Abstract
Growth factor signaling is mediated through Class IA phosphatidylinositol 3-kinases (PI3Ks). Among this class of enzymes, only p110α, encoded by the PIK3CA gene, has been found to be mutant in human cancers. To determine the specific functions of p110α, we generated mice carrying a conditionally targeted allele of the PIK3CA gene. Here, we report that PIK3CA-knockout mouse embryonic fibroblasts are deficient in cellular signaling in response to various growth factors, unable to differentiate into adipocytes, and resistant to oncogenic transformation induced by a variety of oncogenic receptor tyrosine kinases, indicating a fundamental role for p110α in these biological processes.
Keywords: adipocyte differentiation, cancer therapy, conditional knockout, PIK3CA
Class IA PI3Ks are heterodimeric lipid kinases consisting of a p110 catalytic subunit complexed to one of a family of regulatory subunits (p85α, p55α, p50α, p85β, p55γ), collectively called p85 (1, 2). In response to growth factor stimulation and the subsequent activation of receptor tyrosine kinases (RTKs), class IA PI3Ks are recruited to the membrane via interaction of the p85 subunit with phosphotyrosine-containing motifs on the activated receptor. The p110 catalytic subunit of PI3K then catalyzes the phosphorylation of phosphatidylinositol 4,5-bisphosphate (PIP2) to form phosphatidylinositol 3,4,5-trisphosphate (PIP3). The lipid second messenger PIP3 in turn activates the Ser/Thr kinase AKT and other downstream effectors to regulate multiple cellular functions, including proliferation, survival and migration. There are three isoforms of the p110 catalytic subunit (α, β, and δ) that are encoded by the PIK3CA, PIK3CB, and PIK3CD genes, respectively. Both p110α and p110β isoforms are ubiquitously expressed, whereas the p110δ subunit is primarily found in leukocytes (3, 4). Notably, among these isoforms, only p110α has been found to be frequently mutated in human tumors (5), suggesting distinct roles for the individual p110 isoforms in both normal signaling and oncogenic transformation. Previous efforts using gene targeting in mice have revealed that p110δ plays essential roles in the development and function of lymphocytes, mast cells and possibly neutrophils (6–10). However, mice lacking p110α or p110β die early in embryonic development (11, 12), which has precluded delineating the functions of the individual p110α and p110β isoforms by genetic ablation. Recent articles have used either a mouse heterozygous for the knockin of a kinase dead allele of p110α (13) or small molecule inhibitors of PI3K-p110α (14) to study its role in insulin signaling. Here, we report, for the first time, the effects of complete genetic ablation of PI3K-p110α on signaling elicited by a panel of growth factors, adipocyte differentiation and oncogenic transformation. Strikingly, we observe that knockout of this single isoform is capable of blocking both normal and oncogenic growth factor signal pathways.
Results and Discussion
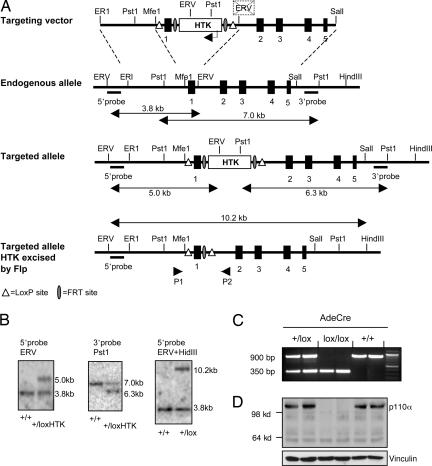
To investigate the specific role of p110α in signaling and to examine it as a potential therapeutic target in oncogenic growth factor signaling, we exploited the Cre-loxP mediated recombination system to conditionally inactivate the PIK3CA gene. A targeting vector was constructed in which exon1 of the mouse PIK3CA gene is flanked by loxP sites, with an FRT-flanked selection cassette inserted between exon1 and the left loxP site (Fig. 1A). In choosing this strategy we targeted the same region that had been deleted in the earlier traditional knockout (11). After homologous recombination of the targeting construct into the PIK3CA locus in ES cells, clones harboring recombinants were transiently transfected with a plasmid expressing the FLP recombinase to remove the FRT-flanked selection cassette (Fig. 1A). The resultant ES clones carrying one floxed allele of PIK3CA were injected into mouse blastocysts to generate chimeric mice. Male chimeras were bred with C57BL/6 females, and germ line transmission was confirmed both by Southern blot analysis (Fig. 1B) and genomic PCR (data not shown). The resulting floxed line was intercrossed to generate homozygous floxed mice.
