Abstract
Maintaining muscle size and fiber composition requires contractile activity. Increased activity stimulates expression of the transcriptional coactivator PGC-1α (peroxisome proliferator-activated receptor γ coactivator 1α), which promotes fiber-type switching from glycolytic toward more oxidative fibers. In response to disuse or denervation, but also in fasting and many systemic diseases, muscles undergo marked atrophy through a common set of transcriptional changes. FoxO family transcription factors play a critical role in this loss of cell protein, and when activated, FoxO3 causes expression of the atrophy-related ubiquitin ligases atrogin-1 and MuRF-1 and profound loss of muscle mass. To understand how exercise might retard muscle atrophy, we investigated the possible interplay between PGC-1α and the FoxO family in regulation of muscle size. Rodent muscles showed a large decrease in PGC-1α mRNA during atrophy induced by denervation as well as by cancer cachexia, diabetes, and renal failure. Furthermore, in transgenic mice overexpressing PGC-1α, denervation and fasting caused a much smaller decrease in muscle fiber diameter and a smaller induction of atrogin-1 and MuRF-1 than in control mice. Increased expression of PGC-1α also increased mRNA for several genes involved in energy metabolism whose expression decreases during atrophy. Transfection of PGC-1α into adult fibers reduced the capacity of FoxO3 to cause fiber atrophy and to bind to and transcribe from the atrogin-1 promoter. Thus, the high levels of PGC-1α in dark and exercising muscles can explain their resistance to atrophy, and the rapid fall in PGC-1α during atrophy should enhance the FoxO-dependent loss of muscle mass.
Keywords: denervation, fasting, muscle fiber, energy metabolism, mitochondria
The mass and functional capacity of skeletal muscle are tightly regulated by contractile activity, nutrient supply, and hormones (1). Contractile activity is necessary for postnatal muscle growth and for the maintenance of muscle mass in adults, and increased work can cause fiber hypertrophy (2). Conversely, disuse or denervation causes rapid atrophy (3). Skeletal muscle also serves as the organism's major protein reservoir from which amino acids can be mobilized for gluconeogenesis, new protein synthesis, or as an energy store (4). Consequently, during food deprivation and in many systemic disease states, including sepsis, cancer, burn injury, diabetes, and cardiac and renal failure, there is a generalized muscle wasting, which results primarily from increased breakdown of muscle proteins, although protein synthesis also falls in most of these conditions (5). In all of these systemic catabolic states, the loss of muscle mass involves a common pattern of transcriptional changes, including induction of genes for protein degradation and decreased expression of various genes for growth-related and energy-yielding processes (5–7). We have termed this group of coordinately regulated genes “atrogenes.” Recent work indicates that the same transcriptional program occurs during atrophy induced by denervation and disuse as occurs in these catabolic states (8).
In all of these types of atrophying muscle, the ubiquitin–proteasome system is activated, and it catalyzes the degradation of the bulk of muscle proteins, especially myofibrillar components (5). In addition, there is a dramatic (8- to 40-fold) induction of two muscle-specific ubiquitin ligases, atrogin-1/MAFbx and MuRF-1, whose induction occurs before the onset of muscle weight loss (7, 9) and which is necessary for rapid atrophy (10). On the other hand, the expression of these ubiquitin ligases and the enhancement of overall protein breakdown are blocked by the IGF-1/insulin/phosphatidylinositol 3-kinase (PI3K)/AKT signaling pathway (11, 12), which also activates protein synthesis and net growth of these muscles. The key mediators of this catabolic response during atrophy are the FoxO family of transcription factors, whose activity is suppressed during growth by phosphorylation by AKT (12, 13) but whose expression and dephosphorylation rises in these catabolic states (7, 13). Activation of FoxO3 promotes the expression of atrogin-1 and other atrogenes, leading to a dramatic loss of muscle mass (13). On the other hand, when FoxO3 function is blocked, atrogin-1 expression and the muscle atrophy induced by fasting or glucocorticoids are prevented (13).
The mechanisms by which contractile activity preserves muscle mass, even in the face of catabolic signals (14–17), have long been a mystery. Muscle wasting does not occur similarly in all types of muscle fiber. During fasting (18) or exposure to glucocorticoids (19, 20), sepsis (21), and cancer cachexia (22, 23), type II glycolytic muscle fibers show greater atrophy than the type I oxidative fibers. On the other hand, during unloading or denervation, the fatigue-resistant, slow-contracting, dark muscles show more pronounced atrophy than fast-contracting, glycolytic, pale ones (24). These differences appear to be caused by greater activity of neurons innervating the dark muscles (19). In fact, increased activity protects pale muscles from glucocorticoid-induced atrophy, and disuse sensitizes dark muscles to this catabolic hormone (19, 25).
