Abstract
p53 is essential for the cellular responses to DNA damage that help to maintain genomic stability. Protective p53-dependent cell-cycle checkpoints are activated in response to a wide variety of stresses, including not only DNA damage but also arrest of DNA synthesis and of mitosis. In addition to its role in activating the G1 and G2 checkpoints, p53 also helps to protect cells in S phase when they are starved for DNA precursors by treatment with the specific aspartate transcarbamylase inhibitor N-phosphonacetyl-l-aspartate (PALA), which blocks the synthesis of pyrimidine nucleotides. Even though p53 is activated, PALA-treated cells expressing low levels of p53 or lacking expression of p21 do not arrest in G1 or G2 but are blocked in S phase instead. In the complete absence of p53, PALA-treated cells continue to synthesize DNA slowly and eventually progress through S phase, suffering severe DNA damage that in turn triggers apoptosis. Expression of the secreted protein macrophage inhibitory cytokine 1 (MIC-1), a member of the TGF-β superfamily, increases substantially after PALA treatment, and application of exogenous MIC-1 or its constitutive expression from a cDNA provides remarkable protection of p53-null cells from PALA-mediated apoptosis, arguing that the p53-dependent secretion of MIC-1 provides a major part of such protection. Stimulation of MIC-1-dependent S phase arrest in normal gut epithelial cells might help to revitalize the clinical use of PALA, which has been limited by gut toxicity.
Keywords: apoptosis, cell-cycle arrest, TGF-β
In about half of all human tumors, p53 is inactive or mutant, and its normal negative regulatory functions in tumors with wild-type p53 are likely to be compromised by other mechanisms. p53 is essential for the normal responses to DNA damage that help to maintain genomic stability. Extensive DNA damage leads to the irreversible arrest or apoptosis of normal cells, preventing cells in which mutations are likely to have occurred from propagating. In contrast, arrest of DNA or RNA synthesis or depletion of pyrimidine nucleotide pools does not lead to DNA damage directly. In these cases, reversible arrest is appropriate to protect the stressed cells, rather than disposing of them to protect the organism. Cell-protective p53-dependent cell-cycle checkpoints are activated in response to arrest of DNA synthesis (1, 2) or of mitosis (3, 4). Fibroblasts from normal individuals or Li-Fraumeni patients arrest stably when treated with N-(phosphonacetyl)-l-aspartate (PALA), a potent and reversible inhibitor of pyrimidine nucleotide synthesis in cells (5), and these cells remain viable for several weeks (6). However, this protective arrest is lost in postcrisis Li-Fraumeni fibroblasts when the remaining wild-type p53 allele is lost. We showed previously that restoration of wild-type p53 to the p53-null Li-Fraumeni cell line MDAH041 restored a normal response to PALA (7). As shown by Yin et al. (6), Agarwal et al. (7), and Livingstone et al. (8), the ability of human or mouse cells to give rise to PALA-resistant colonies depends upon inactivation of the p53 response pathway. PALA-treated cells that have functional p53 arrest stably and reversibly and do not give rise to resistant colonies, whereas those that do not arrest stably give rise to rare PALA-resistant cells (8).
Normal cells do not incur double-strand breaks in their DNA upon treatment with PALA (1). Rodent and human fibroblasts with wild-type functional p53 arrest in the cell cycle, preventing them from synthesizing DNA under adverse conditions (9, 10). Previously, work from our laboratory has shown that a p53-dependent arrest within S phase is revealed in several cell lines that fail to arrest in G1 or G2 because of low levels of p53 or because they lack the expression of p21 (7, 11). In contrast, as shown here, cells completely lacking p53 continue DNA synthesis, thereby incurring DNA damage that leads to their death. The mode of death in these cells has not been studied previously nor is it known how p53 is activated under conditions of starvation for pyrimidine nucleotides or how activated p53 mediates reversible protective cell-cycle arrest. Furthermore, it is not known how cells other than fibroblasts behave in PALA, and it is especially important to investigate the responses of epithelial cells, which are involved in forming most tumors.
We now show that the secreted protein macrophage inhibitory cytokine 1 (MIC-1), whose expression is increased after PALA treatment in cells that express p53, mediates a protective S phase arrest of epithelial cells. MIC-1 inhibits tumor cell growth through a TGF-β-like signaling pathway (12). It is also known as prostate differentiation factor, placental TGF-β (PTGF-β), growth differentiation factor 15 (GDF-15), and nonsteroidal anti-inflammatory (NSAID) drug-activated gene-1. Sequence analysis indicates that MIC-1 is a member of a new subgroup within the TGF-β superfamily (13). Although it shares only 25% overall sequence identity with other superfamily members, MIC-1 has several major conserved elements, including a signal peptide, a consensus RXXRA/S cleavage signal for processing to the mature form, and seven conserved cysteine residues near the C terminus (14, 15). MIC-1 is synthesized as a 62-kDa intracellular precursor, cleaved by a furin-like protease, and secreted as a 25-kDa disulfide-linked dimmer (15, 16). We now provide evidence for a novel role for MIC-1 as a major mediator of p53-induced S phase cell-cycle arrest.
