Abstract
Precise regulation of MHC gene expression is critical to vertebrate immune surveillance and response. Polymorphisms in the 5′ proximal promoter region of the human class II gene HLA-DQA1 have been shown to influence its transcriptional regulation and may contribute to the pathogenesis of autoimmune diseases. We investigated the evolutionary history of this cis-regulatory region by sequencing the DQA1 5′ proximal promoter region in eight nonhuman primate species. We observed unexpectedly high levels of sequence variation and multiple strong signatures of balancing selection in this region. Specifically, the considerable DQA1 promoter region diversity was characterized by abundant shared (or trans-species) polymorphism and a pronounced lack of fixed differences between species. The majority of transcription factor binding sites in the DQA1 promoter region were polymorphic within species, and these binding site polymorphisms were commonly shared among multiple species despite evidence for negative selection eliminating a significant fraction of binding site mutations. We assessed the functional consequences of intraspecific promoter region diversity using a cell line-based reporter assay and detected significant differences among baboon DQA1 promoter haplotypes in their ability to drive transcription in vitro. The functional differentiation of baboon promoter haplotypes, together with the significant deviations from neutral sequence evolution, suggests a role for balancing selection in the evolution of DQA1 transcriptional regulation in primates.
Keywords: balancing selection, cis-regulatory evolution, major histocompatibility complex, primate molecular evolution
Changes in cis-regulatory DNA sequence can alter the timing, level, and tissue distribution of gene expression, potentially modifying the emergence and expression of key phenotypic traits (1, 2). However, despite its biological importance, relatively little is known about the nature and patterns of extant cis-regulatory variation, the evolutionary forces acting on such variation, or the potential functional consequences of that diversity.
The MHC is a multigene family essential to the development and activation of vertebrate adaptive immunity. Within the MHC, the class II region contains the classical genes that encode the distinct α (A) and β (B) subunits that constitute functional class II molecules (in humans, HLA-DR, HLA-DQ, and HLA-DP). A primary function of these heterodimeric cell-surface molecules is to present short peptides from extracellular pathogens to helper T cells, an interaction that is essential for the initiation and regulation of adaptive immune responses against pathogens (3). Because of this, the class II region is genetically associated with more diseases than any other comparably sized region in the human genome (4). Furthermore, there is substantial evidence, across multiple taxa, that class II gene coding sequence has evolved in response to natural selection (5–7).
Cell surface expression of class II molecules is finely and tightly regulated to balance an appropriately vigorous response to pathogens with minimal collateral damage to self tissues (4). Constitutive expression is normally restricted to specialized antigen presenting cells, e.g., thymic epithelial cells, macrophages, and B cells, but expression on other cell types can be induced by immune activators (3, 8). Class II expression is also developmentally regulated during immune cell maturation (3, 8). Precise regulation of class II expression is essential for normal immune function; defects in proper regulation can have profound immune consequences (9, 10).
Expression of class II molecules is regulated primarily at the level of transcription (3, 4, 10). Conserved cis-regulatory sequence elements in the 5′ proximal promoter region, i.e., the 300-bp region immediately upstream of translation start, direct most aspects of constitutive and inducible expression in class II genes (3, 8, 11). These elements are short sequences, typically 7–14 bp, that function as transcription factor binding sites. Additional gene-specific cis-regulatory elements further affect expression phenotypes (12) and may provide additional levels of regulatory control. Overall, the proximal promoter region behaves as a single functional unit, with the sequence, spacing, and orientation of the regulatory elements synergistically contributing to the expression phenotype (10).
Mutations in the proximal promoter region of the human class II gene HLA-DQA1 (Online Mendelian Inheritance in Man database accession no. 146880) significantly affect constitutive and inducible expression in reporter assays (13). Furthermore, DQA1 promoter region polymorphisms are associated with quantitative expression differences in cell lines (14), and they affect the strength of transcription factor binding (15). Moreover, specific human DQA1 proximal promoter alleles may be associated with increased susceptibility to a number of autoimmune and infectious diseases (16–18).
The overall importance of proper immune regulation, the observed population genetic diversity of the DQA1 promoter in humans (19–21), and the demonstrated functional significance of this diversity make the proximal promoter region of DQA1 a likely target of natural selection (22). To explore this possibility, we investigated the evolutionary history of the DQA1 proximal promoter region in primates and characterized cis-regulatory variation at both the nucleotide and functional levels. To assess the functional importance of observed diversity, we used a cell line-based reporter assay to quantify expression differences between DQA1 promoter region haplotypes observed in the wild Amboseli baboon population (23).
Results
Extensive Polymorphism but Not Fixed Divergence Characterizes the DQA1 Promoter Region in Primates.