Fig. 1.
Targeting vector and Cre-mediated deletion of p110α. (A) Diagram of the targeting vector and deletion strategy. The coding exons encoding p110α are shown as filled boxes. HTK denotes the hygromycin and thymidine kinase selection cassette, flanked by FRT recombination sites. HTK expression is driven by the phosphoglycerol kinase (PGK) promoter transcribing in the opposite direction to transcription of p110α as drawn. The positions of 5′ and 3′ external probes and PCR primers (P1 and P2) for genotyping are indicated. (B) Southern blot analysis of genomic DNA extracted from wild-type and positive recombinant ES cells. Hybridization of 5′ probe on EcoRV digests or 3′ probe on PstI digests detected 5.0- and 6.3-kb fragments, respectively, in recombinant ES cells carrying one allele of floxed p110α with HTK cassette still present. After excision of the FRT-flanked HTK cassette in recombinant ES clones, genomic DNA was extracted and digested with EcoRV and HindIII. Hybridization of the 5′ probe on the digested DNA detected a 10.2-kb fragment in the recombinant ES cells, as expected. (C) PCR analysis of Genomic DNA extracted from p110α (+/lox), (lox/lox) and (+/+) MEFs: Floxed MEFs were treated with Ade-Cre using the pair of primers P1 and P2 as indicated above in (A) The smaller band of 350-bp represents the deleted allele of the p110α gene. (D) Western blot analysis of p110α ablation: Western blots probed with p110α-specific antibodies show the absence of both full-length p110α and a potential truncated form of the protein.
To examine the effect of p110α ablation on cellular responses to different growth factor signals, we prepared cohorts of p110α (+/+), p110α (+/lox) and p110α (lox/lox) mouse embryonic fibroblasts (MEFs) from embryos obtained by intercrossing p110α (+/lox) mice. Cells at passage 1 were infected with an adenovirus expressing Cre recombinase (Ade-Cre), and the Cre-mediated excision of exon 1 in MEFs harboring p110α (+/lox) or p110α (lox/lox) was monitored by genomic PCR (Fig. 1C). We next examined the expression of p110α by Western blotting, looking both for the expression of full-length protein and a potential truncated form. As described in ref. 11, a downstream start codon residing within exon 2 could potentially allow the translation of the deleted allele which could produce a truncated form (∼97 KD) of p110α lacking the N-terminal p85-binding domain. Our Western blot analysis using an antibody raised against an epitope mapping downstream of the p85 binding domain of p110α failed to detect both full-length and the theoretical truncated form of p110α in homozygous knockout MEFs (Fig. 1D).
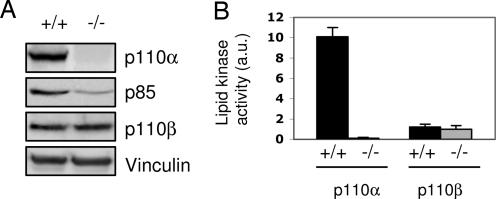
We also tested the effects of p110α ablation on expression of other components of the class IA PI3K family. The protein levels of p110β in p110a-null (−/−) MEFs are equivalent to those in wild-type p110α cells (+/+) (Fig. 2A). In contrast, the expression level of p85 was reduced in the absence of p110α (Fig. 2A). Conventional knockout of p110α yielded embryos which died too early to allow isolation of MEFs, but p85 was found to be overexpressed in the early embryos that could be obtained (11). However, a reduction of p85 levels was also found in B cells derived from p110δ-knockout mice (8) and in liver and muscle from mice doubly heterozygous for loss of p110α and p110β (15). The lipid kinase activity in immunoprecipitates of p110α is completely abolished in p110α-knockout MEFs, whereas the activity of immunoprecipitates of p110β remains unchanged (Fig. 2B).