Results
p53 Protects Epithelial Cells from Death upon Treatment with PALA.
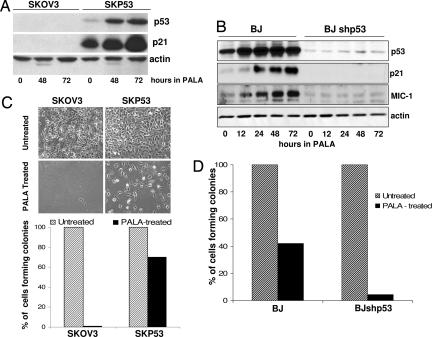
Previously, we established tetracycline-regulated expression of wild-type p53 in the p53-null human fibroblast cell line MDAH041 (16). Using a similar approach, we restored p53 expression in SKOV3 epithelial cells, derived from an ovarian carcinoma. Clone SKP53 expresses constitutively a level of p53 comparable to that of normal human fibroblasts, and p53 can be induced and activated by treatment with PALA (Fig. 1A and B) or adriamycin (data not shown). We also knocked down p53 expression in BJ cells by stable infection with p53 shRNA (BJshp53 cells). Treatment with PALA induced the expression of p53 and p21 in BJ and SKP53 but not BJshp53 or SKOV3 cells (Fig. 1 A and B), indicating that the p53 in SKP53 cells is functional, and that the low levels of p53 in BJshp53 cells cannot activate downstream p53 target genes. When BJ and BJshp53 cells, along with SKOV3 and SKP53 cells, were treated with 250 μM PALA (3 × ID50), the p53-null BJshp53 and SKOV3 cells began to die after 9 days, as observed previously for p53-null fibroblasts, and most were dead after 12 days in PALA. In contrast, the BJ and SKP53 cells remained attached, with no apparent death (Fig. 1 C and D). More than half of the PALA-treated BJ cells and 70% of the SKP53 cells were still able to form colonies (Fig. 1 C and D). Furthermore, 40% of the SKP53 cells left in PALA for 20 days still formed colonies (data not shown), showing that the protective arrest of growth can be maintained for quite a long time.
Fig. 1.
p53 protects PALA-treated cells. Cells were treated with 250 μM PALA for 12–72 h. (A and B) Expression of p53, p21, and MIC-1, analyzed by the Western blot method. (C and D) Colony formation for cells treated with 250 μM PALA for 12 days. Cells plated for colony formation were assayed after an additional 15 days.
Cells Expressing p53 Cease DNA Synthesis in the Presence of PALA, Whereas Cells Lacking p53 Continue to Cycle.
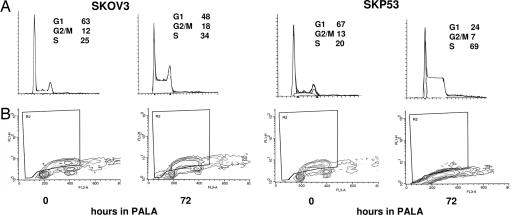
SKOV3 and SKP53 cells were left in 250 μM PALA for 24–96 h, stained with propidium iodide, and analyzed by FACS. Profiles of DNA content revealed an initial increase in S phase cells for both cell types (Fig. 2A and Table 1). However, the S phase population of SKP53 cells continued to increase over 96 h, reaching 86%. To investigate the status of DNA synthesis, SKP53 and SKOV3 cells were treated with 250 μM PALA for 72 h and then pulse-labeled with BrdU for 4 h (Fig. 2B). The S phase population of SKOV3 cells actively incorporated BrdU, whereas there was little if any incorporation in the SKP53 cells. These observations indicate that PALA-treated SKP53 cells arrest in S phase, do not synthesize DNA, and are protected, whereas upon treatment with PALA, SKOV3 cells actively synthesize DNA, traverse slowly through S phase, and die.
Fig. 2.
Cells expressing p53 stop making DNA in PALA, whereas those lacking p53 continue to cycle. (A) FACS analysis of SKOV3 and SKP53 cells treated with 250 μM PALA for 72 h. The cells were stained with propidium iodide to measure DNA content. Percentages of cells in G1, G2/M, and S are shown in each image. (B) Incorporation of BrdU. Untreated and treated SKOV3 and SKP53 cells were labeled with BrdU for 4 h, fixed in methanol, stained with anti-BrdU, and measured by FACS.