The present studies were undertaken to clarify how exercise can retard muscle wasting and why fiber types differ in their susceptibility to atrophy. One attractive possibility is that the effects of activity on muscle size are linked to the peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1 (PGC-1) family of coactivators (26–28), which are the prime regulators of mitochondrial content and oxidative metabolism, and they are critical in the maintenance of glucose, lipid, and energy homeostasis in muscle and other tissues. PGC-1α was identified as a PPARγ-interacting protein (29), and two other family members, PGC-related coactivator (PRC) and PGC-1β have also been identified (for review, see ref. 26). In skeletal muscle, PGC-1α functions as a critical metabolic sensor of motor neuron-induced calcium signaling (30–32). Its expression is induced by both short-term and chronic exercise in rodents and humans (33, 34). Transgenic expression of PGC-1α in fast-twitch, glycolytic muscles promotes mitochondrial biogenesis and oxidative metabolism (35), and importantly, it transforms the type IIb muscle fibers into a more oxidative phenotype. PGC-1α therefore appears to be an important mediator of exercise and motor-nerve activity in skeletal muscle.
In this work, we demonstrate that PGC-1α is an important factor opposing the effects of FoxO3 on muscle mass. Elevated levels of PGC-1α through transgenic expression reduce muscle atrophy during denervation or fasting and the atrophy-promoting effects of FoxO3 apparently by suppressing the associated changes in transcription of key atrogenes. Moreover, we show that PGC-1α expression falls dramatically after denervation and in various other types of muscle wasting, which enhance the FoxO-induced loss of muscle mass.
Results
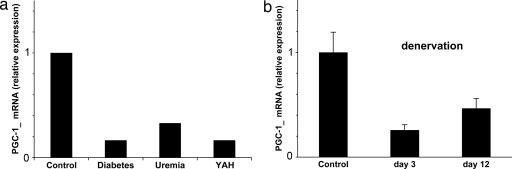
To investigate whether a decrease in PGC-1α activity might be important in the transcriptional changes during muscle wasting, we initially examined how PGC-lα levels might vary in different experimentally induced rat models of atrophy that mimic major human diseases (7). The level of PGC-1α mRNA, as measured by real-time PCR, fell dramatically in gastrocnemius muscles undergoing rapid atrophy caused by untreated streptozotocin-induced diabetes (36), cancer cachexia induced with Yoshida ascites hepatoma (22), and uremia induced by subtotal nephrectomy (37) (Fig. 1a). At the times studied, muscle protein degradation is markedly accelerated in all of these conditions. Similarly, the loss of neural activity after section of the sciatic nerve also caused a sharp fall in PGC-1α expression (Fig. 1b). This marked reduction in PGC-1α mRNA occurred early, within the 1st day after the operation, and it was maximal at 3 days (data not shown). In related studies in rat, we observed a similar rapid fall in PGC-1α mRNA after denervation or pure disuse of the gastrocnemius muscle induced by spinal isolation (8). Thus, its fall preceded the large decrease in muscle mass, and it persisted for at least 2 weeks (Fig. 1b) (8), after which time atrophy continues at a slower rate (8).
Fig. 1.
Inhibition of PGC-1α expression in various types of muscle atrophy. (a) Rat models of muscle atrophy: acute streptozotocin-induced diabetes mellitus, chronic renal failure induced by subtotal nephrectomy, and cancer cachexia induced by Yoshida ascities hepatoma. Animal models were described in depth elsewhere (22, 36, 37). Samples were taken at times when the muscles were undergoing rapid weight loss. PGC-1α was assayed by quantitative PCR. (b) Denervation by unilateral transection of the sciatic nerve in mouse. Results are expressed relative to mRNA levels in the contralateral enervated muscle.
Transgenic Expression of PGC-1α Protects Muscles Against Atrophy.