Fig. 2.
Expression and activities of PI3K subunits in the absence of p110α. (A) Western blotting analysis of pan-p85 and p110β expression in wild-type (+/+) and p110α-null (−/−) MEFs. Note that in this and all following figures the control cells (+/+) and the p110α-knockout cells (−/−) are the isogenic pair of p110α (lox/lox) cells without (+/+) or with (−/−) Ade-Cre treatment. (B) In vitro lipid kinase assay: Immunoprecipitates were prepared from p110α (+/+) and (−/−) MEFs with antibodies specific to p110α or p110β and assayed for associated lipid kinase activity by using phosphatidylinositol phosphates as substrates. Average results from three experiments are presented as arbitrary units (a.u.). Error bars indicate SD.
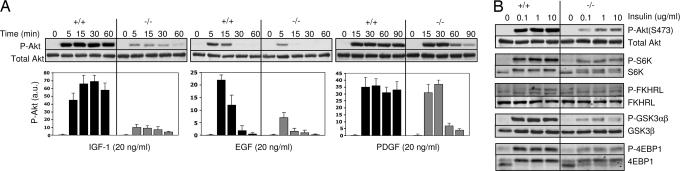
To assess the effect of p110α ablation on PI3K signaling in response to growth factor stimulation, we measured phosphorylation of Akt at both Ser-473 (Fig. 3) and Thr-308 (data not shown) as a signaling readout for PI3K activity at various times after addition of a given growth factor to quiescent cells. We first studied the effects of insulin and IGF-1, the pair of growth hormones most closely tied to PI3K signaling in evolutionary studies. A severe reduction (up to 90%) of phospho-Akt levels was observed in p110α-knockout MEFs when quiescent cells were stimulated with 10 μg/ml of insulin (data not shown) or 20 ng/ml of insulin-like growth factor 1 (IGF-1) (Fig. 3A). A similar loss of insulin/IGF-1 responsiveness was seen when p110α-knockout cells were stimulated with various concentrations of insulin (Fig. 3B). In line with this observation, phosphorylation levels of several components downstream of PI3K/Akt were also reduced in response to insulin stimulation in the absence of p110α (Fig. 3B). In addition to insulin signaling, we also monitored the effects of p110α ablation on PI3K signaling downstream from the receptors for EGF and PDGF, two other commonly studied RTKs in fibroblasts. The induction of phospho-Akt was also compromised in p110α-knockout cells after EGF stimulation (Fig. 3A). Phospho-Akt levels in p110α deficient cells were as high as in wild-type cells at early time points afterPDGF treatment, but markedly diminished in knockout cells relative to control cells at 60 min after PDGF stimulation (Fig. 3A). Therefore, selective impairment of p110α while sparing other p110 isoforms significantly diminishes insulin, EGF, and sustained PDGF signaling.
Fig. 3.
Signaling responses to growth factor stimulation in the absence of p110α. (A) The levels of phospho-Akt at Ser-473 are shown in wild-type (+/+) and p110α-knockout (−/−) MEFs stimulated with IGF1, EGF and PDGF as indicated. Bar graphics reflect the average ± SD of three independent experiments. (B) The phosphorylation levels of several components downstream of PI3K (Akt, S6K, FKHRL, GSK, and 4EBP1) were analyzed in response to insulin stimulation for15 min in both wild-type (+/+) and p110α-knockout (−/−) MEFs.