Table 1.
Cell-cycle profiles of PALA-treated SKOV3, SKP53, and SKMIC-1 cells
| Percentage of cells in |
|||
|---|---|---|---|
| Hours of treatment | G1 | G2/M | S |
| SKOV3 | |||
| 0 | 63 | 12 | 25 |
| 24 | 48 | 15 | 47 |
| 48 | 55 | 13 | 32 |
| 72 | 48 | 18 | 34 |
| 96 | 43 | 20 | 37 |
| SKP53 | |||
| 0 | 67 | 13 | 20 |
| 24 | 47 | 8 | 45 |
| 48 | 36 | 18 | 46 |
| 72 | 24 | 7 | 69 |
| 96 | 11 | 3 | 86 |
| SKMIC-1 | |||
| 0 | 57 | 11 | 32 |
| 24 | 54 | 5 | 41 |
| 48 | 22 | 4 | 74 |
| 72 | 30 | 10 | 60 |
| 96 | 38 | 7 | 55 |
Data are shown from a FACS analysis of cells treated with 250 μM PALA for 24–96 h and stained with propidium iodide to measure DNA content.
Previously, we reported that the p53-dependent protective S phase arrest of fibroblasts in response to PALA was independent of p21 (1, 7). To explore the role of p21 in the p53-dependent S phase arrest of epithelial cells, we used human colon cancer HCT116 cells, which have wild-type p53, and derivatives of HCT116 cells that lack either p53 or p21 (7, 17). HCT116 cells expressing p53 accumulated in G1 over a 96-h period when treated with PALA, whereas cells lacking p53 continued to cycle and died. However, cells lacking p21 accumulated much more in S phase (Table 2), and many cells survived for up to 20 days in PALA (data not shown). These observations, together with our previous data (7), indicate that S phase arrest in response to PALA is independent of p21 in cells of both fibroblast and epithelial origin.
Table 2.
Cell-cycle profiles of PALA-treated HCT116 cells and p53- and p21-null derivatives
| Percentage of cells in |
|||
|---|---|---|---|
| Hours of treatment | G1 | G2/M | S |
| SKOV3 | |||
| 0 | 50 | 24 | 26 |
| 24 | 48 | 5 | 47 |
| 48 | 77 | 12 | 11 |
| 72 | 76 | 12 | 12 |
| 96 | 87 | 6 | 7 |
| p53-null | |||
| 0 | 37 | 19 | 44 |
| 24 | 40 | 8 | 52 |
| 48 | 23 | 20 | 57 |
| 72 | 37 | 19 | 44 |
| 96 | 45 | 20 | 35 |
| p21-null | |||
| 0 | 37 | 21 | 42 |
| 24 | 24 | 17 | 59 |
| 48 | 23 | 13 | 64 |
| 72 | 18 | 11 | 71 |
| 96 | 21 | 13 | 66 |
Data are shown from a FACS analysis of cells treated with 250 μM PALA for 24–96 h and stained with propidium iodide to measure DNA content.
PALA-Treated Cells Lacking p53 Undergo Apoptosis.
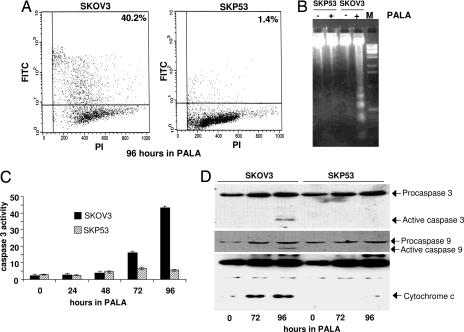
Cell-cycle profiles revealed that SKOV3 but not SKP53 cells have a DNA content of <2 N, suggesting DNA breakdown and cell death, after 96 h or more in PALA (data not shown). To investigate the mode of cell death, several different assays were used (18). When treated with 250 μM PALA, cells lacking p53 readily incorporated acridine orange, an indicator of apoptosis, whereas cells expressing p53 did not (data not shown). After treatment with PALA, p53-null cells exhibited distinct DNA laddering and stained positively in the TUNEL assay, whereas cells expressing wild-type p53 did not (Fig. 3A and B). To investigate whether p53-null cells undergoing apoptosis activate caspases and whether p53 protects cells by blocking this signal, we measured the activity of caspase 3, a crucial component in apoptosis induced by diverse stimuli, in SKOV3 and SKP53 cells treated with 250 μM PALA. After PALA treatment, a cleaved product of caspase 3 was observed, and caspase 3 activity was induced significantly (8-fold by 72 h) in cells lacking p53 (Fig. 3 C and D). These observations suggest that cells lacking p53 undergo apoptosis through caspase 3 activation when starved for pyrimidine nucleotides. The data also suggest that p53 inhibits the activation of caspase 3 in the same cells. Because the activation of caspase 3 depends on the activation of caspase 9, we analyzed caspase 9 from untreated and PALA-treated cells (Fig. 3D). Procaspase 9 is cleaved and activated only in PALA-treated cells lacking p53. Cytochrome c is released into the cytosol from mitochondria in response to a number of apoptotic stimuli, triggering a cascade that activates both caspases 9 and 3. SKOV3 cells release cytochrome c after PALA treatment, but SKP53 cells do not (Fig. 3D).