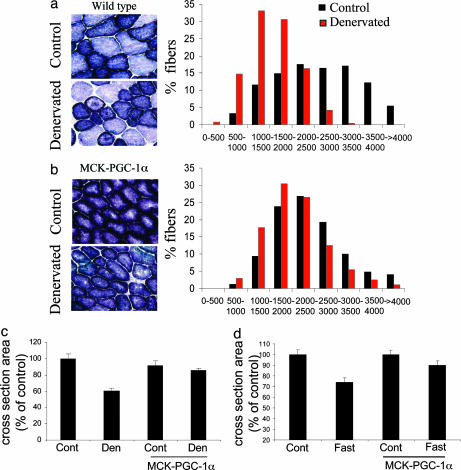
This rapid suppression of PGC-1α expression in many types of atrophy suggests that the decrease in this coactivator might contribute to the loss of muscle mass. To determine whether PGC-1α affects atrophy, we used our strain of transgenic mice with muscle-specific PGC-1α expression (35) under the control of muscle creatine kinase (MCK) promoter. The MCK promoter is particularly active in glycolytic, type IIb muscle fibers (e.g., the tibialis anterior). Thus, in the transgenic animals, the level of PGC-1α expression in these fibers resembles the high level typically seen in the soleus, which contains primarily oxidative type IIa and type I fibers. Fibers from these transgenic animals have a more oxidative phenotype, as shown by greater content of succinic dehydrogenase and a shift from type IIb to type IIa and I fibers (Fig. 2b) (35). Section of the sciatic nerve was performed in the transgenic and control mice. By 12 days, the denervated tibialis anterior in wild-type animals showed >40% reduction in mean fiber diameter. This decrease was mainly evident in large glycolytic type IIb fibers (Fig. 2 a and c). By contrast, in the transgenic animals, although the fibers were generally smaller, denervation of the tibialis anterior caused much less atrophy. Only a 10% decrease in cross-sectional area and a minor shift in fiber size distribution were seen (Fig. 2 b and c).
Fig. 2.
PGC-1α-transgenic mice are protected from denervation- and fasting-induced muscle atrophy. (a) Fiber size in control and denervated wild-type mice. (Left) Succinate dehydrogenase staining of mock-transected (Upper) and denervated (Lower) tibialis anterior muscle. (Right) Fiber size distribution of tibialis anterior muscles. Red bars, denervated; black bars, mock-transected. (b) As in a, using MCK–PGC-1α-transgenic mice. (c) Mean cross-sectional area of denervated and control wild-type and MCK–PGC-1α transgenic tibialis anterior muscles. (d) Mean cross-sectional area of tibialis anterior muscles from food-deprived and fed wild-type and MCK–PGC-1α transgenic animals.
To determine whether PGC-1α can also protect against other types of muscle wasting, we studied the susceptibility of muscles from these transgenic mice to fasting-induced atrophy, which is signaled by a fall in insulin and by glucocorticoids (38, 39) rather than by inactivity. By 48 h after food was removed from the cages, the tibialis muscles from wild-type animals showed a 25% reduction in mean fiber size, in accord with prior findings (40). However, the muscles of PGC-1α-transgenic mice were relatively resistant to atrophy, losing only 10% of their cross-sectional area (Fig. 2d). Together, these experiments indicate that elevated levels of PGC-1α protect muscles against various types of atrophy, including that induced by disuse of a specific muscle or by endocrine changes; thus, the fall in PGC-1α in the atrophying muscles (Fig. 1) is probably a key factor contributing to the loss of muscle mass.
PGC-1α Alters Expression of Key Atrophy-Specific Genes.
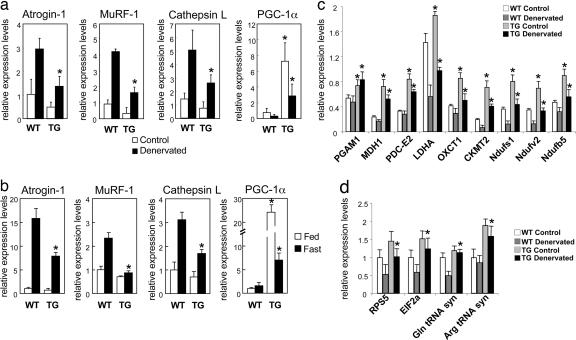
We have previously shown that various types of muscle atrophy occur through a common program of transcriptional changes (7, 8). Because of the capacity of PGC-1α to protect against atrophy, we investigated whether transgenic expression of PGC-1α blocked the induction or action of genes critical in the atrophy process. The expression of three atrogenes involved in protein breakdown, the ubiquitin ligases MuRF-1 and atrogin-1 and the lysosomal hydrolase cathepsin L are dramatically induced in atrophying muscles (7). As shown by real-time PCR (Fig. 3a), mRNAs for each were strongly induced after denervation in wild-type control mice, as described previously (8). However, the increases in mRNA for atrogin-1, MuRF-1, and cathepsin-L after denervation were blunted by 40% in muscles expressing PGC-1α transgenically. In addition, the contralateral innervated muscles of the transgenic animals showed lower expression of these three genes. Interestingly, the activity of the MCK promoter also appears to be modulated by denervation because in the transgenic mice PGC-1α mRNA levels in the denervated muscles were lower than in the innervated controls.