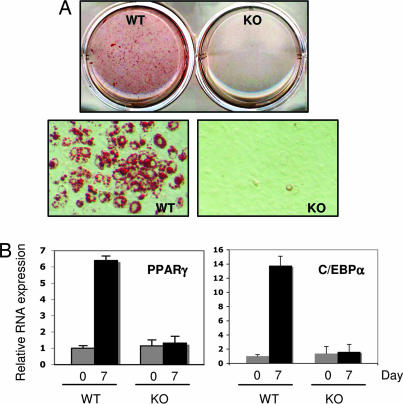
The insulin/IGF-1 receptors and their downstream substrates, IRS-1 and IRS-2, have been reported to play important roles in adipocyte differentiation by regulating the expression of two key transcriptional factors for adipogenesis, PPARγ and C/EBPα (16). Akt1/2 are also known to be required for the induction of PPARγ and C/EBPα and adipocyte differentiation (17). Both of the recent studies using a “knockin” of a kinase-dead allele of p110α (13) and chemical inhibitors specific for various PI3k isoforms (14) showed that p110α is the primary insulin/IRS responsive PI3K in the liver and adipocytes. However, the role of p110α in adipocyte differentiation has not been reported. To investigate whether p110α might be the molecular link between IRS-1/IRS-2, and Akt1/2 in adipogenesis, we used a 3T3 protocol to establish immortalized wild-type and p110α-knockout MEFs and subjected them to a standard regimen for the induction of adipocyte differentiation. The ability of p110α-knockout cells to differentiate into adipocytes was markedly impaired as shown in Fig. 4A as was the expression of PPARγ and C/EBPα (Fig. 4B). These results provide strong genetic evidence that the p110α isoform is required for adipocyte differentiation.
Fig. 4.
Impaired adipocyte differentiation in p110α-knockout MEFs. Wild-type (+/+) and p110α-knockout (−/−) 3T3 MEFs (passage 26) were grown to confluence and induced with IBMX/DEX/insulin/rosiglitazone for differentiation. (A) Cells were stained with Oil red O after 7 days of induction. (B) The mRNA levels of PPARγ and C/EBPα of both wild-type and knockout cells before (day 0) and after (day 7) induction were analyzed by using real-time RT-PCR, with TBP as an internal control. The data represent the mean ± SD from three independent experiments.
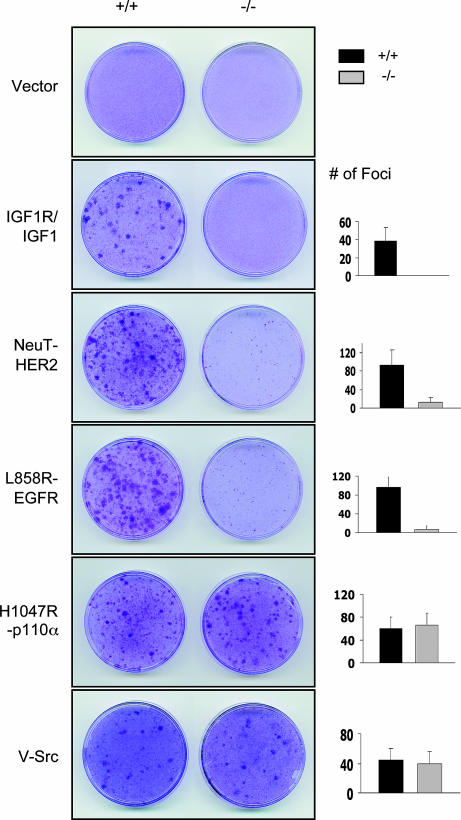
Previous attempts to determine the biological effects of p110α loss on growth factor signaling have focused almost exclusively on metabolic responses to insulin. Because nature seems to have singled out p110α for activation in cancer, we wondered whether it also plays an important role in mediating oncogenic growth factor signaling. Thus, we investigated the effect of ablation of p110α on oncogenic transformation driven by constitutive activation of the insulin-like growth factor 1 receptor (IGF-1R) and EGF receptor (EGFR). Primary murine cells can be transformed in vitro by two cooperating oncogenic events (18). The inactivation of p53 pathway is commonly the first of these with activation of an oncogene occurring subsequently. We first immortalized both wild-type and floxed p110α MEFs by stably introducing dominant negative p53 mutant (p53DD) via retroviral mediated gene transduction. To generate the experimental cells, adenovirus expressing Cre was introduced to ablate p110α expression in the p53DD immortalized p110α (lox/lox) MEFs, whereas the parental p53DD immortalized p110α (lox/lox) or wild-type MEFs at an equivalent passage served as controls. To evaluate oncogenic IGF-1 signaling, we transduced monolayers of p53DD immortalized control and p110α-knockout MEFs with wild-type IGF-1R by retroviral infection. These MEFs were cultured in medium with reduced serum (2% FBS) but in the presence of elevated IGF-1 (50 ng/ml) or insulin (30 μg/ml) to allow foci to arise. The combination of overexpressing IGF-1R with the addition of either IGF-1 or insulin was sufficient to promote focus formation in wild-type MEFs but not in p110α-knockout cells (Fig. 5). Because we have observed that p110α-deficient cells have increased