Abstract
Drosophila has a primitive yet highly effective innate immune system. Although the infection-dependent activation mechanisms of the Drosophila immune system are well understood, its inhibitory regulation remains elusive. To find novel suppressors of the immune system, we performed a genetic screening for Drosophila mutants with hyperactivated immune responses and isolated a loss-of-function mutant of caspar whose product is homologous to Fas-associating factor 1 in mammals. Interestingly, caspar mutant flies showed increased antibacterial immune responses including increased resistance to bacterial infection and a constitutive expression of diptericin, a representative antibacterial peptide gene. Conversely, ectopic expression of caspar strongly suppressed the infection-dependent gene expression of diptericin, which allowed bacterial outgrowth. Consistent with these physiological phenotypes, Caspar negatively regulated the immune deficiency (Imd)-mediated immune responses by blocking nuclear translocation of Relish, an NF-κB transcription factor. In addition, we further demonstrated that Dredd-dependent cleavage of Relish, a prerequisite event for the nuclear entry of Relish, is the target of the Caspar-mediated suppression of the Imd pathway. Remarkably, Caspar was highly specific for the Imd pathway and did not affect the Toll pathway, which is crucial for antifungal immunity. Collectively, our elucidation of an inhibitory mechanism of the Imd pathway by Caspar will provide a valuable insight into understanding complex regulatory mechanisms of the innate immune systems in both Drosophila and mammals.
Keywords: immune deficiency, Relish
Over the past decade, Drosophila has been developed as a highly attractive model for the study of innate immunity (1, 2). Drosophila is able to fight invading microorganisms via the innate immune reaction in at least three ways (3): a melanization response that exposes microbes to reactive oxygen; phagocytosis or encapsulation of invaders; and massive synthesis of cationic antimicrobial peptides. The synthesis of antimicrobial peptides is induced in the fat body (a functional analog of the mammalian liver) within a few hours after injury or microbial infection, and the synthesized peptides are immediately secreted into the insect hemolymph in high concentrations. Molecular genetics studies have shown that two distinct pathways, the immune deficiency (Imd) pathway and the Toll pathway, govern the infection-dependent synthesis of these antimicrobial peptides (1, 2, 4–6). The Imd pathway controls acute expression of most antibacterial peptide genes like diptericin through the mammalian p105/p110 NF-κB homolog Relish (7), whereas the Toll pathway regulates gene expression of antifungal peptides such as drosomycin through the mammalian p65 NF-κB homologs Dorsal and Dorsal-related immunity factor (DIF) (8).
The Imd pathway was initially characterized by a mutation in the imd gene, which blocks infection-dependent induction of antibacterial peptide genes and renders flies highly susceptible to Gram-negative bacterial infection (4, 6). Extensive genetic screening of Drosophila mutants with phenotypes similar to imd led to the identification of a number of other genetic components related to imd, and additional genetic and biochemical studies suggested a mechanistic model of signal flow in the Imd pathway that is similar to the mammalian TNF receptor signaling pathway (1, 2). The pattern recognition receptors (9) stimulated by peptidoglycans activate the Imd protein (4, 6), possibly through DTRAF2 (10). The Imd protein in turn activates the DmIKK complex, Ird5/Kenny (11, 12), through dTAK1 (13) and dFADD (14, 15). Subsequently, the DmIKK complex phosphorylates Relish (11, 16), and Dredd caspase cleaves Relish into the N-terminal Rel homology domain and the C-terminal inhibitory domain (17–19). This cleavage leads to nuclear translocation of the Rel homology domain, which is required for the induction of antibacterial gene transcription (18, 19).
Because host antibacterial–immune responses are necessary only with microbial infection, negative regulation of the immune signaling pathway is crucial for preventing deleterious side effects of unnecessarily induced or hyperactivated immune responses in an organism. Previously, Khush et al. (20) showed that Skp1/Cullin/F-box components in the ubiquitin–proteasome pathway have a repressive role for antibacterial immune responses in Drosophila, demonstrating the existence of a negatively regulating mechanism in the immune signaling pathway.