Fig. 3.
PGC-1α reduces transcription of key atrogenes involved in protein degradation. (a) Expression of ubiquitin-ligases atrogin-1, MuRF-1, and lysosomal hydrolase cathepsin L after denervation for 12 days, analyzed by real-time PCR. (b) Expression of ubiquitin-ligases atrogin-1, MuRF-1, and lysosomal hydrolase cathepsin L, after food deprivation for 2 days, analyzed by real-time PCR. (c) Expression of genes involved in energy metabolism after 12 days of denervation. ∗, P < 0.05 between control and transgenic mice, by Student's t test.
Similarly, when the expression of these key atrogenes was monitored during muscle wasting induced by fasting, the marked induction of atrogin-1 after food deprivation was much smaller in the transgenic animals, and no induction of MuRF-1 was evident at all (Fig. 3b). Because the induction of these genes appears essential for rapid atrophy (7, 10), the ability of PGC-1α to reduce muscle atrophy likely occurs in part by suppressing the transcriptional program activated in these muscles. In addition to the content of the key ubiquitin ligases atrogin-1 and MuRF-1, the major lysosomal protease cathepsin L, which also contributes to the loss of muscle protein (ref. 41; J. Zhao, et al., unpublished work; C. Mammacuri, unpublished work) is regulated similarly.
Expression of a variety of genes for enzymes important in glycolysis and oxidative phosphorylation is suppressed coordinately in many forms of muscle wasting. These enzymes include lactic dehyrodrogenase, malate dehydrogenase, the pyruvate dehydrogenase E2 component, oxoacid CoA transferase, mitochondrial creatine kinase II, NADH dehydrogenase (ubiquinone) FeS protein 1, NADH dehydrogenase (ubiquinaone) flavoprotein 2, and NADH dehydrogenase (ubiquinone) 1β subcomplex 5 (7, 8). In microarrays from skeletal muscles of mice lacking PGC-1α, we found that 53% (nine) of these atrogenes were also suppressed compared with wild-type animals (data not shown). We also measured the expression of these genes by real-time PCR in the denervated and contralateral innervated PGC-1α-transgenic animals (Fig. 3c). In each case, the mice producing PGC-1α transgenically had higher levels of these mRNAs. Their expression fell after denervation, but it still remained significantly higher than in controls. Together, these findings strongly suggest that the suppression of PGC-1α expression may underlie many of the transcriptional changes found in atrophying muscle, especially the decreased transcription of genes for ATP production.
Interestingly, the expression of several genes that may promote protein synthesis at the translational level, e.g., initiation factor α2 (E1Fα2), the ribosomal protein S5, and both arginine and glutamine tRNA synthase, also decreased during denervation atrophy, but they were increased in the PGC-1α-transgenic mouse (Fig. 3d). Although overall rates of protein synthesis are not significantly altered after denervation (42) or by PGC-1α expression in cultured myotubes (data not shown), these observations are further evidence that the fall in PGC-1α production helps determine the pattern of transcriptional changes seen during atrophy.
PGC-1α Reduces FoxO3-Dependent Transcription of Atrogin-1 and Muscle Atrophy.
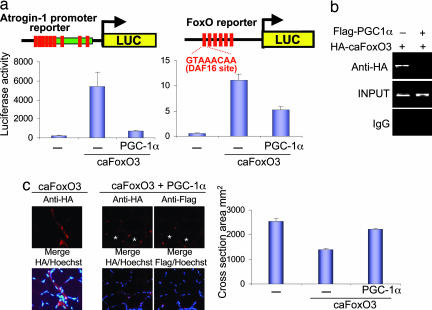
Because mRNAs for key atrogenes including atrogin-1 were suppressed in the PGC-1α-transgenic animals, we examined in greater depth the effect of PGC-1α on the transcription of the atrogin-1 gene. In fasting and various catabolic states, where there is decreased insulin/IGF-1 or insulin resistance, the forkhead family of transcription factors, including FoxO3, become dephosphorylated, enter the nucleus, and activate transcription (12, 13, 43). FoxO3 activation alone is sufficient to cause dramatic muscle wasting in vivo (13). To investigate whether PGC-1α might affect FoxO3-dependent transcription, we analyzed the effect of PGC-1α on the atrogin-1 promoter in adult mouse muscles. Tibialis anterior muscle fibers were electroporated with an atrogin-1 promoter–reporter construct (13) with or without constructs expressing PGC-1α and a constitutively active form of FoxO3, c.a.FoxO3A. This mutant FoxO3 cannot be phosphorylated/inactivated by AKT because it carries mutations in the three AKT phosphorylation sites (T32A, S253A, and S315A). As shown in Fig. 4a and as reported previously (13), c.a.FoxO3 caused a dramatic (30- to 50-fold) induction of the atrogin-1 promoter, but coexpression of PGC-1α in these cells markedly suppressed this induction by FoxO3. Further strong evidence of the ability of PGC-1α to suppress FoxO3-dependent transcription was obtained in similar experiments using a synthetic FoxO3 reporter containing six concatemerized DAF16-binding sites (Fig. 4a). The ability of c.a.FoxO3 to bind to the atrogin-1 promoter was tested by chromatin immunoprecipitation (ChIP) assays in nuclei isolated from mouse skeletal muscle. As shown in Fig. 4b, c.a.FoxO3 electroporated into mouse muscle was found in complex with the atrogin-1 promoter. However, PGC-1α strongly suppressed c.a.FoxO3 binding at this site (Fig. 4b).
