sites in the overlaid structures of the Gαt(GDP) (4) and Gαt(GTPgS) (5) subunits (Fig. 1A). Sites 206, 209, 211, 214, and 217 are in switch II, and site 300 is in α4. The spin-labeled derivatives of Gαi1(GDP) bind to Gβγ (see below) and show essentially WT activity with respect to R* binding and nucleotide exchange (Fig. 5, which is published as supporting information on the PNAS web site). To follow structural/dynamical changes in Gα, a series of EPR spectra were collected after sequential sample additions to each R1-labeled Gαi1 mutant as indicated in the top row of Fig. 2. Spectral simulations are used in some cases to estimate the orders and rates of nitroxide motion, given, respectively, by the order parameter (S) and effective correlation time (τ) (21, 23).
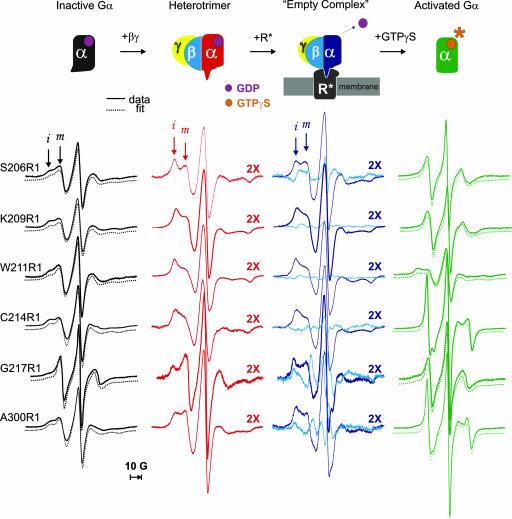
Fig. 2.
Monitoring conformational changes in Gα. The top row shows the states of Gαi1 investigated along the activation pathway; below each state are shown the corresponding EPR spectra at the sites investigated, color-coded according to the Gαi1 state. All spectra were normalized to the same number of spins. MOMD simulations of GDP and GTPγS bound forms are shown as offset dotted lines. Arrows highlight immobilized (i) and mobile (m) spectral components. The spectra for the heterotrimer and empty complex are multiplied by 2. Difference spectra (empty complex − heterotrimer, cyan, third column) illustrate the EPR changes due to R* binding.
The Gαi1(GDP) Inactive State.
The EPR spectra for the Gαi1(GDP) state are shown in the left column of Fig. 2. The spectra of R1 in α2 and at site 300 in α4 are remarkably similar to each other and are reasonably well fit to a model with two components (dashed traces in Fig. 2), one of which corresponds to an R1 population with weakly ordered and fast anisotropic motion (S ≈ 0.25, τ = 2–3 ns) and the other with isotropic slow motion (S ≈ 0; τ ≈ 8.5 ns) (m and i arrows, respectively, in Fig. 2). Site 217R1, in a loop after α2, is also fit with a two-component model but is dominated by a mobile population with fast isotropic motion (S ≈ 0; τ = 1.5 ns), consistent with a flexible loop structure (26). The switch II sequence is not resolved in the crystal structure of Gαi1(GDP) but is helical in the homologous Gαt(GDP) crystal structure with average thermal factors. The similar EPR spectra along the sequence are not compatible with the expected site-to-site variations in R1 motion along the helix, particularly for 214R1, which is located at a buried site and would be strongly immobilized. Implications of the EPR spectra regarding the probable solution structure in Gαi1(GDP) will be considered in Discussion.
Helix α4 is well ordered in crystal structures of Gαi1(GDP), where its N terminus resides at a contact site with a symmetry-related molecule. Residue Ala-300 is on the buried surface of the helix (solvent accessibility 4%), and R1 cannot be modeled without severe atomic overlaps. However, the high reactivity of the Ala300Cys mutant with the spin labeling reagent (see Materials and Methods) suggests high accessibility in solution, consistent with the presence of a component in the EPR spectrum reflecting a mobile state of R1. These results imply a structure in solution different from that in the crystal or that the structure is in equilibrium with other conformations in which residue 300 has a larger solvent accessibility.
The Gαi1(GDP)βγ Heterotrimer.
