Abstract
Whether the mate sampling and choice performed by females in nature influences offspring performance is a controversial issue in theory and an open empirical question. Pronghorn (Antilocapra americana) females engage in an obvious and energetically expensive mate sampling process to identify vigorous males. Although individual females sample independently, their choices converge on a small proportion of males that sire most young. Offspring of attractive males were more likely to survive to weaning and to age classes as late as 5 years, resulting in a selection differential, calculated by expected differences in lifetime number of offspring weaned, of 0.32 against random mating. Enhanced survival to weaning appeared to be accomplished by faster growth rates. Females compensated for matings with a less attractive mate by elevating rates of milk delivery to their young. Because pronghorn males do not have costly ornaments, we conclude that female choice for good genes can exist in the absence of ornaments. Furthermore, female choice may be important and unrecognized as a force that can lower population genetic load.
Keywords: female mate choice, pronghorn
The reality of active female mate sampling and mate choice is now unquestioned. In some species, females gain direct benefits, such as resources or safety, through the exercise of choice. However, in many other species, direct benefits are not detectable, and thus an indirect benefit, the creation of genetically superior offspring, is implicated. Several experimental studies on captive domestic or laboratory species reported indirect benefits (1–6), but a demonstration that the free choices of females in nature influence offspring survival by means of a sire effect has been a more elusive goal. Indeed, the magnitude of indirect effects, if they exist, is an open empirical question (7). Here, we show that the free mate choices that pronghorn (Antilocapra americana) females make by means of an energetically expensive mate sampling process have a strong effect on offspring survival.
We studied pronghorn at the National Bison Range, an enclosed 7,504-hectare National Wildlife Refuge in northwestern Montana. In this and other populations, all females 1 year of age and older enter estrus in September of each year and bear dizygotic twins the next May or June. In the 2 weeks preceding estrus, each female visits several potential mates that hold widely spaced harems. Males cannot coerce or force copulation and do not offer resources. Each female moves independently and switches between harems at an increasing rate as she approaches estrus, always leaving males that fail to show effective harem defense (8). Multiple repeat visits to the same male are common. This female sampling behavior is energetically expensive (9). As a consequence of independent female sampling, harem composition is fluid, and male yearly mating success is closely correlated with the yearly sum of harem sizes across rut (10). Female sampling produces a kind of winnowing process in which a small population subset of males gains most of the available copulations in any year. These are the males that, by sustained harem defense over many consecutive days, have demonstrated vigor (8). Our study was designed to test the hypothesis that this seemingly honest indicator of male quality was indeed a predictor of offspring quality.
Results
During the years of our study (2000–2003), all individual pronghorn in the National Bison Range population were ear-tagged or individually recognizable, and all were genotyped at 10 microsatellite loci. We captured fawns at a median age of 1 day, inserted plastic ear tags, retained pinna tissue from the hole punched for tags, weighed the fawn to the nearest 0.10 kg, and measured the length of the right tarsus to the nearest millimeter. We recorded dam identity of each fawn and assigned paternity genetically. Here, we confined analysis to 164 fawns for which the sire was assigned with strict confidence by using Cervus (11), which is the most appropriate paternity assignment program for our population (12).
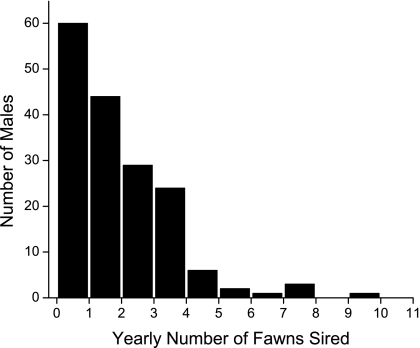
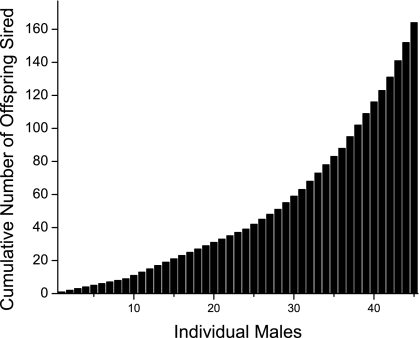
As predicted by behavioral observation, paternity in any rut year was highly skewed (Fig. 1), and consistency in the year-to-year choices of females created a small group of males (14 of 45) that, across the 4 years of our study, sired 59% of all fawns (Fig. 2). In analysis, each male's attractiveness was assigned as the total number of offspring that he sired across all 4 years.