Fig. 3.
Cells lacking p53 undergo apoptosis in PALA. SKOV3 and SKP53 cells were treated with 250 μM PALA for the times indicated. (A) Fraction of apoptotic cells, determined by the TUNEL assay. (B) DNA laddering in untreated and PALA-treated SKOV3 and SKP53 cells. (C) Activation of caspase 3 in PALA-treated cells. (D) Activation of caspases 9 and 3 and release of cytochrome c in PALA-treated cells. Proteins were measured by the Western blot method.
Expression of p53 Target Genes in Response to PALA.
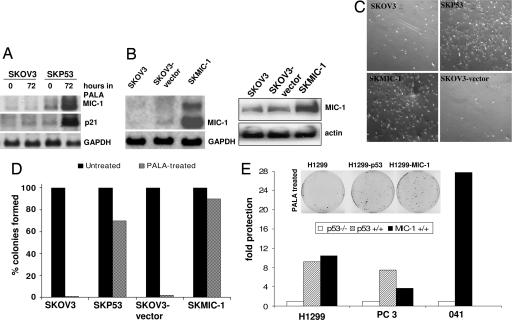
To identify molecular targets of p53 involved in the protective S phase arrest induced in response to PALA, we used an array of p53-dependent cDNAs (19). The accumulation of p53 starts ≈24 h after PALA treatment and reaches a level comparable to that induced by other DNA-damaging agents (for example, camptothecin) after 48 h (data not shown). Therefore, we collected total RNAs from untreated and PALA-treated SKOV3 and SKP53 cells 48 h after treatment, labeled them with Cy5 or Cy3, and hybridized them to an array of p53-induced cDNAs. In this analysis, the expression of only a few p53-regulated genes was markedly different after PALA treatment of the two cell lines. For example, the levels of MIC-1, α-crystallin, and EST1037 increased after PALA treatment of SKP53 but not SKOV3 cells (data not shown). Northern blot analysis confirmed that the PALA-induced levels of MIC-1 and p21 mRNAs are seen only in SKP53 cells (Fig. 4A).
Fig. 4.
Constitutive expression of MIC-1 protects PALA-treated cells lacking p53. (A) Analysis of MIC-1 and p21 mRNA expression. The Northern blot method was used. (B) Analysis of expression of MIC-1 in cells transfected with MIC-1 cDNA by the Northern (Left) and Western (Right) blot methods. (C) Growth of cells in PALA. SKOV3, SKP53, SKMIC-1, and SKOV3 vector. Cells were treated with 250 μM PALA, and photographs were taken 7 days later. (D) Colony formation in SKOV3, SKP53, and SKMIC-1 cells treated with 250 μM PALA for 12 days. The percentage of colonies formed with individual cloned cell lines is shown. Similar results were obtained with pools (not shown). (E) Colony formation in H1299, PC3, 041, and its counterparts expressing MIC-1 or p53, treated with 250 μM PALA for 12 days. Fold protection was calculated by dividing the number of colonies formed in p53- or MIC-1-expressing cells by the number formed in parental p53-null cells.
MIC-1 Protects Cells Lacking p53 from Apoptosis.