We sequenced the 270-bp region immediately upstream of the DQA1 translation start site (i.e., the DQA1 proximal promoter region) in 80 individuals from eight nonhuman primate species (n = 10 chimpanzees, 3 bonobos, 1 gorilla, 5 orangutans, 3 siamangs, 53 baboons, 4 rhesus macaques, and 1 pigtailed macaque) (Table 1, which is published as supporting information on the PNAS web site). Combining the 10 published human sequences (19–21) with our sequencing results, we identified 47 unique DQA1 proximal promoter region haplotypes across the nine primate species (Fig. 4, which is published as supporting information on the PNAS web site). We observed 125 mutations, occurring at 113 nucleotide sites in these 47 haplotypes, yielding an average density of one mutation every 2.2 bp. The proportion of sites that varied within or between species (≈42%) and the mean number of nucleotide differences per site between the 47 unique haplotypes (0.071) were considerable, in contrast to previous descriptions of primate class II promoter region diversity (24, 25).
The extensive allelic and nucleotide diversity we observed in the DQA1 promoter region was characterized by a profound lack of fixed divergence between species. For example, we identified 34 mutations in the 10 human DQA1 promoter haplotypes and 26 mutations in the five chimpanzee haplotypes but found no fixed differences between the two species (Table 2, which is published as supporting information on the PNAS web site). In contrast, recent analyses of the initial chimpanzee genome sequence estimated that fixed differences accounted for 78–86% of the genome-wide divergence between humans and chimpanzees (26). The lack of fixed divergence in the DQA1 promoter region is also apparent in comparisons of more distantly related primate species. Remarkably, we found no fixed differences between human haplotypes and those from three Old World monkey species (baboon, rhesus, and pigtailed macaque) (Table 2). This is unexpected given that the four species last shared a common ancestor ≈25 million years ago (27, 28). In fact, the only fixed differences we observed involved species for which we had extremely small sample sizes (siamang, n = 2 unrelated individuals; gorilla, n = 1) and thus may be due to sampling effects.
Under the neutral model, levels of divergence between species and polymorphism within species should be correlated, because both are related to the neutral mutation rate (29). The lack of fixed differences in the DQA1 promoter region, particularly in light of the substantial polymorphism observed in these species, therefore suggests balancing selection. Because it maintains allelic lineages for long periods of time, balancing selection tends to enhance levels of polymorphism relative to divergence (29). However, nonselective forces may also influence the ratio of polymorphism to divergence (30).
Primate DQA1 Promoter Haplotypes Exhibit Widespread Trans-Species Polymorphism.
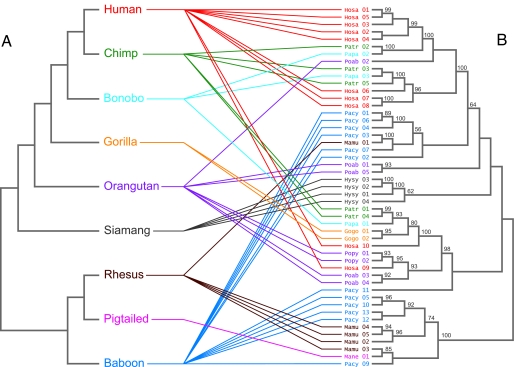
The elevated diversity of the DQA1 promoter region in primates may be due to long-term maintenance of ancestral polymorphism or to diversification after species divergence. To differentiate between these possibilities, we compared promoter sequences from the nine primate species (Fig. 1A) and used Bayesian methods to infer the phylogenetic relationships of the DQA1 promoter region haplotypes (Fig. 1B). The most probable DQA1 promoter tree shows several well supported clusters consisting of haplotypes from different species (Fig. 1B). This extensive trans-species polymorphism is highly unlikely under neutrality and is expected only after long-term maintenance of allelic lineages, e.g., across speciation events (31–33). Thus, the occurrence of trans-specificity provides strong evidence of long-term balancing selection (32–34). Although it is frequently observed in phylogenies of MHC exon 2 coding sequence (35), the trans-species polymorphism we observe in the DQA1 promoter region is one of the first examples of trans-specificity in a primate cis-regulatory region (36, 37).
Fig. 1.
Incongruity of primate species tree and DQA1 promoter region gene tree. (A) Species tree of the nine species from which DQA1 promoter sequences were obtained. (B) The most probable DQA1 promoter tree topology from the Bayesian analysis, with posterior probabilities of branch support. Branch lengths for both trees are arbitrary. Species and gene tree were reconciled by using Page's method (38) for reconciling phylogenetic trees (69).