Fig. 1.
The yearly distribution of number of offspring sired by the total population of breeding-age pronghorn males.
Fig. 2.
The cumulative number of offspring sired across the 4 study years by all males that sired at least one offspring. Each bar represents a single male.
To examine the effect of sire attractiveness on offspring survival, we used a GLLAM (general linear latent and mixed models) program (13) to construct a model of the effects of nested independent variables (female individual identity nested within sire attractiveness nested within birth year) on the binomial survival variable. Within the GLLAM program, we then used the model to predict individual offspring probability of survival.
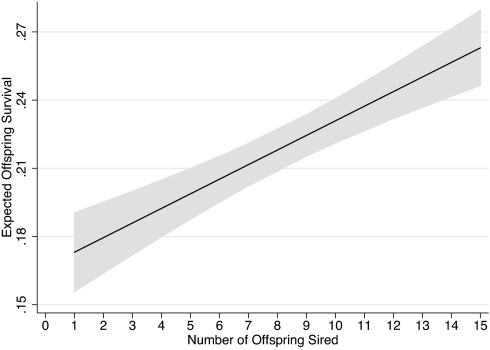
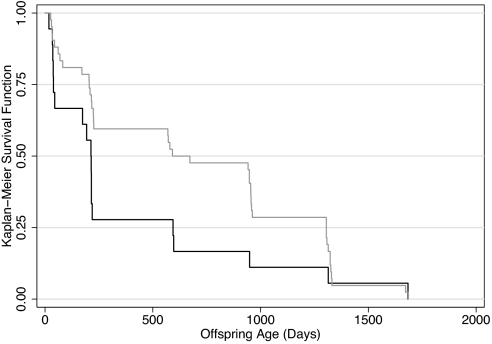
Offspring survival to weaning (12 weeks after birth, the age at which recruitment into the population is certain) was significantly associated with sire attractiveness (Fig. 3; F 1,166 = 36.1, P = 0.001, r2 = 0.18). Fawn death in this and in most North American populations is caused by coyote predation (10). Fawns become invulnerable to coyotes at ≈50 days of age and likely become so when they are large enough to sprint quickly. Thus, differences in growth rates of fawns could influence survival. We were able to measure skeletal growth (hind foot length) in offspring of eight preferred (attractiveness ≥5, the top 14 of 45 males) and four nonpreferred males. Offspring of preferred males had significantly faster growth (2.04 ± 0.12 mm per day versus 1.07 ± 0.10 mm per day: t = 2.52, P = 0.03). In addition, offspring of preferred males had greater probabilities of survival to age classes as far as year 5 (Fig. 4; Tarone–Ware log rank test, χ2 = 4.88, df = 1, P = 0.027).
Fig. 3.
Expected offspring survival to weaning (line indicates prediction; shaded area indicates 95% confidence interval) as a function of sire attractiveness as predicted by a generalized linear latent and mixed model program that removed the effects of maternal identity and birth year.
Fig. 4.
Kaplan–Meier survival functions for offspring of attractive (upper line) versus nonattractive (lower line) males. See text for the definition of attractiveness.