population-doubling time compared with control cells, we purposely looked for focus formation over an extended period of culture but failed to observe the appearance of any foci in the p110α-knockout cells. Separate measurements showed that p110α-knockout cells expressed equivalent levels of IGF-1R to control cells (data not shown). Notably, the p110α deficient cells were not impervious to transformation. Both a tumor derived activating mutant version of p110α, H1047R, and an oncogenic allele of src, v-src, were able to raise foci in both wild-type and p110α-knockout cells at equivalent levels (Fig. 5). The latter result was predicted by published experiments showing that v-src mediated transformation of NIH 3T3 cells could not be blocked by expression of a dominant negative form of p85 (19). In addition, a recent study by Vogt's group showed that Rapamycin blocked cellular transformation induced by active PI3K mutants, but failed to interfere oncogenic transformation induced by v-src (20).
Fig. 5.
The ablation of p110α impairs transformation induced by various oncogenic signals. Both p53DD immortalized p110α (+/+) and (−/−) MEFs were infected with a control empty-vector virus, a virus overexpressing wild-type IGF1R, or viruses carrying various oncogenes: mutant alleles of EGFR, L858R-EGFR, NeuT-HER2, or the tumor mutant allele of p110α, H1047R, as indicated. Cells were transfected with pSG5 expressing v-src. Cells were then cultured in the medium containing 5% FBS or containing 2% FBS but with addition of insulin (30 μg/ml) or IGF1 (50 ng/ml) in the case of cells infected with IGF1R. Foci were scored when wild-type MEFs were cultured for 3 weeks, p110α-knockout (−/−) cells for 4.5 weeks.
The EGFR is frequently mutated in many types human cancer, including breast, colon, lung and brain tumors. Enhanced expression or activation of HER2/Neu has been observed in 20%–30% of primary human breast tumors (21). Earlier work has demonstrated that the PI3K pathway is important for HER2-mediated cellular signaling and transformation (22), but did not demonstrate which PI3K isoform(s) are required for HER2/Neu mediated oncogenic signaling. Whereas NeuT, the constitutively activated allele of HER2/Neu (23), raised foci efficiently in control cells, it formed only mini foci in the p110α-knockout cells (Fig. 5). Our results suggest that p110α is important for oncogenic HER2/Neu mediated transformation.
We have also chosen two somatic activating EGFR mutants found in non-small-cell lung cancers for our analysis: a small in-frame deletion mutant allele (L747_E749del, A750P) and a missense substitution within ATP-binding pocket of EGFR (L858R) (24). Previous work has demonstrated that the Akt pathway is preferentially activated downstream of these mutant alleles of EGFR (25). Interestingly, we found that both EGFR tumor mutants were able to develop large foci in wild-type MEFs but induced only minute foci in p110α-deficient cells (Fig. 5 and data not shown). These mini foci remained small even upon extended culture. Once again, separate analyses showed that the EGFR mutants were equally well expressed in both p110α-knockout and control cells (data not shown). In regarding to the importance of the PI3K/Akt pathway in EGFR mutant-transformed cells, the results are in excellent agreement with the studies presented by Engelman et al. (26). Taken together, our data strongly suggest that ablation of the p110α isoform impedes oncogenic transformation arising from overactivation of IGF1, insulin, or EGF signaling, despite the continued expression of the other p110 isoforms.