A full-length cDNA encoding MIC-1 was cloned into an expression vector, and SKOV3 cells were infected either with this construct or with an empty vector control. Twelve hours later, these populations of cells were treated with 250 μM PALA, which was maintained for 5 days. As expected, cells infected with the empty vector were readily killed by PALA. However, a significant proportion of the cells infected with the MIC-1 construct behaved similarly to PALA-treated SKP53 cells (data not shown). To confirm that exogenous expression of MIC-1 in p53-null cells protects them, we generated individual stable clones and stable pools of SKOV3 cells expressing MIC-1 and treated them with 250 μM PALA for 12 days. Two such clones were studied and behaved similarly; data for only one of these are shown (Fig. 4B), together with data from pools of transfected cells. Cells expressing MIC-1 remained attached to the plates and looked similar to SKP53 cells treated with PALA. Of course, these cells do not grow in the presence of PALA, because they cannot make DNA. In contrast, cells not expressing MIC-1 were killed in PALA, similarly to cells lacking p53 (Fig. 4C). To investigate their long-term viability in PALA, SKOV3 cells expressing MIC-1 were left in PALA for 12 days and subsequently plated for colony formation. More than 80% of the cells from the clone and half of the cells from the stable pools still formed colonies (Fig. 4D), showing that the protective arrest of growth rendered by MIC-1 is reversible. We next investigated whether MIC-1 is capable of protecting other p53-null cell lines from PALA-mediated death. We stably infected p53-null H1299, PC3, or 041 cells with empty vector (control) or vector expressing p53 or MIC-1. Pools of H1299 cells and the p53 and MIC-1 derivatives were treated with 600 μM PALA for 12 days, whereas pools of PC3 and 041 cells and their derivatives were treated with 250 μM PALA for 12 days. The concentrations used (3 × ID50) reflect different ID50 values for the different cell types. H1299 cells expressing either p53 or MIC-1 formed many more viable colonies than control-infected p53-null H1299 cells (Fig. 4E). Pools of cells expressing p53 or MIC-1 formed at least five times more colonies than parental p53-null cell lines. Thus, MIC-1-mediated protection from PALA is not a cell-type-specific phenomenon, because all of the cells tested here were protected significantly.
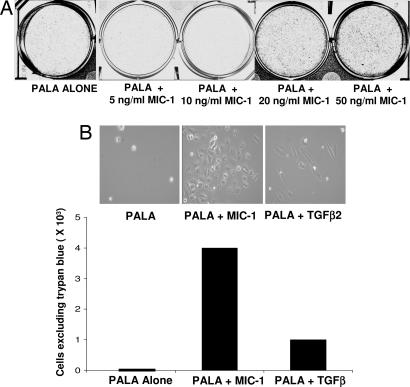
We also tested whether exogenous MIC-1, added to the culture medium, would protect cells from PALA-mediated toxicity in a manner similar to the protection afforded by endogenous expression of MIC-1. SKOV3 cells were treated daily with 5, 10, 20, or 50 ng/ml pure MIC-1 or TGF-β2, together with 250 μM PALA, and assayed after 12 days (Fig. 5A). Treatment with 5 or 10 ng/ml MIC-1 did not confer substantial protection but, at higher concentrations, there was dose-dependent protection of up to 12-fold (Fig. 5 A and B, and data not shown). The protective effect of TGF-β2 was slightly less than that afforded by treatment with MIC-1, indicating that these two cytokines probably act through a similar mechanism (Fig. 5B).
Fig. 5.
Treatment with MIC-1 protects p53-null SKOV3 cells from PALA. (A) Cells in 250 μM PALA were treated with various concentrations of MIC-1 (replenished daily), and after 12 days, the plates were stained with methylene blue and photographed. (B) Cells were treated with a 50 ng/ml concentration of either MIC-1 or TGF-β2 and 250 μM PALA. Twelve days later, the cells were photographed, and their viability was measured by trypan blue exclusion.
Discussion
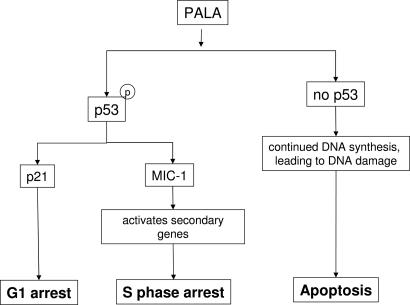
PALA, a potent and reversible inhibitor of aspartate transcarbamylase, blocks de novo pyrimidine nucleotide biosynthesis and induces growth arrest of normal mammalian fibroblasts (reviewed in refs. 7 and 8). A signal activated in response to PALA leads to the rapid induction and activation of p53, leading in turn to reversible cell- cycle arrest, even after continuous exposure to PALA for several weeks. This arrest takes place in the absence of detectable double-strand breaks in the DNA (1). Three major questions are evident: How is p53 activated in response to starvation for DNA precursors? What p53-dependent signal triggers protective arrest in S phase? How is replication arrested for many days without irreversible damage to DNA? The results of the current study deal with the second question and are summarized in Fig. 6. The mechanism of activation of p53 is a subject matter of another study (K.H., unpublished work), and that of protective arrest remains to be explored in future work.
Fig. 6.
p53-dependent effects on cell-cycle checkpoints in response to PALA.