To uncover other inhibitory mechanisms of antibacterial immunity, we screened for Drosophila mutants with hyperactivated immune responses using GenExel (Taejon, Korea) EP lines and subsequently identified a mutant, caspar, which displayed a constitutive expression of antibacterial peptides and showed resistance to bacterial infection. Interestingly, there is a single homolog of Caspar in the mammalian genome, namely Fas-associating factor 1 (FAF1), which has been previously reported to associate with various components of the TNF/NF-κB signaling pathway, such as FAS, FADD, caspase-8, and NF-κB (21–23). However, the precise in vivo roles of FAF1 have remained elusive because of the absence of a genetic model. Here, using various biochemical and genetic techniques, we clearly demonstrated that Caspar specifically suppresses the Imd-mediated immune response by preventing Dredd-dependent nuclear translocation of Relish in Drosophila.
Results
Isolation of caspar Mutants.
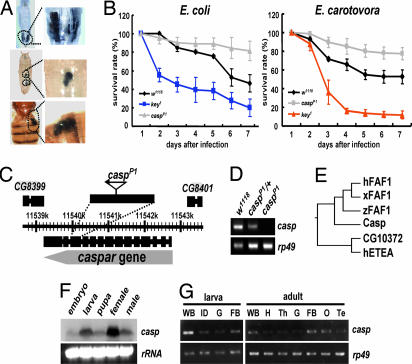
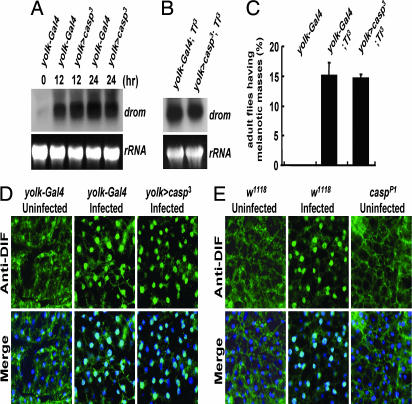
To discover suppressors of the Drosophila immune system, we screened mutants with ectopic melanization, which is generally observed in various mutants suffering from a hyperactivation of immune responses and is easily detectable (24, 25). Among 15,000 independent nonlethal EP lines from the GenExel library, we isolated a mutant line showing a high rate of melanization (15% for larvae, 14% for pupae, and 17.5% for adults) and named the putatively affected gene as “caspar” (named after one of the three Magi, “Caspar”) and the mutant allele as “casparP1.” The melanization of casparP1 flies was conspicuous around internal organs such as the gut and fat body (Fig. 1A).
Fig. 1.
Identification and characterization of caspar. (A) Ectopic melanization phenotypes of casparP1 larva (Top), pupa (Middle), and adult (Bottom). The areas of ectopic melanization are indicated by black dotted lines and are magnified in Right. (B) The survival rate of w1118, key1, and casparP1 adults at 1–7 days after E. coli (Left) or Er. carotovora (Right) infection. The survival rate of casparP1 mutants is significantly different from that of w1118 at 7 days after infection (P = 5.35 × 10−4 for E. coli infection, and P = 6.16 × 10−3 for Er. carotovora infection). (C) Schematic genomic organization of the caspar locus and transcripts. An EP-element in the casparP1 mutant (triangle) is inserted in the same direction as the caspar gene at the 11,540,319th base pair of Drosophila melanogaster chromosome 2R sequence (release v. 4.3). (D) caspar expression in w1118, casparP1/+ (caspP1/+), and casparP1/casparP1 (caspP1). The amount of caspar transcripts was visualized by RT-PCR between exons 13 and 14. rp49 was used as a loading control. (E) Molecular phylogenic tree of the Caspar family proteins from various species: hFAF1, Xenopus FAF1 (xFAF1), zebrafish FAF1 (zFAF1), Drosophila Caspar (Casp), and human ETEA (hETEA). Drosophila CG10372 (CG10372), previously inaccurately annotated as a fly FAF1, was revealed to be a Drosophila homolog of ETEA proteins, constituting a protein family distinct from FAF1s. (F) caspar expression at different stages of development. Northern blot analysis was performed by using a 600-bp DNA fragment encompassing the central portion of caspar coding region as probe. Ribosomal RNA (rRNA) was used as a loading control. (G) Tissue-specific distribution of caspar transcripts. The semiquantitative RT-PCR analyses of caspar were performed in various tissues. rp49 was used as a loading control. WB, whole body; ID, imaginal discs; G, gut; FB, fat body; H, head; Th, thorax; O, ovary; Te, testis.
casparP1 Is Resistant to Bacterial Infection.