To quantify the degree of trans-specificity observed in the primate DQA1 promoter tree, we estimated the minimum number of “deep coalescent” events necessary to reconcile a gene tree with the accepted species tree (38). A deep coalescent event, in this case, describes an interspecific coalescence of alleles. We compared the most probable DQA1 promoter tree from the Bayesian analysis (Fig. 1B) to the primate species tree (Fig. 1A) and found that 37 deep coalescent events are required to account for the incongruence between the trees. Because there is uncertainty in the phylogenetic reconstruction of any single tree, we analyzed the 100 most frequently sampled DQA1 promoter topologies from the Bayesian analysis and obtained a mean of 37.8 deep coalescent events (95% credible interval of 32–44 events). Under the simplest models of neutrality and drift, there should be complete congruence between gene and species trees, and a set of species separated by >4 Ne generations should have (n − 1) interspecific coalescent events, where n is the number of species sampled (7, 39). Therefore, the mean of 37.8 interspecific coalescent events observed in our DQA1 analysis is nearly 5-fold higher than that expected when there is complete congruence between the gene and species trees (n − 1 = 8). Our estimate should be taken with caution, however, because it does not factor in the effect of recombination, which may lead to an overestimate of the number of trans-specific lineages (40).
This pattern of trans-species polymorphism is particularly evident in pairwise species comparisons of individual mutations. For example, 18 of the 34 mutations observed in human DQA1 promoter haplotypes are shared with chimpanzees (Table 2). Because neutral polymorphism is expected to persist for ≈4 Ne generations, the probability of observing polymorphisms shared between humans and chimpanzees, two species that diverged ≈20 Ne generations ago, is extremely low (41, 42). This prediction is supported empirically: a large-scale comparison of 6,700 chimpanzee SNPs and 10,000 human SNPs identified only eight trans-species polymorphisms (42), and a microarray analysis of 397 human SNPs failed to find any polymorphisms shared with common chimpanzees (43). Furthermore, shared polymorphisms in the DQA1 promoter region were not restricted to closely related species like humans and chimpanzees. Multiple shared polymorphisms were observed in nearly all pairwise species comparisons (Table 2), including the comparisons of humans and Old World monkeys, species that diverged ≈25 million years ago (27, 28). This maintenance of substantial levels of ancestral polymorphism across multiple speciation events, occurring at different times in primate evolution, supports the hypothesis of long-term balancing selection on the DQA1 promoter region.
Transcription Factor Binding Sites Are Polymorphic Within and Shared Between Species.
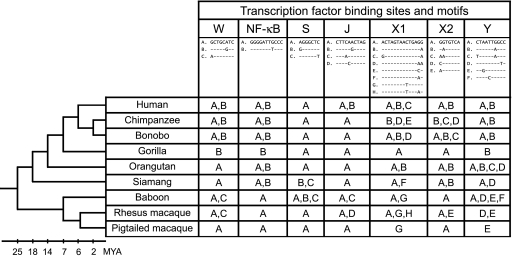
The functional importance of the cis-regulatory elements in the proximal promoter region of human MHC class II genes has been examined in great detail (reviewed in refs. 3 and 8–12). The DQA1 proximal promoter region contains four essential transcription factor binding sites (the S box, X1 box, X2 box, and Y box) that are conserved across class II genes (3). In addition, this region contains three DQA1-specific binding sites [the NF-κB box (44), the W box (45), and the J box (46)] that affect DQA1 transcription. To elucidate the evolutionary history of these cis-regulatory elements, we analyzed patterns of intraspecific and interspecific diversity in the seven binding sites (Fig. 2).
Fig. 2.
Transcription factor binding site motifs in the DQA1 proximal promoter in primates. Letters A–H refer to unique transcription factor binding site motifs, with the corresponding sequence represented above. W, NF-κB, S, J, X1, X2, and Y refer to binding sites identified in the DQA1 promoter (3, 11, 12). Branch lengths of the species phylogeny on the left (27, 28) are roughly proportional to the timeline indicated below it. MYA, million years ago.
There was considerable intraspecific diversity in transcription factor binding site sequence in the DQA1 promoter region. In the seven species represented by more than one individual (humans, chimpanzees, bonobos, orangutans, siamangs, baboons, and rhesus macaques), most binding sites were polymorphic; on average, five of seven binding sites were variable within a given species (Fig. 2). Most binding sites were dimorphic, but there were many examples of three or four different sequence motifs segregating at a single binding site. Considering just the baboon haplotypes, for example, there were two invariant binding sites (NF-κB and X2), three dimorphic binding sites (W, J, and X1), one binding site polymorphic for three motifs (S), and one binding site polymorphic for four motifs (Y) (see Fig. 2). Given the empirically documented importance of binding site sequence in transcriptional regulation, this segregating diversity has the potential to seriously affect the regulation of DQA1 expression.
Transcription factor binding site polymorphisms were often shared among several species (Fig. 2). In some cases, sharing extended across tens of millions of years of primate evolution. For example, the same two Y box motifs (A and B in Fig. 2) were found in four hominoid species (humans, chimpanzees, bonobos, and orangutans), a group that last shared a common ancestor ≈12–15 million years ago (27, 28). Interestingly, these two Y box motifs were shown to have different transcription factor binding affinities and were associated with divergent DQA1 expression profiles in humans (15). Even more striking is the presence of two NF-κB binding site motifs (A and B in Fig. 2) in humans, chimpanzees, bonobos, orangutans, and siamangs, a collection of species that last shared a common ancestor ≈18 million years ago (28).