Comparing the cell-cycle responses of several different cell lines, we found earlier that PALA-treated fibroblasts lacking p53 continue to cycle, incur DNA damage, and are readily killed (7). Reintroduction of p53 protected these cells by using a cell-cycle checkpoint that functions during S phase. This checkpoint is p21-independent, because fibroblasts lacking p21 were also arrested in S phase in PALA and were protected (7). In the present work, the responses of p53-positive human colon epithelial HCT116 cells were compared with those of derivatives lacking either p53 or p21. As shown in Table 2, the parental cells accumulate in G1 upon treatment with PALA, as observed in normal fibroblasts, whereas cells lacking p53 continue to cycle and then die. However, cells lacking p21 bypass the G1 checkpoint and accumulate in S phase, where they remain for long periods and are protected similarly to fibroblasts. We propose that the level of p53 is a critical determinant of whether fibroblasts or epithelial cells arrest in G1 or S, and that the S phase checkpoint is present in normal cells but is more apparent when the G1 and G2 checkpoints are missing, as in cells lacking p21 or with low levels of p53.
When starved for pyrimidine nucleotides, epithelial cells lacking p53 enter S phase and make DNA, as revealed by strong incorporation of BrdU. The great majority of cells lacking p53 traverse through S phase and gradually accumulate in G2. However, the progression is slow, because it takes several days for cells to traverse from G1 to G2. At the end of 3 days, most of the cells are still in S or G2. With low pyrimidine nucleotide pools in the absence of p53, cells continue DNA synthesis and are very likely to incur more DNA damage. In cells where the degree of DNA damage is beyond the capacity for repair, a signal is generated that leads to apoptosis. In the presence of functional p53, the great majority of cells starved for pyrimidine nucleotides eventually enter S phase and arrest there, as revealed by propidium iodide staining. These cells fail to synthesize DNA, as revealed by their very low incorporation of BrdU, and thus are protected from the damage caused by misincorporation of nucleotides into DNA from the highly unbalanced pools that result from the action of PALA.
The existence of a p53-dependent S phase arrest may be important in several different situations. For example, when conditions for growth are unfavorable or when cells are exposed to a stress, DNA may be damaged if cells attempt to replicate it. A p53-mediated blockade of DNA synthesis may prevent the generation and propagation of additional DNA damage. After arrest, the cells may either repair the damaged DNA or undergo apoptosis before entering the next cell cycle. p53-dependent checkpoints in both S phase and G2/M also may help to protect cells in which a mutation has inactivated a protein required for G1 arrest, such as Rb. Multiple checkpoints in different parts of the cell cycle are likely to be important in realizing the full ability of p53 to maintain genomic stability.
To elucidate the basis of the p53-dependent differential cellular responses to S phase arrest or cell death in response to starvation for pyrimidine nucleotides, we compared the expression of p53 target genes known to mediate different biological activities of p53. The comparative expression of genes from PALA-treated p53-null SKOV3 and p53 wild-type SKP53 cells surprisingly revealed only a few mRNAs whose expression was different. Most interestingly, the expression of the secreted protein MIC-1 was much higher in cells expressing wild-type p53 than in cells lacking p53. MIC-1 has been reported to be a p53 target gene that inhibits tumor cell growth by a TGF-β-like signaling pathway (12). Overexpression of MIC-1 from a recombinant adenoviral vector led to both G1 cell-cycle arrest and apoptosis in breast tumor cells (20). However, overexpression of a cDNA encoding MIC-1 in ovarian cancer SKOV3 cells did not result in negative growth regulation, assayed either by cell- cycle arrest or apoptosis. The cells we studied tolerated high levels of MIC-1 (Fig. 4).
The properties of PALA are ideal for a potential antitumor drug, because virtually all cancer cells have some disruption of p53 function. During the late 1980s, studies in cell culture and animal models revealed PALA to be highly promising as an antitumor drug, and it was tested in phase I, II, and III clinical trials (21). However, PALA did not prove to be useful in patients because of dose-limiting gut toxicity. Unfortunately, normal gut epithelial cells are not protected as well as normal fibroblasts from PALA-induced starvation for pyrimidine nucleotides. The basis of this difference is not understood. However, our identification of MIC-1 as a mediator of p53-dependent protective cell-cycle arrest opens a possible new approach to mitigating the gut toxicity that has limited the use of PALA in treating cancer. Furthermore, MIC-1 overexpression from a recombinant adenoviral vector led to both arrest in the G1-phase of the cell cycle and apoptosis in breast tumor cells (20), reinforcing the possibility that stimulation of reversible S phase arrest in cells of gut epithelium by providing MIC-1 exogenously or by stimulating its synthesis might help to revitalize the clinical use of PALA.
Materials and Methods
Cell Lines and Treatments.