The switch II sequence makes direct contact with the Gβ subunit upon heterotrimer formation (Fig. 1B). R1 side chains in switch II at Gβ interface would thus be expected to become immobilized upon heterotrimer formation. This is indeed the case, as shown by the greatly increased intensity of a spectral component corresponding to a strongly immobilized state of R1 upon addition of Gβγ (i component) (Fig. 2, second column). The presence of substantial mobile components in the spectra of 206R1 and 217R1 (m component), located at the N terminus of α2 and a loop after the C terminus, respectively, is compatible with the crystal structure of the heterotrimer; at these sites, the R1 side chain can project away from the interface, removing constraints on the motion (see, for example, the model of 217R1 in Fig. 1B). Interestingly, the spectrum of 300R1 in α4, distant from the binding interface, also shows significant immobilization, indicating allosteric changes propagated through Gα upon heterotrimer formation. Complexation with Gβγ increases the rotational correlation time of Gαi1 by a factor of ≈2, but this cannot account for the changes in R1 mobility. For example, increasing the viscosity of the medium by a factor of 2 with Ficoll increases the rotational correlation time of Gαi1 by the same factor but has only minor effects on the spectrum of 214R1 (data not shown).
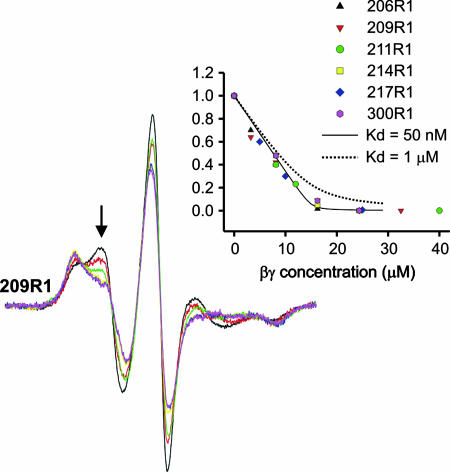
To evaluate the level of perturbation caused by introduction of R1 at the subunit interface, Gβγ was titrated into solutions containing a fixed concentration of the individual spin-labeled Gαi1 subunits. In these experiments, changes in normalized EPR spectral intensity served as an indicator of heterotrimer formation. Fig. 3 shows example spectra for 209R1 and a plot of the fractional change in intensity [(I − Ifinal)/(Iinitial − Ifinal)] at the indicated field position versus the concentration of Gβγ added to the solution. Similar data are included for the other mutants investigated. The solid line in the plot is calculated for Kd = 50 nM, the dissociation constant for the Hexa I base mutant of Gαi1 used in the present experiments (28) (see Materials and Methods). As is evident, the presence of R1 at the interaction interface apparently has remarkably little effect on the binding.
Fig. 3.
Titration of spin-labeled Gαi1 (15 μM) with Gβγ (see text). Example spectra are shown for 209R1. (Inset) The fractional change in spectral intensity at the indicated field position as a function of added Gβγ for all sites in Switch II. Several titration points (4–6) were collected for each spin-labeled mutant. The solid and dotted lines are calculated for Kd values of 50 nM and 1 μM, respectively.
The Gαi1(0) βγ–Receptor Complex (the “Empty Complex”).
Binding of the heterotrimer to R* in the absence of GTP results in the release of GDP from Gα and the formation of a high-affinity “empty” (no nucleotide bound) complex at the membrane surface (29). Under the conditions of the experiments, all spin-labeled Gαi1βγ mutants show essentially WT levels of binding to R* (≈50–80% bound) (Fig. 5). The EPR spectra for the heterotrimer show little change upon addition of rod outer segment (ROS) membranes in the dark (data not shown). However, upon photoactivation, a distinctive pattern of EPR spectral changes relative to the heterotrimer alone is observed that is seen in the difference of the normalized EPR data (empty complex − heterotrimer) (column 3, cyan traces) or in direct spectral overlays of the two states (Fig. 6, which is published as supporting information on the PNAS web site). The presence of some fraction of unbound heterotrimer (20–50%) will not affect the shape of the difference spectra but will underestimate the magnitude. In the difference spectra, positive and negative amplitudes in the low-field region reflect increases and decreases, respectively, in populations of mobile (m) and immobile (i) states that are reciprocally related. As is evident, there is a substantial shift in the population toward more mobile states for 206R1, a subtle change at 209R1 in the same direction, very small changes at 211R1 and 214R1, and a large opposite shift toward more immobilized states for 217R1. The changes at 206 and 209 reveal structural rearrangements at the Gα–Gβγ interface due to complex formation, but the immobilization of 217R1 likely reflects direct receptor contact, because it projects directly toward the putative binding interface (Fig. 1B). Once again, the spectrum of 300R1, distant from both the Gα–Gβ interface and the putative receptor contact surface, reports a change triggered by R* interactions.