The recent finding that some 30% of brain, breast and colon tumors carry activating mutations in the PIK3CA gene has underscored the importance of the p110α isoform in tumorigenesis, and intensified interest in targeting PI3K with small molecule inhibitors. It is hoped that these compounds will have clinical benefit in tumors that contain mutations in p110α itself, in PTEN, the tumor suppressor that negatively regulates the PI3K pathway, and in the growth factor receptors that activate it. Recent data with chemical inhibitors of the PI3K family have suggested that a dual inhibitor of p110α and mTOR can block growth of a tumor cell line as a xenograft (27). The data presented here demonstrates that genetic ablation of the p110α isoform alone can significantly impair the oncogenic action of a number of activated RTKs and oncogenic Ras. Thus, isoform-specific inhibition of p110α may be sufficient to interrupt oncogenic signaling arising from multiple upstream events in cancers.
Materials and Methods
Generation of Conditional Targeted p110α Mice and MEFs.
The PIK3CA gene-targeting construct was assembled by isolating an 8.1-kb genomic fragment encompassing exons 1–5 from a 129/SvJ mouse genomic library and subcloning it into pKO915-DT (Lexicon Genetics, Woodlands, TX). A loxP site was inserted upstream of the exon 1. The FRT-flanked hygromycin and thymidine kinase (HTK) selection cassette followed by a second loxP site was then inserted into the EcoRV site downstream of the exon 1, thus destroying this restriction site. The construct was electroporated into embryonic stem (ES) cells, and correctly targeted recombinant ES clones were identified by Southern blot analyses with probes outside the regions of homologous recombination. 11 of 192 ES cell clones analyzed had undergone homologous recombination at the correct locus. Four recombinant ES clones were transiently transfected with pCAGGS-FLPe (a gift from S. M. Dymecki, Harvard Medical School) to remove the FRT-flanked HTK cassette. ES clones that had lost HTK cassette were identified by both Southern blotting and genomic PCR. Two correctly targeted and karyotypically normal ES clones were used to generate chimeric floxed p110α mice. Male chimeras were bred to C57BL/6 females. Transmission of the germ line was identified by PCR and confirmed by Southern blotting.
Animal care and protocols were approved by the Institutional Animal Care and Use Committees of Dana–Farber Cancer Institute and Harvard Medical School.
MEF Cultures and Genotyping.
Mouse embryonic fibroblasts (MEFs) were prepared from day 13.5 embryos from p110α (lox/+) intercrosses. MEFs from littermate embryos were used for our experiments to minimize genetic variability. MEFs were infected twice with Adenovirus CMV-Cre (Ade-Cre) (Gene Transfer Vector Core, Iowa City, IA) at passage 1 or immortalized with p53DD before Ade-Cre infection for the focus formation analyses. Genotyping by PCR using the primer pair P1 (5′-CTG TGT AGC CTA GTT TAG AGC AAC CAT CTA) and P2 (5′-ACA GCC AAG GCT ACA CAG AGA AAC CCT GTC TTG) amplifies a 900-bp fragment from the wild-type allele, a 1,100-bp fragment from the loxP allele, and a 350-bp fragment from the deleted allele. Cre-mediated deletion of exon 1 of the PIK3CA gene was verified by Southern blotting and sequencing. Both primary MEFs and p53DD immortalized MEFs were cultured in DMEM supplemented with 10% FCS and 10 units/ml penicillin and 10 μg/ml streptomycin.
Growth Factor Stimulation, Cell Lysates, and Western Blotting.