Functionally important cis-regulatory sequence may be more conserved than surrounding sequence because of purifying selection (2). To test whether nucleotide diversity differs between cis-regulatory elements and intervening sequence in the DQA1 promoter, we compared the number of mutations per nucleotide in the experimentally validated transcription factor binding sites to the number of mutations per nucleotide in adjacent nonbinding site sequence. We estimated total mutations per nucleotide because there are 12 instances of multiple mutations at a single nucleotide, the majority of which (11 of 12) occur in nonbinding site sequence. Comparing across the 47 unique DQA1 promoter region haplotypes, the number of mutations per binding site nucleotide (17/60 = 0.28) is significantly lower (Fisher's exact test, P = 0.028) than the number of mutations per nonbinding site nucleotide (108/210 = 0.51). The preferential conservation of transcription factor binding sites suggests functional constraint due to purifying selection.
Baboon DQA1 Promoter Polymorphisms Have Functional Effects on Transcription in Vitro.
Changes in cis-regulatory sequence affect phenotype in highly variable and often unpredictable ways (2). Some changes are silent, others affect transcription factor binding and/or transcriptional output, and a small subset induce change in phenotypes related to physiology, behavior, disease resistance, and life history (47). To assess the functional consequences of mutations in cis-regulatory sequence, we used a reporter assay system to experimentally determine the effect of naturally occurring baboon DQA1 promoter region diversity on transcriptional activation in vitro. We cloned each of the 12 baboon DQA1 promoter haplotypes (size = 310 or 318 bp) upstream of a luciferase reporter gene. The resulting constructs contained all seven of the transcription factor binding sites (Fig. 2) found in the DQA1 proximal promoter region. The 12 baboon DQA1 promoter constructs were transiently transfected into an immortalized baboon lymphocyte cell line, and the resulting luciferase activity was quantified.
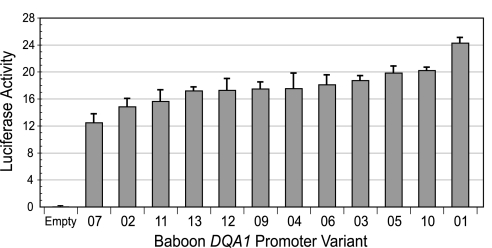
The baboon DQA1 promoter constructs differed significantly in their ability to drive expression of the reporter gene in baboon lymphoblast cells (Fig. 3) (ANOVA, P = 0.00017, F = 6.07, df = 11). Although many of the constructs exhibited similar mean expression levels, there was a 2-fold range of variation between the lowest (variant 07) and highest (variant 01) mean expression values (Fig. 3). This suggests that the intraspecific DQA1 promoter diversity represented in the constructs is both visible to and differentially utilized by the transcriptional machinery of the cells.
Fig. 3.
Luciferase activity induced by baboon DQA1 promoter haplotypes in a transient transfection reporter assay. Bars represent luciferase activity of 12 different baboon DQA1 promoter constructs calculated as the ratio of firefly to Renilla (coreporter) luminescence. Error bars represent standard error of the mean estimated from three replicate experiments. Numbers on the x axis correspond to the unique baboon DQA1 promoter haplotypes, and “Empty” refers to the promoter-less pGL3 basic reporter construct.
Discussion
We have documented elevated diversity, a lack of fixed differences, considerable trans-species polymorphism, and extensive sharing of transcription factor binding site motifs in the DQA1 5′ proximal promoter region in primates. These results suggest nonneutral evolution and are characteristic of genes evolving under balancing selection. The demonstrated functional effect of promoter polymorphism in the reporter assay is also consistent with the action of balancing selection, inasmuch as the result provides evidence for the functional differentiation of alleles, a necessary condition for natural selection to act. The preferential conservation of transcription factor binding site sequence, however, suggests that purifying selection may be acting in addition to balancing selection, eliminating deleterious mutations. Evidence of balancing selection shaping patterns of 5′ cis-regulatory diversity has recently been documented in human population genetic studies of other immune system genes, such as the class Ib gene HLA-G (48), the class II genes HLA-DPA1 and HLA-DPB1 (49), and the cell surface chemokine CCR5 (50). The HLA-G and CCR5 promoter regions, for example, showed similar signatures of balancing selection in human populations: elevated diversity, more polymorphism relative to divergence, an excess of intermediate frequency alleles, and a deep divergence between allelic lineages (48, 50).