The Gαi1(GTPγS)-Activated State.
Addition of GTPγS to the spin-labeled empty complex results in dissociation of the activated Gαi1(GTPγS) subunit from Gβγ and the receptor (29) (Fig. 5), with concomitant changes in the corresponding EPR spectra (Fig. 2, last column). For all but 217R1, the EPR spectra of the activated subunit Gαi1(GTPγS) are dramatically different from those of Gαi1(GDP) and identify structural and dynamical changes in both switch II and in α4 that are directly coupled to the change in nucleotide (see Fig. 6 for a direct spectral comparison).
The crystal structure of Gαi1(GTPγS) is shown in Fig. 4 along with models of the R1 side chain in a favored rotameric state (see Materials and Methods) at selected sites in α2. At the N terminus of α2, 206R1 and 209R1 are located at the solvent-exposed surface of the helix where little contact with the surrounding structure is expected. In accord with this expectation, the spectra of both 206R1 and 209R1 are well fit with a dominant component of weakly ordered and fast motion (S ≈ 0.2, τ ≈ 1.6 ns) (dashed curves in Fig. 2, last column), characteristic of R1 residues in a flexible helical structure (24). For reference, R1 in a well ordered helix at a noninteracting solvent exposed site has S ≈ 0.45, τ ≈ 2 ns (23). The spectrum of 211R1 contains a dominant immobilized population, again consistent with the crystal structure, where it is located at a relatively buried site (position marked by a sphere at Cα).
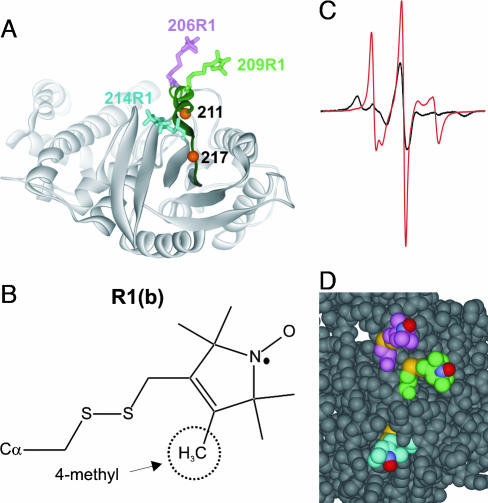
Fig. 4.
Models of R1 at sites 206, 209, and 214 in Gαi1(GTPγS) (Protein Data Bank ID code 1GIA). (A) Ribbon diagram with R1 as stick models; location of sites 211 and 217 are shown as orange spheres. (B) Structure of R1b. (C) EPR spectra of 214R1 (red trace) and 214R1b (black). (D) Space-filling model showing R1 at 206, 209, and 214; the nitrogen, oxygen, and sulfur atoms of R1 are colored blue, red, and yellow for reference.
The EPR spectrum of 214R1 is striking and has been previously observed only for R1 at a single site in a short helix of CRBP (30); the spectrum of 300R1 is similar. Fitting these spectra (dashed traces) reveals a single dynamic mode with high ordering and fast motion (S ≈ 0.6, τ = 0.6 ns) about the nitroxide x axis (along the N—O bond) (Fig. 1C). This is in contrast to the z axis ordering typical of R1 at most helical sites, including those at 206 and 209 in Gαi1(GTPγS) discussed above. This unusual motion, confirmed by simulation of spectra collected at three microwave frequencies (S, X, and Q bands) (Fig. 7, which is published as supporting information on the PNAS web site), could in principle arise from either backbone fluctuations or internal motions in R1. Addition of a 4-methyl group in the nitroxide ring (R1b) (Fig. 4B) has previously been shown to selectively restrict torsional oscillations about X5 and provides a means of identifying a contribution due to that motion (24). The spectrum of 214R1b is strongly immobilized (Fig. 4C), demonstrating that the motion arises primarily within the side chain, although some contribution from the flexible backbone is likely. The terminal bond vector (X5) (Fig. 1C) has a substantial projection along the N—O bond, and rapid motion about X5 could give rise to the observed x axis anisotropy. The space-filling model of Fig. 4D suggests how this situation may occur; the side chain of R1 at 214 is entirely sequestered in the structure up to the nitroxide ring, allowing free motion only about X5. In contrast, the 206R1 and 209R1 side chains are unrestricted with respect to X4 and X5, giving rise to the z axis anisotropy previously described (Fig. 4D) (23). The essentially identical motion of 300R1 in α4 likely arises from a similar unique environment, which is not provided by the crystal structure; as pointed out above, 300R1 cannot be modeled in the crystal structures of Gα. Thus, as for Gαi1(GDP), the solution structure of Gαi1(GTPγS) must be different from that in the crystal in the neighborhood of residue 300 in the spin-labeled protein.