For growth factor stimulation, cells were starved in DMEM (Invitrogen, CA) without serum for 2 h, followed by stimulation with insulin, IGF1, EGF or PDGF as indicated in Fig. 3. Cells were lysed and processed for Western blotting as described (28). Anti-p110α, anti-phospho-Akt (Ser-473 or Thr-308), anti-Akt, anti-p70 S6 kinase, anti-phosphop70 S6 kinase (Thr-389), anti-GSK-3b, anti-phopspho-GSK-3a/b(Ser-21/9), anti-phosphop-4E-BP1 (Ser-65), and anit-4E-BP1 anti- antibodies were obtained from (Cell Signaling Technology, Beverly, MA). Anti-p85 antibody, anti-phospho-FKHRL1(Thr-32), and anti-FKHRL1 were obtained from Upstate Biotechnology (Lake Placid, NY). This anti-p110α antibody was raised against a peptide matching a region downstream of p85-binding domain (as stated by customer support at Cell Signaling Technology). The anti-vinculin antibody was obtained from Sigma (St. Louis, MO).
In Vitro Lipid Kinase Assay.
In vitro lipid kinase assays were carried out basically as described (15). Briefly, immunoprecipitates made with anti-p110α or p110β from freshly prepared cell lysates were subjected to an in vitro lipid kinase assay using phosphoinositide (PI; Avanti Polar Lipids, Alabaster, AL) as a substrate. The phosphorylated lipids were resolved by a tin-layer chromatography (TLC) for 6 h. The radioactivity of phosphotidylinositol 3-phophate (PIP) was visualized and quantified by using a PhosphorImager (Molecular Dynamics).
Adipocyte Differentiation.
The induction of adipoctye differentiation was performed by using a standard protocol with addition of rosiglitazone as described (16).
Focus Formation Assays.
For focus formation, cell monolayers were infected with the various retroviruses, pBabe-IGF1R (kindly provided by C. R. Kahn, Harvard Medical School), pBabe-EGFR mutants, L747_E749del, A750P and L858R (kindly provided by M. Meyerson, Harvard Medical School), pBabe-NeuT, pBabe-MT and pBabe-H1047R, or transfected with pSG5-v-src (kindly provided by C. Chen, Boston University, Boston, MA). The dilutions used for infection were determined according the number of foci developed cells in pilot experiments. To determine expression levels for the transduced proteins, parallel plates were subjected to drug selection and analyzed by Western blotting. In all cases protein expression levels were comparable in control and experimental cells. MEFs were cultured in DMEM with 5% FCS (2% FCS with addition of 30 μg/ml insulin or 50 ng/ml IGF1R in the case of IGF1R infected cells) without splitting for 3 weeks for wild-type cells and 4.5 weeks for p110a knockout cells after transduction. Medium was changed every 3 days. Confluent monolayer cultures with foci were stained with 0.5% crystal violet in 10% ethanol.
Acknowledgments
We thank Drs. L. Cantley, C. D. Stiles, and J. D. Iglehart for advice; Drs. S. H. Lee and S. Brachmann and Z. Liu for help on the lipid kinase assays; Drs. G. Girnun, C. Walkey, and M. Uldry for help on the adipocyte differentiation experiment. This work was supported by National Institutes of Health (NIH) Grants P01-CA50661 (to T.M.R.), P01-CA089021 (to T.M.R.), and CA30002 (to T.M.R.); NIH Specialized Program of Research Excellence Award 5P50CA090381-05 (to J.J.Z.); a Claudia Barr Award (to J.J.Z.); and Department of Defense Grant BC051565 (J.J.Z.).
Abbreviations
- EGFR
EGF receptor
- HTK
hygromycin thymidine kinase
- MEF
mouse embryonic fibroblast
- PIP
phosphotidylinositol 3-phosphate
- RTK
receptor tyrosine kinase.
Footnotes
Conflict of interest statement: In compliance with Harvard Medical School guidelines, we disclose that T.M.R. has consulting relationships with Novartis Pharmaceuticals, Inc., and Upstate Biotechnology.