In addition to selection acting directly on the DQA1 proximal promoter region, other evolutionary forces may have contributed to the observed levels and patterns of diversity. First, the evolution of the DQA1 promoter region may have been influenced by its proximity, i.e., physical linkage, to the highly polymorphic second exon of the gene, a coding sequence located 4 kb downstream that is known to be evolving under balancing selection (7). Balancing selection on one locus increases levels of neutral variation at closely linked sites, as a function of the physical distance between the two loci, intensity of selection, recombination rate, effective population size, and mutation rate (33, 34, 51, 52). In the human genome, this genetic hitchhiking has the potential to impact neutral diversity tens of kilobases away from a selected site (33, 53) and has been proposed to explain the elevated silent site diversity (53) and increased polymorphism in a pseudogene (54) in the human MHC region. Second, epistatic selection may have generated linkage disequilibrium between nucleotides in the proximal promoter region and the second exon. In this case, the signal of hitchhiking emerges because recombination between epistatic loci produces relatively unfit haplotypes that are eliminated by selection (55). Selection against recombinants has been proposed to explain the strong, long-range linkage disequilibrium between neutral and selected sites in 100-kb haplotypes of the human MHC class II region (56). Examples of epistatic interactions between cis-regulatory variants and coding polymorphisms were quite common in a recent survey of human cis-regulatory evolution (47).
Several lines of evidence suggest that the evolution of the primate DQA1 promoter region is not simply due to linkage disequilibrium with the second exon. First, although hitchhiking may have some influence on neutral diversity over a large chromosomal region, theory predicts that the strength of the effect decreases rapidly with increasing chromosomal distance (51, 52, 55). Likewise, empirical studies have shown that the elevated diversity and signatures of selection found in the second exon of MHC loci decay rapidly in flanking intronic sequence [i.e., within a few hundred base pairs in human class I (57) and class II loci (58)] and neighboring exons (52). This rapid decay was also apparent in the MHC genes of fish and birds, taxa with very different demographic, selective, and recombination histories (59, 60). Second, recent models of trans-species polymorphism (and their application to empirical data sets) demonstrated that the probability of observing a trans-species polymorphism at neutral sites located >1 kb away from the selected site is very low and that regions of trans-specificity are quite short, at most a few hundred base pairs (32). This suggests that the strong signal of trans-species polymorphism we observe in the DQA1 promoter region is not likely to be a result of hitchhiking, considering the nearly 4 kb of sequence that separates the promoter and second exon. Third, although the linkage between DQA1 promoter and second exon is often described as strong, there is considerable evidence for recombination, both historical and recent, between these loci in human populations (19, 20). These recombination events have the potential to break down linkage disequilibrium and mitigate the hitchhiking effect (7, 52, 53, 55). Finally, DQA1 promoter sequence variation has clear functional consequences on transcriptional regulation in vitro, as shown in human (13–15) and baboon (Fig. 3) experiments. Considering the strong signatures of balancing selection in the DQA1 promoter region sequence, the demonstrated functional significance of diversity in the region, and the dire consequences of aberrant class II expression, it is unlikely that this cis-regulatory region is evolving as a neutral locus and that the patterns we report are due to linkage effects.
The model of balancing selection operating in the DQA1 promoter region is presumably pathogen-driven and most likely involves a combination of heterozygote advantage (overdominance), frequency-dependent selection, and/or selection that varies in time or space. Of these, overdominant selection for multiple expression phenotypes has been a primary hypothesis for the maintenance of MHC class II promoter region diversity (22, 61). This is based on the idea that functional cis-regulatory diversity within an individual confers additional range and flexibility in important parameters of transcriptional output (e.g., site, timing, and responsiveness to cytokines), which facilitates a more precise, context-specific immune response. Overall, this would translate into a relative fitness benefit for the heterozygote (22, 61). The plausibility of tissue-type overdominance has been demonstrated in the HLA-DQB1 promoter region, where the relative expression levels associated with two promoter haplotypes were reversed in different cell types (62).
The present study is among the first to demonstrate a role for balancing selection in cis-regulatory evolution in primates and adds to a growing list of studies linking cis-regulatory mutations and phenotypic variation. Future studies integrating molecular evolutionary and functional analyses will greatly enhance our understanding of cis-regulatory structure, function, and evolution.
Materials and Methods
Primate Sequences.
Human DQA1 promoter sequences came from diversity surveys of European and North American populations (19, 20, 63). Genomic DNA samples for eight nonhuman primate species were obtained from several sources (Table 1). The DQA1 proximal promoter region, i.e., from −282 to −13 upstream of the translation start site, was PCR-amplified by using a touchdown program. Two forward primers, DQApFor (5′-CAGACATGCACACACCAGAGAA-3′) and DQApForII (5′-TGCACACACCAGAGAAGATTCC-3′), were used in separate reactions with the reverse primer, DQApRev (5′-GGATCATCYTCTTCCCAAGG-3′). PCR product was purified and sequenced directly. Heterozygous PCR product was cloned, and plasmid DNA was isolated from multiple positive transformants. Sequences were produced by using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) and analyzed on an ABI 3700 capillary sequencer. Multiple identical clones were obtained for each putative DQA1 promoter haplotype to reduce PCR artifact. Mendelian inheritance was confirmed in parent–offspring dyads. No more than two sequence variants were identified per individual, confirming that a single locus was amplified.
Sequence Analysis.