Discussion
The function of G proteins is based on guanine nucleotide-driven structural rearrangements in the switch regions of Gα. In the inactive, Gα(GDP)βγ heterotrimer, Gβγ significantly alters the conformation of switches I and II, thereby dismantling the binding sites for Mg2+ and the GTP γ-phosphate, explaining the observation that GTP and Gβγ binding to Gα are negatively cooperative (31). Receptor-mediated GDP release leads to an empty GDP-binding pocket and a cooperative high-affinity interaction with the receptor that is released when GTP binds. NMR studies have begun to identify structural changes in Gα that are related to guanine nucleotide exchange. However, at this point only four resonance assignments have been made in 15N filtered HSQC experiments (32, 33), and no data have yet been reported on the structure of the important rhodopsin bound complex. The SDSL study reported here is the first to examine molecular changes underlying the complete G protein cycle in solution. A significant finding is that switch II in the active Gα(GTPγS) state is more flexible, not less, as was thought from the crystal structures, than the inactive Gα(GDP) form. Implications for function are discussed below.
Solution Structure and Dynamics in Gαi1(GDP) and Gαi1(GTPγS).
The two-component EPR spectra of R1 at sites 206, 209, 211, and 214 in Gαi1(GDP) could in principle arise from two rotamers of R1 in which the nitroxide experiences two environments (22), but it could also arise from two conformations of switch II in slow exchange (microseconds to milliseconds) on the EPR time scale. Given the facts that switch II is disordered in the Gαi1(GDP) crystal structure and that slow conformational exchange (milliseconds) is observed in switch II in solution structures of small G proteins (10, 13), a plausible interpretation of the EPR data are that switch II in Gαi1(GDP) is also in slow conformational exchange between at least two states in solution. This interpretation is further supported by the striking similarity of the spectra at all sites, which is not expected for a uniquely ordered structure, such as that seen in the homologous Gαt(GDP) structure.
Upon formation of the activated Gαi1(GTPγS) state, the dramatic changes in the EPR spectra signal a change in conformation of switch II to a single ordered state compatible with that observed in the crystal structure. The structure offers a natural explanation for the unique EPR line shape of 214R1 and accounts for the lack of change upon activation at 217R1, which is located at a hinge for α2 motions (Fig. 1A). A feature not appreciated from the crystal structure is the apparent flexibility on the nanosecond time scale of at least the N-terminal portion of α2 in the activated state; the crystallographic thermal factors in this region are average for the structure. Interestingly, the C terminus of Gαi and the “finger loop” of arrestin, both interaction sequences in the binding to activated rhodopsin, are also highly flexible in solution (34, 35). Moreover, the third cytoplasmic loop of rhodopsin, involved in both interactions (36, 37), is itself highly flexible (25). The flexibility in each case is on the nanosecond time scale, as compared with the microseconds to milliseconds time scale for conformational exchange. These data suggest that enhanced backbone flexibility may be a general feature of protein–protein interaction sequences in signal transduction and that part of the trigger involved in activation of Gα may be the release of constraints on α2 to generate flexibility.
A finding of this study is the conformational change in α4 detected by 300R1 upon Gαi1 activation, which is not predicted from the essentially identical conformation around this site in all crystal structures of Gα, irrespective of the identity of the bound nucleotide. The difference between the solution and crystal structures may be due to the contact of α4 with a symmetry-related molecule in the crystal lattice. The structural origin of the EPR spectral change cannot be inferred from data on a single site; nevertheless, the unspecified change supports previous data that α4 may be involved in effector protein interactions triggered by Gα activation (17).