Fig. 4.
PGC-1α suppresses FoxO3 action. (a Left) Plasmids bearing a luciferase reporter driven by the atrogin-1 promoter, c.a.FoxO3A, and PGC-1α were electroporated into the intact tibialis anterior muscles of adult mice, and luciferase activity was measured in extracts from the muscles 1 week later. (a Right) As in a Left, but using a canonical FoxO sequence (DAF16) driving the luciferase gene. (b) c.a.FoxO3 binding to a FoxO response element in the atrogin-1 promoter is blocked by PGC-1α. Muscles were collected 8 days after c.a.FoxO3 transfection with or without FLAG-PGC-1α, and ChIP assays were performed as described. (c) Constitutively active HA-tagged FoxO3A with or without FLAG-PGC-1α was electroporated into the intact tibialis anterior muscles of adult mice. Muscles were harvested after 8 days, serially sectioned, and subjected to immunohistochemistry (Left) or fiber size quantification (Right).
As found in the reporter studies (Fig. 4a) and with ChIP (Fig. 4b), electroporation of PGC-1α together with c.a.FoxO3 caused a marked reduction in FoxO-dependent transcription.
Because of this ability to block the transcription by FoxO3, we analyzed whether PGC-1α expression could also inhibit the rapid loss of muscle mass induced by FoxO3. Adult tibialis anterior muscles were electroporated with c.a.FoxO3 with or without PGC-1α. The muscle fibers overexpressing c.a.FoxO3 underwent marked atrophy, losing >40% of their cross-section area in 8 days, in accord with prior findings (13). In contrast, the coexpression of PGC-1α with FoxO3 blocked this response, and it maintained the fiber diameter close to that in the surrounding untransfected fibers (Fig. 4c). These results together demonstrate that PGC-1α protects against muscle atrophy at least in part by suppressing FoxO-dependent transcription of critical atrophy-related genes.
Discussion
It is now clear that a specific program of transcriptional changes underlies the rapid atrophy of muscle seen in a wide variety of pathological states, ranging from denervation or disuse of a specific muscle to fasting and systemic diseases (5, 7). Based on the findings presented here, it is clear that the PGC-1α coactivator is an important determinant of this transcriptional program and the extent of fiber atrophy. First, the level of PGC-1α was shown to decrease sharply in muscles early during atrophy induced by section of the motor neuron in mice, as well as by diabetes, renal failure, and cancer cachexia in rats. This fall in PGC-1α expression with disuse is the mirror image of the increased expression after exercise (32–34), believed to be signaled by Ca2+ influx and calcineurin (30, 31). However, the mechanism for the surprising fall in PGC-1α in diverse catabolic states is unclear, and it represents an important question for study. The systemic muscle wasting in these conditions appears to be triggered by insulin resistance and/or insulin deficiency (44), glucocorticoids (45), and/or various monokines (46). The marked reduction in PGC-1α mRNA in atrophying muscles occurs where FoxO factors are activated (13) and expressed at high rates (7). In related experiments, we observed a large fall in PGC-1α mRNA in rat muscle by 1 day after nerve section or complete inactivation with neurons intact (8). In these different situations, the decrease in PGC-1α expression occurs early, and it precedes the rise in protein degradation and marked weight loss; thus it is sufficiently rapid to help signal the atrophy process.