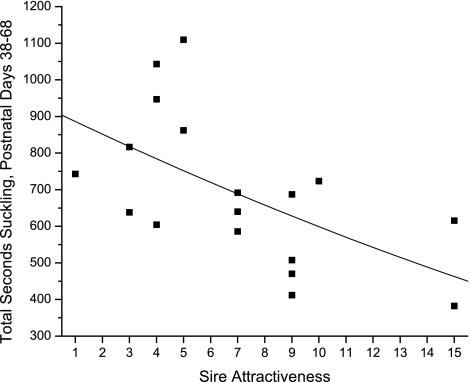
The differential allocation hypothesis (14) predicts that females that have mated with a preferred male should elevate their parental investment in offspring of that mating. Although we controlled for female identity in our analysis of fawn survival, we considered it prudent to check for differential allocation by the measurement of suckling rates of fawns. All observations of suckling were completed before paternities were assigned, so observers did not know the attractiveness of the sires of the fawns they observed. Our estimates of total seconds suckling during peak lactation, the period when mothers exercise complete control of suckling frequency and duration (10), showed that differential allocation did not occur (Fig. 5). In fact, suckling rate was negatively associated with sire attractiveness (F 116 = 7.92, P = 0.01, r2 = 0.33), a result that is predicted by the compensation hypothesis (15). This unexpected evidence for compensation yields further support for the conclusion that there is a strong sire effect on offspring performance.
Fig. 5.
Estimates of total suckling time of pronghorn fawns during peak lactation as a function of the total number of offspring sired in the 4-year study period by the sire.
Discussion
The magnitude of the sire effect on offspring survival creates a strong selective advantage for female mate sampling. We assigned fitness as expected lifetime number of offspring surviving to weaning and calculated fitness for a female that mated randomly and for a female that actively sampled and mated with a preferred male in each of the 12 bouts of reproduction expected in this population (10). Fitness for random mating was 3.53 and fitness for sampling and active choice was 5.15, yielding a selection differential of 0.32 against random mating. The size of the selection differential explains why energetically expensive mate sampling (9) is maintained in pronghorn.
Genetic quality in a sire may be viewed as genes that are compatible with individual maternal genotype or as genes that have a population wide advantage (16). The convergence of most females in all years on a small subset of males supports the latter interpretation in this population. Preferred males, those that have demonstrated impressive vigor under the relentless scrutiny of the female mate sampling process, likely are those with relatively low numbers of small-effect deleterious mutations (17). In support of this conclusion, preferred males did not have levels of expected heterozygosity (18) or mean d2 (19) different from those of nonpreferred males (percent loci heterozygous: 55.7 vs. 54.5, t = −0.21, df = 21, P = 0.83; mean d2: 15.0 vs. 12.9, t = −0.60, df = 19, P = 0.56). Thus, rather than a process that results in cumbersome male ornaments, female choice in pronghorn and likely in many other polygynous mammals may act as a force that reduces population genetic load. The male characteristics that are favored by females (running speed, endurance, agility, and tactical spatial sense) are essentially the same that were favored by the formidable suite of North American predators that shaped much of pronghorn biology (10). We suggest that there has been an overemphasis on female choice as a mechanism to explain male ornaments. For example, a recent review (20) used the following definition of good genes: “Models of sexual selection that assume extreme ornaments indicate the genetic quality of the bearer, defined as breeding value for fitness.” Our results show that females can use information other than that in ornaments to select genetically superior mates and raise the question of whether ornaments might represent the minority information gathering process. We suggest that that Trivers' (21) prediction, that the strength of female mate choice should be related to the degree of asymmetry between the sexes in reproductive expenditure in offspring, merits renewed attention. Female choice for good genes in a mate can exist in the absence of male ornaments, and the diversity of ways in which females evaluate genetic quality of mates remains unexplored.
Methods
Genotypes.