Heterotrimer Formation and the Receptor Empty Complex.
All mutants of Gαi1(GDP) bearing R1 in switch II form heterotrimers with Gβγ with a Kd similar to that for the WT subunits (28), despite the presence of R1 directly at the Gαi1(GDP)–Gβγ interface (Figs. 1B and 3). This result joins earlier observations that R1 is tolerated at contact surfaces in arrestin–rhodopsin interactions (35) and in SecA–SecB interactions (19). This result may not be general, but as long as highly specific polar interactions are not perturbed, the nonpolar and flexible nature of the R1 side chain apparently allows such substitutions to be made with retention of function.
The EPR spectra of R1 at all sites along switch II in the Giα1(GDP)βγ heterotrimer are compatible with the corresponding crystal structure. On the other hand, the decrease in mobility of 300R1 upon heterotrimer formation reveals a conformational transition in α4 not obvious from an alignment of the nucleotide binding domains of the Gαi1(GDP) and Gαi1(GDP)βγ crystal structures. This interesting result implies that long-range structural changes are propagated through Gαi1 from the Gα–Gβγ interface to α4 upon heterotrimer formation.
Complex formation between the Gαi1(GDP)βγ heterotrimer and photoactivated rhodopsin triggers changes in the heterotrimer that result in dissociation of GDP to form the empty complex. Current models for the complex involve structural changes propagated from the receptor interaction surface through the heterotrimer to reach the nucleotide binding site some 30 Å away (Fig. 1B). Recently, SDSL studies have identified one such change, propagated by rigid body motions of helix α5 directly to the nucleotide binding site in Gαi1 (34). The results presented here identify another allosteric change wherein the Gαi1–Gβγ interface is modulated by receptor activation, perhaps coupled to GDP release.
Mechanisms whereby receptor interaction directly modulates the Gα–Gβγ interface to cause GDP release have been proposed. In a “lever-arm” model, both the C and N termini of Gα interact with the receptor in such a way as to rotate Gβγ away from Gα, pulling switches I and II with it and opening a door for escape of the nucleotide (38, 39). Conversely, the “gearshift” model proposes that a motion occurs in the opposite direction, i.e., rotation of Gβγ into Gα (40). Although the R1 side chains in switch II clearly identify changes at this interface, the data are too sparse to evaluate the models.
In summary, this study illustrates the capability of SDSL to map structural and dynamical changes in a G protein along the activation pathway using nanomole quantities of protein at ambient temperature without restrictions regarding molecular weight or complexity of the species involved. Crystal structures play a key role in providing a starting point for interpretation of the EPR data, which in turn provide a means for evaluating the influence of the crystal environment on structure as well as a new dimension of dynamic information on the protein and its complexes in solution. The important findings observed here are the increase in dynamics in switch II upon Gα activation and the identification of an allosteric change at the Gα–Gβγ interface triggered by interactions with activated receptor; the strong immobilization of 217R1 at the C terminus of switch II likely reflects direct receptor contact.
Materials and Methods
Materials.
GDP and guanosine 5′-O-(3-thiotriphosphate) (GTPγS) were from Sigma (St. Louis, MO). The sulfhydryl spin-label reagent, S-(1-oxy-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate, was a generous gift from Kalman Hideg (University of Pecs, Pecs, Hungary). All other reagents and chemicals were of the highest available purity.
Preparation of ROS Membranes and Gβγ Subunits.
Urea-washed ROS membranes and Gb1γ1 were prepared as previously described (28) and stored at −80°C. All ROS and Gβγ samples were buffer-exchanged into 20 mM Mes (pH 6.8), 100 mM NaCl, 2 mM MgCl2, and 10% glycerol before EPR experiments.
Construction, Expression, and Purification of Mutant Proteins.
Briefly, a plasmid encoding Gαi1 was used which contained six amino acid substitutions at solvent-exposed cysteines (C3S-C66A-C214S-C305S-C325A-C351I) and a hexahistidine tag between amino acid residues Met-119 and Thr-120 (28). This construct served as a template for introducing individual cysteine substitutions using the QuikChange system (Stratagene, La Jolla, CA). All mutations were confirmed by DNA sequencing (DNA Sequencing Facility, Vanderbilt University, Nashville, TN). The mutant constructs were then transformed in Escherichia coli BL21–Gold (DE3) (Stratagene), expressed, and purified as previously described (28).