Importantly, we have found that when the levels of PGC-1α are maintained, either by use of transgenic expression or by electroporation of this cDNA into adult muscle fibers, muscles are protected to a large extent from the atrophy induced by denervation, fasting, or expression of FoxO3. This protective effect can explain how exercise, by inducing expression of PGC-1α, can maintain muscle mass and retard atrophy, even in the face of circulating catabolic factors (14). This role for PGC-1α in influencing fiber size and blocking atrophy complements its well established role in determining fiber type (35), mitochondrial content and oxidative capacity (47–49), and other exercise-induced adaptations (32). PGC-1α functions as a key sensor of muscular activity; and as shown here, its fall appears to play a key role in allowing the large induction of the critical ubiquitin ligases, atrogin-1 and MuRF-1, as well as another major atrogene, cathepsin L, the lysosomal hydrolase that presumably is important in the enhanced lysosomal proteolysis also seen in atrophying muscle (41; Zhao et al., in preparation) (Fig. 5).
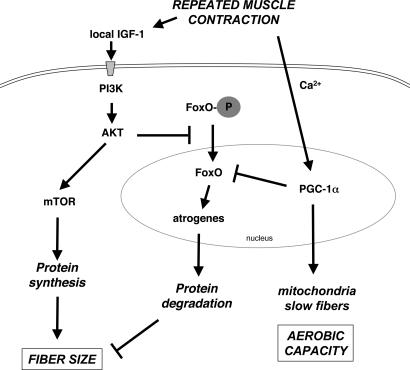
Fig. 5.
Mechanisms for inhibition of atrophy and growth promotion by muscle activity. With repeated muscle contractions, IGF-1 production by the muscle increases, which stimulates protein synthesis and fiber hypertrophy through activation of PI3K and AKT kinases. AKT also causes phosphorylation and nuclear exclusion of FoxO 1, 3, and 4, which suppresses atrogene expression and proteolysis. In addition, PGC-1α is induced, leading to increased production of mitochondria and a shift to slow-twitch, oxidative fibers. PGC-1α also inhibits transcriptional activity of FoxO3, which suppresses atrogene expression and protein degradation.
The major signal that regulates postnatal growth of muscle is the IGF-1/PI3K/AKT pathway, which promotes protein synthesis by activating translation generally, but it also retards protein degradation and the expression of various atrogenes (12, 13, 50) (Fig. 5). Overproduction of active AKT prevents atrophy especially through its ability to block the activation of FoxO1, 3, and 4 (2, 13). Unlike IGF-1/AKT, PGC-1α, if overproduced, does not lead to a significant increase in muscle mass in transgenic mice (35); and when transfected into cultured muscle cells, it does not stimulate overall protein synthesis (W.Y., unpublished observations). Nevertheless, PGC-1α clearly inhibits the atrophy induced by denervation or fasting, both of which involve FoxO-dependent transcription. The inhibition of the profound atrophy and atrogene expression induced by electroporation of c.a.FoxO3 is noteworthy because this effect cannot be mediated by changes in AKT, and this action seems sufficient to account for the ability of PGC-1α to retard fiber atrophy. Furthermore, our findings with the promoter of atrogin-1 using ChIP and luciferase reporters indicate that PGC-1α somehow inhibits FoxO-dependent transcription. By contrast, in liver, PGC-1α positively interacts with FoxO1, important in the stimulation of transcription of genes for gluconeogenesis (51). Because PGC-1α is a well established coactivator of transcription, we cannot eliminate the possibility that its inhibitory action on the atrophy process is indirect. For example, PGC-1α may cause expression of an inhibitor of FoxO, or the increased mitochondrial content or enhanced β-oxidative metabolism induced by PGC-1α may indirectly result in protection from atrophy.
The present findings can explain a number of well established observations concerning the interactions of hormones and activity in the control of muscle size. For example, it has long been known that the pale glycolytic fibers found typically in fast- twitch muscles atrophy selectively in response to sepsis (21) or tumors (22, 23), fasting (18), or high levels of glucocorticoids (52). Presumably, their differential susceptibility ensures that in fasting and other stressful states, mobilization of amino acids from muscle protein occurs primarily from easily spared, less frequently used fibers (14). The greater sensitivity of the type IIb fibers can be explained by their lower content of PGC-1α. Accordingly, increased exercise can protect such fibers from glucocorticoids, and it can even induce muscle hypertrophy in fasted animals (14). By contrast, the dark type I fibers high in PGC-1α content become sensitive to glucocorticoid-induced atrophy, and they show the greatest atrophy after denervation, when PGC-1α expression falls (24). Thus, together with the exercise-induced production of IGF-1, the exercise-induced changes in PGC-1α in muscle seem to be critical factors by which contractile activity determines muscle size as well as its enzymatic composition (Fig. 5).
Methods
Animals and in Vivo Transfection Experiments.