Sequences were assembled by using Sequencher 4.2 (Gene Codes, Ann Arbor, MI) and aligned by using ClustalW (64). Estimates of DQA1 haplotype sequence diversity were calculated by using DnaSP 4.00.6 (65). The mean number of nucleotide differences per site between haplotypes (π) was calculated by using the Jukes and Cantor correction for multiple hits per site. The model of sequence evolution that best fit the DQA1 promoter alignment, HKY+G, was determined by using hierarchical likelihood ratio tests in Modeltest 3.07 (66). To infer phylogenetic relationships, Bayesian analysis incorporating the model structure specified by Modeltest was implemented by using MrBayes 3.0b4 (67). The analysis was run for 5 million generations with a sampling interval of 500 generations. Posterior probabilities were derived from trees sampled after the initial burn-in of 1 million generations. Two DQA1 promoter alleles found in both chimpanzees and bonobos were included for both species only in the phylogenetic analysis. To quantify the degree of incongruity between the species tree and DQA1 promoter phylogeny, we used the method of reconciled trees (38, 68) in GENETREE 1.0 (69).
Reporter Assay.
The 12 baboon DQA1 proximal promoter haplotypes were cloned into the pGL3 basic firefly luciferase reporter construct (Promega, Madison, WI). Eleven constructs were 310 bp long (from −303 to +7 after translation start), whereas one (Pacy 02) was 318 bp because of an 8-bp insertion at −22. Construct fidelity was verified by sequencing. For each experiment, 4.5 μg of experimental construct was transiently transfected, along with a Renilla luciferase coreporter (0.5 μg), into 3 million cell aliquots of the baboon lymphoblast cell line 26CB-1 (ATCC CRL-1495) using Nucleofector Kit V (AMAXA Biosystems, Gaithersburg, MD). Cells were maintained in 90% RPMI medium 1640/10% FBS according to American Type Culture Collection instructions. Cells were harvested 24 h after transfection. Firefly and Renilla luminescence were measured in relative light units by using the Dual-Luciferase Reporter Assay System (Promega) on a Turner Designs 20/20 luminometer. Luciferase activity for each construct was calculated as the ratio of firefly to Renilla luciferase luminescence for three replicate experiments. All baboon DQA1 promoter constructs activated reporter gene expression at significantly higher levels (>2 orders of magnitude) than the pGL3 basic construct lacking upstream promoter sequence (“Empty” in Fig. 3). A one-way ANOVA was used to determine whether the mean expression values differed significantly among the 12 experimental constructs.
Supplementary Material
Acknowledgments
We thank the Office of the President of the Republic of Kenya, the Institute of Primate Research, National Museums of Kenya, and the Kenya Wildlife Service for permission to conduct research on the Amboseli baboons. Research was carried out under Duke University Institutional Animal Care and Use Committee protocols A180-0305 and A317-0210 and Princeton University protocol 1456. We thank S. Edwards, C. Ober, R. Haygood, and J. Tung for comments on the manuscript; B. J. Nielsen, H. O'Brien, H. Wieman, and R. Thomason for technical assistance; and Anne Stone (Arizona State University, Tempe, AZ) for providing Pan troglodytes and Pan paniscus DNA samples. Funding was provided by National Science Foundation Grants IOB-0322613, IOB-0322781, BCS-0323553, and BCS-0323596; the Chicago Zoological Society; and the Jane Coffin Childs Memorial Fund.
Footnotes
References
- 1.Davidson EH. Genomic Regulatory Systems: Development and Evolution. San Diego: Academic; 2001. [Google Scholar]
- 2.Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 3.Rohn WM, Lee YJ, Benveniste EN. Crit Rev Immunol. 1996;16:311–330. doi: 10.1615/critrevimmunol.v16.i3.40. [DOI] [PubMed] [Google Scholar]
- 4.Ting JP, Trowsdale J. Cell. 2002;109:S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 5.Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. Crit Rev Immunol. 1997;17:179–224. doi: 10.1615/critrevimmunol.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- 6.Meyer D, Thomson G. Ann Hum Genet. 2001;65:1–26. doi: 10.1046/j.1469-1809.2001.6510001.x. [DOI] [PubMed] [Google Scholar]