Spin-Labeling, EPR Spectroscopy, and Modeling of the R1 Side Chain.
Spin-labeling was carried out in a buffer containing 20 mM MES (pH 6.8), 100 mM NaCl, 2 mM MgCl2, 50 μM GDP, and 10% glycerol (vol/vol). The Gαi1 mutants were incubated with S-(1-oxy-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate at a 1:1 molar ratio at room temperature for 5 min. Under these conditions, only the most reactive Cys residues were modified, and the remaining buried native cysteine residues were unreactive (28). Any excess spin-labeling reagent was removed by extensive washing with buffer using a 30-kDa concentrator. For EPR spectroscopy, a series of spectra were recorded for each spin-labeled mutant. First, Gαi1 mutants (30 μM) were loaded into a sealed quartz flat cell, and spectra were recorded at room temperature on a Bruker E580 spectrometer by using a high-sensitivity resonator at X-band microwave frequency. The data were typically averages of 20–50 scans. Gβγ was then added in a 1:1 molar ratio to form heterotrimers. The diluted samples were concentrated to the same concentration as the initial Gαi1 mutants, and the EPR spectra were recorded both alone in solution and upon addition of urea-washed ROS in the dark (150 μM). The sample was subsequently irradiated for 30 s by using a tungsten lamp (cutoff filter, λ > 500 nm), and the EPR spectra were recorded immediately after bleaching. Finally, GTPγS (200 μM) was added to the samples to form activated Gαi1.
Fitting of spectra to the MOMD model of Freed and coworkers (21) followed previously published methods using principle values for the A and g tensors of Axx = Ayy = 6 G, Azz = 37G, gxx = 2.0078, gyy = 2.0058, and gzz = 2.0023. The R1 side chain was modeled with X1 = −60° and X2 = −60°, a rotamer commonly observed in crystal structures of the side chain in T4 lysozyme (22) (M. R. Fleissner, D. Cascio, and W.L.H., unpublished data). Structures from the unpublished work have been deposited in the Protein Data Bank (PDB ID codes 2CUU and 1ZYT). X3 was selected as ±90 to eliminate steric overlaps.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (to W.L.H. and H.E.H.), a Public Heath Service Award for the Medical Scientist Training Program (to W.M.O.), the Pharmaceutical Research and Manufacturers of America Foundation (W.M.O.), a Ruth L. Kirschstein National Research Service Award (to N.V.E.), and the Jules Stein Professorship (to W.L.H.).
Abbreviations
- ROS
rod outer segment
- SDSL
site-directed spin labeling.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bourne HR. Curr Opin Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 2.Hamm HE. Proc Natl Acad Sci USA. 2001;98:4819–4821. doi: 10.1073/pnas.011099798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamb TD, Pugh EN., Jr Trends Neurosci. 1992;15:291–298. doi: 10.1016/0166-2236(92)90079-n. [DOI] [PubMed] [Google Scholar]
- 4.Lambright DG, Noel JP, Hamm HE, Sigler PB. Nature. 1994;369:621–628. doi: 10.1038/369621a0. [DOI] [PubMed] [Google Scholar]
- 5.Noel JP, Hamm HE, Sigler PB. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 6.Coleman DE, Sprang SR. Biochemistry. 1998;37:14376–14385. doi: 10.1021/bi9810306. [DOI] [PubMed] [Google Scholar]
- 7.Coleman DE, Berghuis AM, Lee E, Linder ME, Gilman AG, Sprang SR. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- 8.Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 9.Slep KC, Kercher MA, He W, Cowan CW, Wensel TG, Sigler PB. Nature. 2001;409:1071–1077. doi: 10.1038/35059138. [DOI] [PubMed] [Google Scholar]
- 10.Loh AP, Guo W, Nicholson LK, Oswald RE. Biochemistry. 1999;38:12547–12557. doi: 10.1021/bi9913707. [DOI] [PubMed] [Google Scholar]
- 11.Feltham JL, Dotsch V, Raza S, Manor D, Cerione RA, Sutcliffe MJ, Wagner G, Oswald RE. Biochemistry. 1997;36:8755–8766. doi: 10.1021/bi970694x. [DOI] [PubMed] [Google Scholar]