We extracted total genomic DNA using Qiagen (Valencia, CA) tissue kits. All samples were genotyped with a panel of 10 polymorphic microsatellite loci,: Aam1, Aam2, Aam5, Aam7, Aam9, Aam10, Aam11, Aam12 (22), ADCYC (22), and PRM605 (23). Amplification and screening procedures were as described by Carling et al. (24) with the exception that nine of the loci were combined into four PCR multiplexes (Aam 1-2, Aam 5-7, Aam 9-10-11, and ADCYC-PRM605). PCR products were visualized on an ABI 377 automated DNA sequencer (Applied Biosystems, Foster City, CA) and analyzed by using Genescan and Genotyper software. Ten percent of samples was reanalyzed to evaluate genotyping error rates by using the methods of Hoffman and Amos (25). Genotyping errors were also minimized by regenotyping all rare alleles and checking inheritance of alleles from mothers to known offspring. Each locus was evaluated for null alleles and deviations from Hardy–Weinberg equilibrium by using Cervus. All loci were in Hardy–Weinberg equilibrium with the exception of Aam-5 (P < 0.001). The estimated frequency of null alleles was <0.01 for all loci with the exception of Aam-5 (0.049) and Aam-2 (0.029).
Paternity.
In the four breeding seasons of our study, ≈510 fawns were born (twice the annual number of breeding-age females). We captured 262 fawns and obtained genotypes for each. In all but a few cases, fawns that we did not capture were those that died (likely killed by coyotes) before we could attempt capture. In Cervus, we entered all breeding age males (2 years of age and older) into the pool of potential sires, set the genotyping error rate at 0.01 (the rate that we measured by reamplification of genotypes of 40 randomly chosen individuals), and set strict confidence at 95%. There were 37, 41, 48, and 43 candidate sires during 2000–2003, and for the most part, the 4-year sample was of the same group of males (the median number of years that individual males were in the candidate pool was 3). Four years represents the entire reproductive lifespan of most pronghorn males (10). We considered paternity confirmed when an assignment was made with strict confidence and we found one or no mismatches between offspring genotype and parental genotypes. Using these standards, we were able to assign a sire to 164 offspring, or 63% of fawns captured and 32% of fawns born.
Growth Rates.
We measured growth of the tarsus by taking photographs of fawns when they stood broadside on a bare substrate. The 35-mm camera and 300-mm lens were fitted with a digital caliper that measured lens extension to 0.01 mm. We recorded fawn identity, age, and lens extension with each photograph. After we finished photographing fawns, we took a series of photographs of a meter stick at a range of lens extensions that comprised the range of those recorded in photographs of fawns. We then printed all photographs on 8- × 10-inch paper with the enlarger head locked at the same height. We scanned all photographs and entered them into Sigma Scan to make measurements. The image size of the meter stick at different lens extensions gave a scale. The image length of the fawn's tarsus divided by the scale gave the actual tarsus length. We subtracted birth tarsus length from the photographic measurement and divided the result by age to obtain growth rate in millimeter per day.
Suckling Rates.
As surviving fawns emerged from hiding, we assigned each to a focal sampling schedule. During focal sampling, we recorded each instance in which a fawn suckled and the duration of the suckling bout. We recorded data during 2,217 focal hours of observation of 34 fawns. We then selected the fawn records for which we had a minimum of 14 h of observation (mean was 32 h), collected between postnatal days 38 and 68. We plotted suckling rate (minutes of suckling per hour of observation) against age for each fawn, then fitted a logarithmic curve to the points, and found the integral of the logarithmic equation between ages 38 and 68. This process gave an estimate of total minutes suckling during the ages of peak lactation.