- 7.Garrigan D, Hedrick PW. Evolution (Lawrence, Kans) 2003;57:1707–1722. doi: 10.1111/j.0014-3820.2003.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 8.Krawczyk M, Reith W. Tissue Antigens. 2006;67:183–197. doi: 10.1111/j.1399-0039.2006.00557.x. [DOI] [PubMed] [Google Scholar]
- 9.Grusby MJ, Glimcher LH. Annu Rev Immunol. 1995;13:417–435. doi: 10.1146/annurev.iy.13.040195.002221. [DOI] [PubMed] [Google Scholar]
- 10.Mach B, Steimle V, Martinez-Soria E, Reith W. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 11.Glimcher LH, Kara CJ. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 12.Benoist C, Mathis D. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- 13.Morzycka-Wroblewska E, Munshi A, Ostermayer M, Harwood JI, Kagnoff MF. Immunogenetics. 1997;45:163–170. doi: 10.1007/s002510050185. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez S, Wassmuth R, Knerr I, Frank C, Haas JP. Eur J Immunogenet. 2003;30:141–148. doi: 10.1046/j.1365-2370.2003.00376.x. [DOI] [PubMed] [Google Scholar]
- 15.Indovina P, Megiorni F, Ferrante P, Apollonio I, Petronzelli F, Mazzilli MC. Hum Immunol. 1998;59:758–767. doi: 10.1016/s0198-8859(98)00082-2. [DOI] [PubMed] [Google Scholar]
- 16.de la Paz Bettinotti M, Kolek A, Brunnler G, Haas P, Paul C, Hochberger M, Bartova A, Kimura A, Sasazuki T, Albert ED. Eur J Immunogenet. 1993;20:339–407. doi: 10.1111/j.1744-313x.1993.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 17.Haas JP, Kimura H, Truckenbrodt H, Suschke J, Sasazuki T, Volgger A, Albert ED. Tissue Antigens. 1995;45:317–321. doi: 10.1111/j.1399-0039.1995.tb02460.x. [DOI] [PubMed] [Google Scholar]
- 18.Yao Z, Kimura A, Hartung K, Haas PJ, Volgger A, Brünnler G, Bönisch J, Albert ED. Immunogenetics. 1993;38:421–429. doi: 10.1007/BF00184522. [DOI] [PubMed] [Google Scholar]
- 19.Petronzelli F, Kimura A, Ferrante P, Mazzilli MC. Tissue Antigens. 1995;45:258–263. doi: 10.1111/j.1399-0039.1995.tb02449.x. [DOI] [PubMed] [Google Scholar]
- 20.Alaez C, Vazquez-Garcia MN, Gorodezky C. Genes Immun. 2001;2:216–221. doi: 10.1038/sj.gene.6363765. [DOI] [PubMed] [Google Scholar]
- 21.Brunnler G, Haas JP, Fan LA, Petzl-Erler ML, Volgger A, Yao Z, Wassmuth R, Albert ED, Middleton D, Barboni F, et al. In: Genetic Diversity of HLA: Functional and Medical Implication. Charron D, editor. France: EDK, Sevres; 1997. pp. 171–175. [Google Scholar]
- 22.Guardiola J, Maffei A, Lauster R, Mitchison NA, Accolla RS, Sartoris S. Tissue Antigens. 1996;48:615–625. doi: 10.1111/j.1399-0039.1996.tb02684.x. [DOI] [PubMed] [Google Scholar]
- 23.Buchan JC, Alberts S, Silk JB, Altmann J. Nature. 2003;425:179–181. doi: 10.1038/nature01866. [DOI] [PubMed] [Google Scholar]
- 24.Gaur LK, Shewey L, Sharkey-Mathis D, Nepom GT. Transgenics. 1994;1:267–276. [Google Scholar]
- 25.Gaur LK, Heise ER, Ting JPY. Immunogenetics. 1992;35:136–139. doi: 10.1007/BF00189524. [DOI] [PubMed] [Google Scholar]
- 26.Mikkelsen TS, Hillier LW, Eichler EE, Zody MC, Jaffe DB, Yang SP, Enard W, Hellmann I, Lindblad-Toh K, Altheide TK, et al. Nature. 2005;437:69–87. [Google Scholar]
- 27.Glazko GV, Nei M. Mol Biol Evol. 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- 28.Wildman DE, Uddin M, Liu GZ, Grossman LI, Goodman M. Proc Natl Acad Sci USA. 2003;100:7181–7188. doi: 10.1073/pnas.1232172100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreitman M, Hudson RR. Genetics. 1991;127:565–582. doi: 10.1093/genetics/127.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fay JC, Wyckoff GJ, Wu CI. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- 31.Klein J. Hum Immunol. 1987;19:155–162. doi: 10.1016/0198-8859(87)90066-8. [DOI] [PubMed] [Google Scholar]
- 32.Wiuf C, Zhao K, Innan H, Nordborg M. Genetics. 2004;168:2363–2372. doi: 10.1534/genetics.104.029488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlesworth D. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahata N. Proc Natl Acad Sci USA. 1990;87:2419–2423. doi: 10.1073/pnas.87.7.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernatchez L, Landry C. J Evol Biol. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- 36.Bontrop RE, Otting N, de Groot N, Doxiadis G. Immunol Rev. 1999;167:339–350. doi: 10.1111/j.1600-065x.1999.tb01403.x. [DOI] [PubMed] [Google Scholar]