- 12.Kraulis PJ, Domaille PJ, Campbell-Burk SL, Van Aken T, Laue ED. Biochemistry. 1994;33:3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- 13.Ito Y, Yamasaki K, Iwahara J, Terada T, Kamiya A, Shirouzu M, Muto Y, Kawai G, Yokoyama S, Laue ED, et al. Biochemistry. 1997;36:9109–9119. doi: 10.1021/bi970296u. [DOI] [PubMed] [Google Scholar]
- 14.Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim S-H. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 15.Kim M, Xu Q, Fanucci GE, Cafiso DS. Biophys J. 2006;90:2922–2929. doi: 10.1529/biophysj.105.078246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wall MA, Coleman DE, Lee E, Iniguez-Liuhi JA, Posner BA, Gilman AG, Sprang SR. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 17.Rarick HM, Artemyev NO, Hamm HE. Science. 1992;256:1031–1033. doi: 10.1126/science.1317058. [DOI] [PubMed] [Google Scholar]
- 18.Downes GB, Gautam N. Genomics. 1999;62:544–552. doi: 10.1006/geno.1999.5992. [DOI] [PubMed] [Google Scholar]
- 19.Crane JM, Mao C, Lilly AA, Smith VF, Suo Y, Hubbell WL, Randall LL. J Mol Biol. 2005;353:295–307. doi: 10.1016/j.jmb.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 20.Kusnetzow AK, Altenbach C, Hubbell WL. Biochemistry. 2006;45:5538–5550. doi: 10.1021/bi060101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budil DE, Lee S, Saxena S, Freed JH. J Magn Reson Ser A. 1996;120:155–189. [Google Scholar]
- 22.Langen R, Oh KJ, Cascio D, Hubbell WL. Biochemistry. 2000;39:8396–8405. doi: 10.1021/bi000604f. [DOI] [PubMed] [Google Scholar]
- 23.Columbus L, Kalai T, Jeko J, Hideg K, Hubbell WL. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 24.Columbus L, Hubbell WL. Biochemistry. 2004;43:7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 25.Columbus L, Hubbell WL. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 26.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 27.Hubbell WL, Cafiso DS, Altenbach C. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 28.Medkova M, Preininger AM, Yu NJ, Hubbell WL, Hamm HE. Biochemistry. 2002;41:9962–9972. doi: 10.1021/bi0255726. [DOI] [PubMed] [Google Scholar]
- 29.Bornancin F, Pfister C, Chabre M. Eur J Biochem. 1989;184:687–698. doi: 10.1111/j.1432-1033.1989.tb15068.x. [DOI] [PubMed] [Google Scholar]
- 30.Lietzow MA. Los Angeles: Univ of California; 1999. PhD dissertation. [Google Scholar]
- 31.Higashijima T, Ferguson KM, Sternweis PC, Smigel MD, Gilman AG. J Biol Chem. 1987;262:762–766. [PubMed] [Google Scholar]
- 32.Abdulaev NG, Ngo T, Zhang C, Dinh A, Brabazon DM, Ridge KD, Marino JP. J Biol Chem. 2005;280:38071–38080. doi: 10.1074/jbc.M505259200. [DOI] [PubMed] [Google Scholar]
- 33.Ridge KD, Abdulaev NG, Zhang C, Ngo T, Brabazon DM, Marino JP. J Biol Chem. 2006;281:7635–7648. doi: 10.1074/jbc.M509851200. [DOI] [PubMed] [Google Scholar]
- 34.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 35.Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Proc Natl Acad Sci USA. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raman D, Osawa S, Gurevich VV, Weiss E. J Neurochem. 2003;84:1040–1050. doi: 10.1046/j.1471-4159.2003.01598.x. [DOI] [PubMed] [Google Scholar]
- 37.Cai K, Itoh Y, Khorana HG. Proc Natl Acad Sci USA. 2001;98:4819–4821. doi: 10.1073/pnas.051632898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iiri T, Farfel Z, Bourne HR. Nature. 1998;394:35–38. doi: 10.1038/27831. [DOI] [PubMed] [Google Scholar]
- 39.Rondard P, Iiri T, Srinivasan S, Meng E, Fujita T, Bourne HR. Proc Natl Acad Sci USA. 2001;98:6150–6155. doi: 10.1073/pnas.101136198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cherfils J, Chabre M. Trends Biochem Sci. 2003;28:13–17. doi: 10.1016/s0968-0004(02)00006-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.