References
- 1.Fruman DA, Meyers RE, Cantley LC. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Annu Rev Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 3.Chantry D, Vojtek A, Kashishian A, Holtzman DA, Wood C, Gray PW, Cooper JA, Hoekstra MF. J Biol Chem. 1997;272:19236–19241. doi: 10.1074/jbc.272.31.19236. [DOI] [PubMed] [Google Scholar]
- 4.Vanhaesebroeck B, Welham MJ, Kotani K, Stein R, Warne PH, Zvelebil MJ, Higashi K, Volinia S, Downward J, Waterfield MD. Proc Natl Acad Sci USA. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, et al. Cancer Biol Ther. 2004;3:772–775. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 6.Okkenhaug K, Bilancio A, Farjot G, Priddle H, Sancho S, Peskett E, Pearce W, Meek SE, Salpekar A, Waterfield MD, et al. Science. 2002;297:1031–1034. doi: 10.1126/science.1073560. [DOI] [PubMed] [Google Scholar]
- 7.Jou ST, Carpino N, Takahashi Y, Piekorz R, Chao JR, Wang D, Ihle JN. Mol Cell Biol. 2002;22:8580–8591. doi: 10.1128/MCB.22.24.8580-8591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clayton E, Bardi G, Bell SE, Chantry D, Downes CP, Gray A, Humphries LA, Rawlings D, Reynolds H, Vigorito E, Turner M. J Exp Med. 2002;196:753–763. doi: 10.1084/jem.20020805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puri KD, Doggett TA, Douangpanya J, Hou Y, Tino WT, Wilson T, Graf T, Clayton E, Turner M, Hayflick JS, Diacovo TG. Blood. 2004;103:3448–3456. doi: 10.1182/blood-2003-05-1667. [DOI] [PubMed] [Google Scholar]
- 10.Ali K, Bilancio A, Thomas M, Pearce W, Gilfillan AM, Tkaczyk C, Kuehn N, Gray A, Giddings J, Peskett E, et al. Nature. 2004;431:1007–1011. doi: 10.1038/nature02991. [DOI] [PubMed] [Google Scholar]
- 11.Bi L, Okabe I, Bernard DJ, Wynshaw-Boris A, Nussbaum RL. J Biol Chem. 1999;274:10963–10968. doi: 10.1074/jbc.274.16.10963. [DOI] [PubMed] [Google Scholar]
- 12.Bi L, Okabe I, Bernard DJ, Nussbaum RL. Mamm Genome. 2002;13:169–172. doi: 10.1007/BF02684023. [DOI] [PubMed] [Google Scholar]
- 13.Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- 14.Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, et al. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brachmann SM, Ueki K, Engelman JA, Kahn RC, Cantley LC. Mol Cell Biol. 2005;25:1596–1607. doi: 10.1128/MCB.25.5.1596-1607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miki H, Yamauchi T, Suzuki R, Komeda K, Tsuchida A, Kubota N, Terauchi Y, Kamon J, Kaburagi Y, Matsui J, et al. Mol Cell Biol. 2001;21:2521–2532. doi: 10.1128/MCB.21.7.2521-2532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, Hay N. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Land H, Parada LF, Weinberg RA. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Viciana P, Warne PH, Khwaja A, Marte BM, Pappin D, Das P, Waterfield MD, Ridley A, Downward J. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 20.Kang S, Bader AG, Vogt PK. Proc Natl Acad Sci USA. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger MS, Locher GW, Saurer S, Gullick WJ, Waterfield MD, Groner B, Hynes NE. Cancer Res. 1988;48:1238–1243. [PubMed] [Google Scholar]
- 22.Yakes FM, Chinratanalab W, Ritter CA, King W, Seelig S, Arteaga CL. Cancer Res. 2002;62:4132–4141. [PubMed] [Google Scholar]
- 23.Bargmann CI, Hung MC, Weinberg RA. Cell. 1986;45:649–657. doi: 10.1016/0092-8674(86)90779-8. [DOI] [PubMed] [Google Scholar]
- 24.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 25.Sordella R, Bell DW, Haber DA, Settleman J. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 26.Engelman JA, Janne PA, Mermel C, Pearlberg J, Mukohara T, Fleet C, Cichowski K, Johnson BE, Cantley LC. Proc Natl Acad Sci USA. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, Shokat KM, Weiss WA. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM. Proc Natl Acad Sci USA. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]