Mice were starved for 48 h, as indicated. Disuse atrophy was induced by cutting the sciatic nerve. After 12 days, the mice were killed. The muscles were collected, serial sectioned, and stained for succinate dehydrogenase. In vivo transfection experiments were performed by intramuscular injection of plasmid DNA in tibialis anterior muscle followed by electroporation as described in ref. 13.
Immunohistochemistry and Fiber Size Measurements.
Mouse muscle fibers expressing HA-tagged or FLAG-tagged proteins were stained in cryocross-sections fixed with 4% paraformaldehyde. Immunohistochemistry with anti-HA polyclonal antibody (Santa Cruz, Santa Cruz, CA) and anti-FLAG polyclonal antibody (Sigma, St. Louis, MO) was as described in ref. 13. Muscle fiber size was measured in transfected fibers and in an equal number of untransfected fibers from the same muscle as described in ref. 13. All data are expressed as the mean ± SEM. Comparisons were made by using Student's t test, with P < 0.05 being considered statistically significant.
ChIP Assay and Promoter Analysis.
ChIP assay was performed by using a kit (Upstate Biotechnology, Lake Placid, NY) according to the manufacturer's instructions. Anti-HA antiserum (Santa Cruz), or an equal amount of IgG, was used. The following primers were used for PCR: atrogin-1 (−1740 to −1537), forward 5′-CTGGCAGGGAGGAGCCTAATGAATC-3′, reverse 5′-GGGAGTGGCAAAGCCGTCTC-3′. The 3.5-kb atrogin-1 and the daf-16 family protein-binding element reporters have been described previously (13). Luciferase assays were performed by standard procedures in muscles removed 8 days after transfection. The Renilla luciferase vector (pRL-TK) was used to normalize for transfection efficiency.
Acknowledgments
This work was supported by National Institutes of Health Grants 2R56DK054477 (to B.M.S.) and R01DK6230701 (to S.H.L.); grants from the Muscular Dystrophy Association (to C.H. and A.L.G.); the National Space Biomedical Research Institute and the Ellison Foundation (A.L.G.); and Telethon Grant S04009, Association Française contre les Myopathies Grant 1026, and a Compagnia San Paolo grant (to M.S.).
Abbreviations
- c.a.FoxO3
constitutively active FoxO3
- MCK
muscle creatine kinase
- PGC-1α
peroxisome proliferator-activated receptor γcoactivator 1α
- PI3K
phosphatidylinositol 3-kinase.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bassel-Duby R, Olson EN. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 2.Glass DJ. Nat Cell Biol. 2003;5:87–90. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- 3.Jackman RW, Kandarian SC. Am J Physiol. 2004;287:C834–C843. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- 4.Lecker SH, Solomon V, Mitch WE, Goldberg AL. J Nutr. 1999;129:227S–237S. doi: 10.1093/jn/129.1.227S. [DOI] [PubMed] [Google Scholar]
- 5.Lecker SH, Goldberg AL, Mitch WE. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 6.Jagoe RT, Lecker SH, Gomes M, Goldberg AL. FASEB J. 2002;16:1697–1712. doi: 10.1096/fj.02-0312com. [DOI] [PubMed] [Google Scholar]
- 7.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 8.Sacheck JM, Hyatt J-P, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. FASEB J. 2006 doi: 10.1096/fj.06-6604com. in press. [DOI] [PubMed] [Google Scholar]
- 9.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Proc Natl Acad Sci USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, et al. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 11.Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. Am J Physiol. 2004;287:E591–E5601. doi: 10.1152/ajpendo.00073.2004. [DOI] [PubMed] [Google Scholar]
- 12.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 13.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg AL. Endocrinology. 1968;83:1071–1073. doi: 10.1210/endo-83-5-1071. [DOI] [PubMed] [Google Scholar]
- 15.Zinna EM, Yarasheski KE. Curr Opin Clin Nutr Metab Care. 2003;6:87–93. doi: 10.1097/00075197-200301000-00013. [DOI] [PubMed] [Google Scholar]
- 16.Alkner BA, Tesch PA. Acta Physiol Scand. 2004;181:345–357. doi: 10.1111/j.1365-201X.2004.01293.x. [DOI] [PubMed] [Google Scholar]
- 17.Fluckey JD, Dupont-Versteegden EE, Montague DC, Knox M, Tesch P, Peterson CA, Gaddy-Kurten D. Acta Physiol Scand. 2002;176:293–300. doi: 10.1046/j.1365-201X.2002.01040.x. [DOI] [PubMed] [Google Scholar]