The human ovarian cancer cell line SKOV3, which lacks p53, was obtained from the American Type Culture Collection (ATCC; Bethesda, MD). SKP53 and SKMIC-1 cells were derived from SKOV3 cells by introducing cDNAs encoding wild-type p53, driven from a tetracyclin-responsive element, or MIC-1, driven from a cytomegalovirus (CMV) promoter, respectively. p53-null H1299 (human lung carcinoma), PC3 (prostate cancer, from ATCC), and MDAH041 cells (human Li-Fraumeni fibroblasts; ref. 18) were infected with retroviruses expressing p53 or MIC-1. Normal human fibroblast BJ cells were obtained from ATCC, and derivative BJshp53 cells were generated by stably infecting them with a lentiviral vector containing short-hairpin RNA directed toward p53 (19). BJ-tert cells were derived by retroviral infection with a construct encoding the TERT subunit of human telomerase. Human colorectal cancer HCT116 cells and derivatives were obtained from the laboratory of Bert Vogelstein (The Johns Hopkins University, Baltimore, MD). Cell lines containing a neo marker were grown in the presence of 600 μg/ml G418 (Sigma–Aldrich, St. Louis, MO). All cells were grown in 10% CO2 in DMEM supplemented with 10% FBS. Purified MIC-1 protein (22) was used to treat 1 × 105 cells grown in individual wells of a six-well plate in the presence of 10% (vol/vol) dialyzed FCS. The cells were treated with 250 μM PALA and varying concentrations of MIC-1 or TGF-β2 (Roche Biochemicals, Indianapolis, IN) for 12 days. Cells that excluded trypan blue were counted. For colony assays, 50,000 cells were treated with PALA for 12 days in dialyzed serum. Viable cells were counted by Trypan blue exclusion, and 1,000 cells were plated in 15-cm plates with DMEM with 10% FBS and 100 mM uridine. After 18–20 days, colonies were stained with methylene blue and counted.
PALA.
PALA (NSC224131), an inhibitor of the aspartate transcarbamylase activity of the multifunctional enzyme CAD, was obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, National Cancer Institute (Bethesda, MD). Treatment with PALA was as described by Agarwal et al. (7). For each cell type, the ID50 was determined, and selection of 2 × 105 cells per 10-cm plate was done in the presence of 10% (vol/vol) dialyzed FCS, using PALA at 3 × ID50. The selective medium was changed every 4–5 days.
Cell-Cycle Analyses.
The DNA content of trypsinized cells was determined by staining with propidium iodide, using the Cycletest kit (Becton Dickinson, Franklin Lakes, NJ). Cell-cycle distribution was determined by using a FACScan instrument (Becton Dickinson), ModFit software (Verity Software House, Topsham, ME), and an HP340 Series 9000 Workstation (Hewlett–Packard, Palo Alto, CA). Dead cells were gated out by using pulse processing. For BrdU labeling, cells were treated with PALA at 3 × ID50 for 72 h, 5 μM BrdU (Sigma, St. Louis, MO) was added, and cells were incubated for 4 h more. Incorporation into DNA was determined by using an FITC-conjugated anti-BrdU antibody (Amersham Biosciences, Piscataway, NJ), as described by the manufacturer.
Array-Based Analyses.
Array-based analyses were performed as described (19). Briefly, cDNAs representing 1,500 p53 target genes were obtained from Research Genetics/Invitrogen (Carlsbad, CA) in the form of bacterial cultures. The cDNAs were amplified and spotted on glass slides in duplicate by using a robotic arrayer. In all, 100 μg of total RNA was labeled by reverse transcription in the presence of Cy5 (red)- or Cy3 (green)-labeled nucleotides. Two labeled RNAs were competitively hybridized to the p53 array, and the signals were analyzed by using a laser scanner. Quantitation was performed by using Genespring software (Agilent Technologies, Santa Clara, CA) and Excel software (Microsoft, Redmond, WA).
Northern and Western Blot Analyses.
Total RNAs were isolated by using the Qiagen (Valencia, CA) RNeasy Mini Kit, and 5–7 μg of total RNA was used for Northern blot analysis, using the digoxigenin (DIG) kit (Roche Biochemicals). Standard protocols for gel electrophoresis, denaturation, and neutralization were used for transferring the RNA to a positively charged nylon membrane. The membrane was hybridized with the DIG-labeled DNA probe (50–100 ng/ml) overnight. After overnight hybridization and washes, the hybridized probes were detected with anti-DIG-AP and visualized with the chemiluminescent substrate CSPD (23).
Total cellular protein was isolated after lysing cells in 20 mM Tris·HCl, pH 7.5/2% (wt/vol) SDS/2 mM benzamidine/0.2 mM phenylmethanesulfonyl fluoride. Protein concentrations were determined by the Bradford method (Bio-Rad, Hercules, CA). Proteins (25 μg per lane) were separated by electrophoresis (SDS–10% PAGE) and electroblotted onto poly(vinylidene difluoride) membranes (Stratagene, La Jolla, CA). The membranes were probed with antibodies against p53, p21 (Santa Cruz Biotechnology, Santa Cruz, CA), MIC-1 (Upstate Biotechnology, Lake Placid, NY), phosphoserine p53, phosphoserine chk1 (Cell Signaling Technology, Beverly, MA), or Actin (Lab Vision, Fremont, CA), followed by incubation with anti-mouse or anti-rabbit secondary antibody–horseradish peroxidase conjugate (Amersham Biosciences). Chemiluminescence was developed by using an ECL kit (Amersham Biosciences).