To test whether casparP1 mutation enhances Drosophila immune responses against bacterial infection, we infected wild-type and casparP1 flies by pricking them with a needle dipped in a concentrated solution of Gram-negative bacteria Escherichia coli and examined their survival rates. Whereas wild-type flies showed a moderate decrease in viability upon infection, key1 flies, which carry a loss-of-function mutation in the gene encoding the Drosophila IKKγ homolog (kenny) (12), showed a dramatically decreased viability (Fig. 1B). However, strikingly, casparP1 mutants showed a much higher survival rate than wild-type flies under identical experimental conditions (Fig. 1B). To further confirm these results, we examined the survival rate upon infection using another type of Gram-negative bacteria, Erwinia carotovora, and obtained results similar to those of E. coli infection (Fig. 1B). From these observations, we deduced that casparP1 mutation enhances resistance against Gram-negative bacterial infection.
caspar Mutation Induces Constitutive Expression of diptericin.
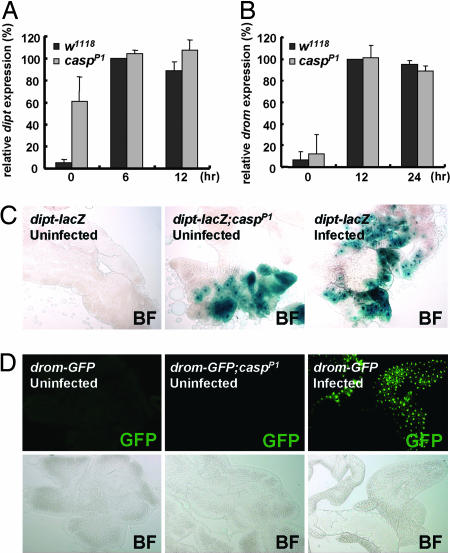
Previous studies showed that resistance of flies against bacterial infection tightly correlates with induction of antibacterial peptides in the fat body (1, 2, 6, 7). Therefore, we questioned whether caspar mutation elevates the expression of antibacterial peptide genes such as diptericin. To test this hypothesis, we monitored diptericin gene expression in uninfected and E. coli-infected flies by Northern blot analyses. Interestingly, uninfected casparP1 adults expressed diptericin modestly at ≈60% of infected wild-type flies (Fig. 2A). A similar level of diptericin expression was also observed in uninfected casparP1 larvae (Fig. 7, which is published as supporting information on the PNAS web site). However, we found no expression of diptericin in uninfected wild-type flies and heterozygous casparP1 mutants (Figs. 2A and 7).
Fig. 2.
Mutation in caspar induces constitutive expression of diptericin. (A and B) Northern blot quantifications of diptericin (dipt) (A) and drosomycin (drom) (B) expression in w1118 and casparP1 adult flies at indicated times after E. coli (A) and B. subtilis (B) infection. (C) Bright-field (BF) images of X-gal-stained larval fat bodies from flies with indicated genotypes and treatments. (D) Fluorescent (GFP) and bright-field (BF) images of larval fat bodies from flies with indicated genotypes and treatments.
To further confirm the constitutive expression of diptericin in casparP1 mutants, we used transgenic fly lines carrying a lacZ reporter gene fused with the diptericin promoter (diptericin–lacZ) (26). As previously reported (10, 27), the diptericin–lacZ reporter activity was dramatically increased by E. coli infection (Fig. 2C). Interestingly, consistent with our results obtained by Northern blot analysis (Fig. 2A), casparP1 mutants showed expression of the diptericin–lacZ reporter even in the absence of bacterial infection (Fig. 2C).
caspar Encodes a Drosophila Homolog of Mammalian FAF1.