- 18.Li JB, Goldberg AL. Am J Physiol. 1976;231:441–448. doi: 10.1152/ajplegacy.1976.231.2.441. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg AL, Goodman HM. J Physiol (London) 1969;200:655–666. doi: 10.1113/jphysiol.1969.sp008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlmann B, Rutschmann M, Reinauer H. Biochem J. 1986;234:659–664. doi: 10.1042/bj2340659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiao G, Lieberman M, Fischer JE, Hasselgren PO. Am J Physiol. 1997;272:R849–R856. doi: 10.1152/ajpregu.1997.272.3.R849. [DOI] [PubMed] [Google Scholar]
- 22.Baracos VE, DeVivo C, Hoyle DH, Goldberg AL. Am J Physiol. 1995;268:E996–E1006. doi: 10.1152/ajpendo.1995.268.5.E996. [DOI] [PubMed] [Google Scholar]
- 23.Acharyya S, Ladner KJ, Nelsen LL, Damrauer J, Reiser PJ, Swoap S, Guttridge DC. J Clin Invest. 2004;114:370–378. doi: 10.1172/JCI20174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbison GJ, Jaweed MM, Ditunno JF. Arch Phys Med Rehab. 1979;60:401–404. [PubMed] [Google Scholar]
- 25.Falduto MT, Czerwinski SM, Hickson RC. J Appl Physiol. 1990;69:1058–1062. doi: 10.1152/jappl.1990.69.3.1058. [DOI] [PubMed] [Google Scholar]
- 26.Lin J, Handschin C, Spiegelman BM. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Finck BN, Kelly DP. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soyal S, Krempler F, Oberkofler H, Patsch W. Diabetologia. 2006;49:1477–1488. doi: 10.1007/s00125-006-0268-6. [DOI] [PubMed] [Google Scholar]
- 29.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 30.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. Proc Natl Acad Sci USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Czubryt MP, McAnally J, Fishman GI, Olson EN. Proc Natl Acad Sci USA. 2003;100:1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor EB, Lamb JD, Hurst RW, Chesser DG, Ellingson WJ, Greenwood LJ, Porter BB, Herway ST, Winder WW. Am J Physiol. 2005;289:E960–E968. doi: 10.1152/ajpendo.00237.2005. [DOI] [PubMed] [Google Scholar]
- 33.Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. FASEB J. 2002;16:1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 34.Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, Meier CA, Bell DR, Kralli A, Giacobino JP, Deriaz O. Diabetes. 2003;52:2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- 35.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 36.Price SR, Bailey JL, Wang X, Jurkovitz C, England BK, Ding X, Phillips LS, Mitch WE. J Clin Invest. 1996;98:1703–1708. doi: 10.1172/JCI118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey JL, Wang X, England BK, Price SR, Ding X, Mitch WE. J Clin Invest. 1996;97:1447–1453. doi: 10.1172/JCI118566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kettelhut IC, Pepato MT, Migliorini RH, Medina R, Goldberg AL. Braz J Med Biol Res. 1994;27:981–993. [PubMed] [Google Scholar]
- 39.Wing SS, Goldberg AL. Am J Physiol. 1993;264:E668–E676. doi: 10.1152/ajpendo.1993.264.4.E668. [DOI] [PubMed] [Google Scholar]
- 40.Medina R, Wing SS, Goldberg AL. Biochem J. 1995;307:631–637. doi: 10.1042/bj3070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Attaix D, Mosoni L, Dardevet D, Combaret L, Mirand PP, Grizard J. Int J Biochem Cell Biol. 2005;37:1962–1973. doi: 10.1016/j.biocel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 42.Furuno K, Goodman MN, Goldberg AL. J Biol Chem. 1990;265:8550–8557. [PubMed] [Google Scholar]
- 43.Lee SW, Dai G, Hu Z, Wang X, Du J, Mitch WE. J Am Soc Nephrol. 2004;15:1537–1545. doi: 10.1097/01.asn.0000127211.86206.e1. [DOI] [PubMed] [Google Scholar]
- 44.Glass DJ. Int J Biochem Cell Biol. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 45.Hasselgren PO. Curr Opin Clin Nutr Metab Care. 1999;2:201–205. doi: 10.1097/00075197-199905000-00002. [DOI] [PubMed] [Google Scholar]
- 46.Reid MB, Li YP. Respir Res. 2001;2:269–272. doi: 10.1186/rr67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 48.Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, et al. Proc Natl Acad Sci USA. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.St Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 50.Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, Lin HC, Yancopoulos GD, Glass DJ. J Biol Chem. 2005;280:2737–2744. doi: 10.1074/jbc.M407517200. [DOI] [PubMed] [Google Scholar]
- 51.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 52.Goldberg AL. J Biol Chem. 1969;244:3223–3229. [PubMed] [Google Scholar]