Acknowledgments
This work was supported by National Institutes of Health Grants GM 49345 (to G.R.S) and CA 98916 (to M.L.A).
Abbreviations
- MIC-1
macrophage inhibitory cytokine 1
- PALA
N-(phosphonacetyl)-l-aspartate.
Footnotes
The authors declare no conflict of interest.
References
- 1.Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 2.Taylor WR, Agarwal ML, Agarwal A, Stacey DW, Stark GR. Oncogene. 1999;18:283–295. doi: 10.1038/sj.onc.1202516. [DOI] [PubMed] [Google Scholar]
- 3.Cross SM, Sanchez CA, Morgan CA, Schimke MK, Ramel S, Idzerda RL, Raskind WH, Reid BJ. Science. 1995;267:1353–1356. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 4.Di Leonardo A, Khan SH, Linke SP, Greco V, Seidita G, Wahl GM. Cancer Res. 1997;57:1013–1019. [PubMed] [Google Scholar]
- 5.Swyryd EA, Seaver SS, Stark GR. J Biol Chem. 1974;249:6945–6950. [PubMed] [Google Scholar]
- 6.Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal ML, Agarwal A, Taylor WR, Chernova O, Sharma Y, Stark GR. Proc Natl Acad Sci USA. 1998;95:14775–14780. doi: 10.1073/pnas.95.25.14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livingstone LR, White A, Sprouse J, Livanos E, Jacks T, Tlsty TD. Cell. 1992;70:923–935. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 9.Khan SH, Wahl GM. Cancer Res. 1998;58:396–401. [PubMed] [Google Scholar]
- 10.Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Stark GR, Taylor WR. Methods Mol Biol. 2004;280:51–82. doi: 10.1385/1-59259-788-2:051. [DOI] [PubMed] [Google Scholar]
- 12.Tan M, Wang Y, Guan K, Sun Y. Proc Natl Acad Sci USA. 2000;97:109–114. doi: 10.1073/pnas.97.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fairlie WD, Moore AG, Bauskin AR, Russell PK, Zhang HP, Breit SN. J Leukocyte Biol. 1999;65:2–5. doi: 10.1002/jlb.65.1.2. [DOI] [PubMed] [Google Scholar]
- 14.Bootcov MR, Bauskin AR, Valenzuela SM, Moore AG, Bansal M, He XY, Zhang HP, Donnellan M, Mahler S, Pryor K, et al. Proc Natl Acad Sci USA. 1997;94:11514–11519. doi: 10.1073/pnas.94.21.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paralkar VM, Vail AL, Grasser WA, Brown TA, Xu H, Vukicevic S, Ke HZ, Qi H, Owen TA, Thompson DD. J Biol Chem. 1998;273:13760–13767. doi: 10.1074/jbc.273.22.13760. [DOI] [PubMed] [Google Scholar]
- 16.Agarwal ML, Agarwal A, Taylor WR, Stark GR. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waldman T, Kinzler KW, Vogelstein B. Cancer Res. 1995;55:5187–5190. [PubMed] [Google Scholar]
- 18.Agrawal S, Agarwal ML, Chatterjee-Kishore M, Stark GR, Chisolm GM. Mol Cell Biol. 2002;22:1981–1992. doi: 10.1128/MCB.22.7.1981-1992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson MW, Agarwal MK, Agarwal ML, Agarwal A, Stanhope-Baker P, Williams BR, Stark GR. Oncogene. 2004;23:4477–4487. doi: 10.1038/sj.onc.1207575. [DOI] [PubMed] [Google Scholar]
- 20.Li PX, Wong J, Ayed A, Ngo D, Brade AM, Arrowsmith C, Austin RC, Klamut HJ. J Biol Chem. 2000;275:20127–20135. doi: 10.1074/jbc.M909580199. [DOI] [PubMed] [Google Scholar]
- 21.Martin DS, Spriggs D, Koutcher JA. Apoptosis. 2001;6:125–131. doi: 10.1023/a:1009692631748. [DOI] [PubMed] [Google Scholar]
- 22.Fairlie WD, Zhang H, Brown PK, Russell PK, Bauskin AR, Breit SN. Gene. 2000;254:67–76. doi: 10.1016/s0378-1119(00)00295-x. [DOI] [PubMed] [Google Scholar]
- 23.Schaap AP, Akhavan H, Romano LJ. Clin Chem. 1989;35:1863–1864. [PubMed] [Google Scholar]