- 37.Vallejo A, Pease L. J Immunol. 1995;154:3912–3921. [PubMed] [Google Scholar]
- 38.Page RDM, Charleston MA. Mol Phylogenet Evol. 1997;7:231–240. doi: 10.1006/mpev.1996.0390. [DOI] [PubMed] [Google Scholar]
- 39.Edwards SV, Chesnut K, Satta Y, Wakeland EK. Genetics. 1997;146:655–668. doi: 10.1093/genetics/146.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schierup MH, Mikkelsen AM, Hein J. Genetics. 2001;159:1833–1844. doi: 10.1093/genetics/159.4.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clark AG. Proc Natl Acad Sci USA. 1997;94:7730–7734. doi: 10.1073/pnas.94.15.7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asthana S, Schmidt S, Sunyaev S. Trends Genet. 2005;21:30–32. doi: 10.1016/j.tig.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 43.Hacia JG, Fan JB, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robbins CM, et al. Nat Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- 44.Dedrick RL, Jones PP. Mol Cell Biol. 1990;10:593–604. doi: 10.1128/mcb.10.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koch W, Benoist C, Mathis D. Mol Cell Biol. 1989;9:303–311. doi: 10.1128/mcb.9.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugawara M, Ponath PD, Shin J, Yang Z, Strominger JL. Proc Natl Acad Sci USA. 1991;88:10347–10351. doi: 10.1073/pnas.88.22.10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rockman MV, Wray GA. Mol Biol Evol. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- 48.Tan Z, Shon AM, Ober C. Hum Mol Genet. 2005;14:3619–3628. doi: 10.1093/hmg/ddi389. [DOI] [PubMed] [Google Scholar]
- 49.Liu X, Fu Y, Liu Z, Lin B, Xie Y, Liu Y, Xu Y, Lin J, Fan X, Dong M, et al. Am J Hum Genet. 2006;78:393–400. doi: 10.1086/500593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bamshad MJ, Mummidi S, Gonzalez E, Ahuja SS, Dunn DM, Watkins WS, Wooding S, Stone AC, Jorde LB, Weiss RB, et al. Proc Natl Acad Sci USA. 2002;99:10539–10544. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hudson RR, Kaplan NL. Genetics. 1988;120:831–840. doi: 10.1093/genetics/120.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahata N, Satta Y. Immunogenetics. 1998;47:430–441. doi: 10.1007/s002510050380. [DOI] [PubMed] [Google Scholar]
- 53.Satta Y, Li Y-J, Takahata N. Front Biosci. 1998;3:459–467. doi: 10.2741/a292. [DOI] [PubMed] [Google Scholar]
- 54.Grimsley C, Mather KA, Ober C. Mol Biol Evol. 1998;15:1581–1588. doi: 10.1093/oxfordjournals.molbev.a025886. [DOI] [PubMed] [Google Scholar]
- 55.Kelly JK, Wade MJ. J Theor Biol. 2000;204:83–101. doi: 10.1006/jtbi.2000.2003. [DOI] [PubMed] [Google Scholar]
- 56.Raymond CK, Kas A, Paddock M, Qiu RL, Zhou Y, Subramanian S, Chang J, Palmieri A, Haugen E, Kaul R, et al. Gemome Res. 2005;15:1250–1257. doi: 10.1101/gr.3554305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cereb N, Hughes AL, Yang SY. Immunogenetics. 1997;47:30–36. doi: 10.1007/s002510050323. [DOI] [PubMed] [Google Scholar]
- 58.Fu YG, Liu ZH, Lin JH, Chen WM, Jia ZJ, Pan DJ, Xu AL. Immunogenetics. 2003;54:761–766. doi: 10.1007/s00251-002-0523-z. [DOI] [PubMed] [Google Scholar]
- 59.Reusch TBH, Langefors A. J Mol Evol. 2005;61:531–541. doi: 10.1007/s00239-004-0340-0. [DOI] [PubMed] [Google Scholar]
- 60.Garrigan D, Edwards SV. Mol Biol Evol. 1999;16:1599–1606. doi: 10.1093/oxfordjournals.molbev.a026072. [DOI] [PubMed] [Google Scholar]
- 61.Cowell LG, Kepler TB, Janitz M, Lauster R, Mitchison NA. Gemome Res. 1998;8:124–134. doi: 10.1101/gr.8.2.124. [DOI] [PubMed] [Google Scholar]
- 62.Beaty JS, Sukiennicki T, Nepom GT. Microbes Infect. 1999;1:919–927. doi: 10.1016/s1286-4579(99)00225-7. [DOI] [PubMed] [Google Scholar]
- 63.Haas JP, Kimura A, Andreas A, Hochberger M, Keller E, Brunnler G, de la Paz Bettinotti M, Nevinny-Stickel C, Hildebrandt B, Sierp G, et al. Hum Immunol. 1994;39:31–40. doi: 10.1016/0198-8859(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 64.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 66.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 67.Huelsenbeck JP, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 68.Page RDM. Syst Biol. 1994;43:58–77. [Google Scholar]
- 69.Page RDM. Bioinformatics. 1998;14:819–820. doi: 10.1093/bioinformatics/14.9.819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.