Acknowledgments
We thank K. Howard, C. Small, D. Vrancken, M. Tommasini, and P. Wiseman for field assistance and the U.S. Fish and Wildlife Service, which manages the National Bison Range, for cooperation. This work was supported by National Science Foundation Grant IBN 0097115 (to J.A.B.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Welch AM, Semlitsch RD, Gerhardt HC. Science. 1998;280:1928–1930. doi: 10.1126/science.280.5371.1928. [DOI] [PubMed] [Google Scholar]
- 2.Moller AP, Alatalo RV. Proc R Soc London Ser B. 1999;266:85–91. [Google Scholar]
- 3.Petrie M. Nature. 1994;371:598–599. [Google Scholar]
- 4.Evans JP, Kelley JL, Bisazza A, Finazzo E, Pilastro A. Proc R Soc London Ser B. 2004;271:2035–2042. doi: 10.1098/rspb.2004.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drickamer LC, Gowaty PA, Holmes CM. Anim Behav. 2000;59:371–378. doi: 10.1006/anbe.1999.1316. [DOI] [PubMed] [Google Scholar]
- 6.Bluhm CK, Gowaty PA. Anim Behav. 2004;68:977–983. [Google Scholar]
- 7.Kirkpatrick M, Barton NH. Proc Natl Acad Sci USA. 1997;94:1282–1286. doi: 10.1073/pnas.94.4.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byers JA, Moodie JD, Hall N. Anim Behav. 1994;47:33–43. [Google Scholar]
- 9.Byers JA, Wiseman PA, Jones L, Roffe TJ. Am Nat. 2005;166:661–668. doi: 10.1086/497401. [DOI] [PubMed] [Google Scholar]
- 10.Byers JA. American Pronghorn: Social Adaptations and the Ghosts of Predators Past. Chicago: Univ of Chicago Press; 1997. [Google Scholar]
- 11.Marshall TC, Slate J, Kruuk LEB, Pemberton JM. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- 12.Jones AG, Ardren WR. Mol Ecol. 2003;12:2511–2523. doi: 10.1046/j.1365-294x.2003.01928.x. [DOI] [PubMed] [Google Scholar]
- 13.Rabe-Hesketh S, Skrondal A, Pickles A. J Econometrics. 2005;128:301–323. [Google Scholar]
- 14.Sheldon BC. Trends Ecol Evol. 2000;15:397–402. doi: 10.1016/s0169-5347(00)01953-4. [DOI] [PubMed] [Google Scholar]
- 15.Bluhm CK, Gowaty PA. Anim Behav. 2004;68:985–992. [Google Scholar]
- 16.Neff BD, Pitcher TE. Mol Ecol. 2005;14:19–38. doi: 10.1111/j.1365-294X.2004.02395.x. [DOI] [PubMed] [Google Scholar]
- 17.Lynch M, Blanchard J, Houle D, Kibota T, Schultz S, Vassilieva L, Willis J. Evolution (Lawrence, KS) 1999;53:645–663. doi: 10.1111/j.1558-5646.1999.tb05361.x. [DOI] [PubMed] [Google Scholar]
- 18.Nei M. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coulson TN, Pemberton JM, Albon SD, Beaumont M, Marshall TC, Slate J, Guinness FE, Clutton-Brock TH. Proc R Soc London Ser B. 1998;265:489–495. doi: 10.1098/rspb.1998.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomkins JL, Radwan J, Kotiaho JS, Tregenza T. Trends Ecol Evol. 2004;19:323–328. doi: 10.1016/j.tree.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 21.Trivers RL. In: Sexual Selection and the Descent of Man, 1871–1971. Campbell B, editor. Chicago: Aldine; 1972. pp. 136–179. [Google Scholar]
- 22.Lou Y. College Station, TX: Texas A&M University; 1998. Ph.D. dissertation. [Google Scholar]
- 23.Stephen CL, Whittaker DG, Gillis D, Cox LL, Rhodes OE. J Wildl Manage. 2005;69:1463–1474. [Google Scholar]
- 24.Carling MD, Passavant CW, Byers JA. Mol Ecol Notes. 2003;3:10–11. [Google Scholar]
- 25.Hoffman JI, Amos W. Mol Ecol. 2005;14:599–612. doi: 10.1111/j.1365-294X.2004.02419.x. [DOI] [PubMed] [Google Scholar]