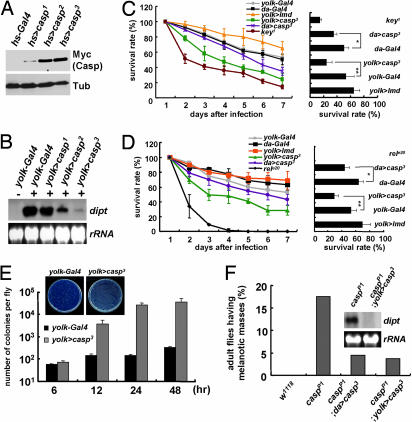
To study the physical nature of the conceptual caspar gene, we performed inverse PCR analysis using casparP1 genomic DNA. Subsequently, we found that casparP1 flies contain an EP-element insertion at the protein-coding sequence of the CG8400 gene (Fig. 1C) that may hamper the gene expression of CG8400. Expectedly, although the 5′ part of CG8400 transcripts was still present in the mutant (data not shown), the 3′ part of the transcripts downstream of the EP-element insertion site was completely abolished by casparP1 mutation (Fig. 1D), demonstrating the absence of a full-length transcription of CG8400 in the mutant. Moreover, casparP1/Df(2R)ED2457 flies carrying a transallelic combination of casparP1 and a CG8400 deficiency allele displayed phenotypes identical to those of casparP1 homozygote mutants (Fig. 8, which is published as supporting information on the PNAS web site). Furthermore, expression of the CG8400 transgene fully suppressed diptericin induction and melanization of uninfected casparP1 mutants (Fig. 3F), demonstrating that the casparP1 mutant phenotypes did indeed result from the loss of CG8400. Collectively, we concluded that CG8400 and caspar essentially refer to the same gene and that casparP1 is a loss-of-function allele of CG8400. Therefore, we named the uncharacterized gene CG8400 as caspar.
Fig. 3.
Ectopic expression of Caspar impairs antibacterial immunity. (A) hs--Gal4-driven expression of Caspar was analyzed by immunoblot analyses using anti-myc (Myc) and anti-tubulin (Tub) antibody. (B) Caspar suppresses diptericin transcription in a dose-dependent manner. Adult female flies after E. coli infection (+) or no infection (−) were subjected to Northern blot analysis for diptericin. rRNA was used as a loading control. (C and D) Survival rates of adult female flies with indicated genotypes at 1–7 days after E. coli (C) and Er. carotovora (D) infection. Bar graphs indicate survival rates at 7 days after infection (in C: ∗, P = 3.49 × 10−2; ∗∗, P = 2.48 × 10−2; in D: ∗, P = 1.50 × 10−2; ∗∗, P = 4.24 × 10−2). The viability of yolk>casp3 or da>casp3 flies was unaffected under uninfected conditions (Fig. 9, which is published as supporting information on the PNAS web site). (E) Bacterial colony counts were assayed from adult female flies at the indicated times after infection with ampicillin-resistant E. coli. Representative plates at 48 h after infection are shown. (F) Suppression of the casparP1 phenotypes by transgenic expression of Caspar. Adult female flies were analyzed for melanization frequency (presented as a bar graph) and for diptericin expression (Northern blot analysis in Inset). Genotypes of the flies are indicated.
Notably, the protein sequence of Caspar was highly similar to that of human FAF1 (hFAF1), and all of the domains previously identified in hFAF1 were conserved in Caspar (Fig. 10, which is published as supporting information on the PNAS web site). Furthermore, through phylogenic analyses of hFAF1 and related proteins, we confirmed Caspar to be the only Drosophila orthologue of hFAF1 (Fig. 1E).
caspar Expression Is Abundant in the Fat Body.
To examine the temporal expression pattern of caspar during Drosophila development, we performed Northern blot analysis using caspar-specific probes. A single ≈2.5-kb transcript was detected in all developmental stages of Drosophila (Fig. 1F). Next, we dissected the larval and adult tissues and subjected them to semiquantitative RT-PCR experiments. Interestingly, caspar transcripts were highly enriched in the fat bodies of larvae and adults (Fig. 1G). Because most antibacterial proteins are expressed predominantly in this organ (1, 2), the abundance of caspar transcripts in the fat body properly supports the notion that Caspar is involved in the immune responses of Drosophila.
Caspar Suppresses the Immune Responses Induced by Bacterial Infection.
To explore the molecular functions of Caspar, we generated flies carrying the caspar transgene. The level of Caspar expression varied among the transgenic alleles (Fig. 3A), allowing dose-dependent studies of Caspar overexpression. Consistent with the loss-of-function studies described above, overexpressed Caspar led to an inhibition of diptericin expression in a dose-dependent manner (Fig. 3B and Fig. 11, which is published as supporting information on the PNAS web site). From these results, we deduced that overexpression of Caspar may inhibit Drosophila immunity against bacterial infection. To prove this hypothesis, we examined whether Caspar overexpression would reduce the survival rate of infected flies. Interestingly, fat body-specific expression of Caspar by yolk–Gal4 and ubiquitous expression by da–Gal4 resulted in a decrease in the viability of flies upon E. coli or Er. carotovora infection, compared with flies overexpressing Imd or containing Gal4 alone (Fig. 3 C and D, respectively). These results were similar to those in key-null (key1) or relish-null (relishe20) flies, which are also highly susceptible to bacterial infection (7, 12) (Fig. 3 C and D, respectively). Furthermore, we observed bacterial colonies in the infected fly extracts to have exponentially increased in Caspar-overexpressing flies but not in control flies (Fig. 3E). Therefore, collectively, we concluded that Caspar inhibits Drosophila immune defenses against Gram-negative bacterial infection.
Caspar Inhibits Nuclear Localization of Relish.
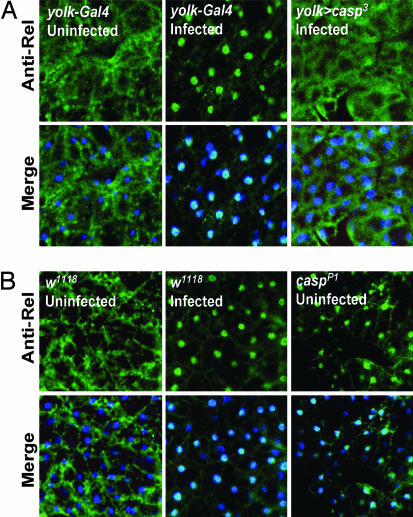
In the fat body, Relish transcription factor migrates into the nucleus upon bacterial infection, and this nuclear translocation is critical for the infection-dependent induction of antibacterial peptide genes such as diptericin (18, 19). Hence, we examined whether Caspar could suppress diptericin induction by preventing the nuclear migration of Relish. As previously reported (18), although Relish was predominantly cytoplasmic in uninfected conditions, bacterial infection strongly induced its nuclear localization in most cells of the fat body (Fig. 4). However, strikingly, overexpression of Caspar completely suppressed this infection-induced nuclear migration of Relish, restraining the protein in the cytoplasmic compartment (Fig. 4A). Conversely, fat body cells of casparP1 flies exhibited ectopic nuclear translocation of Relish even in uninfected conditions (Fig. 4B). Therefore, we concluded that Caspar suppresses the nuclear localization of Relish and the consequent antibacterial gene expression.
Fig. 4.
Caspar inhibits nuclear localization of Relish in Drosophila fat body cells. Fat bodies isolated from adult female flies with the indicated genotypes and treatments were analyzed by anti-N-terminal Relish immunostaining (green, Anti-Rel). DNA was visualized by Hoechst 33258 (blue) in merged images (Merge), showing the location of the nuclei. Fat droplets in these cells generated a mesh-like appearance in the cytoplasm.
Caspar Is Not Involved in the Toll Pathway.
Because Caspar inhibits antibacterial immune responses, we were curious whether Caspar could also inhibit the Toll pathway, an independent immune pathway that regulates antifungal as well as anti-Gram-positive bacterial immunity (1, 2). To address this idea, we examined the expression levels of drosomycin, a representative target of the Toll pathway, both in control and Caspar-overexpressing flies. As previously reported (5, 28), infection with Bacillus subtilis, Gram-positive bacteria, strongly activated drosomycin gene expression (Fig. 5A). Interestingly, overexpression of Caspar did not affect drosomycin expression (Fig. 5A), and casparP1 mutation also was unable to induce drosomycin in uninfected conditions (Figs. 2B and 7). Moreover, the drosomycin–GFP reporter (10, 27, 29) was not activated by casparP1 mutation (Fig. 2D), although it was prominently activated by bacterial infection (Fig. 2D). Furthermore, overexpression of Caspar failed to suppress the elevated transcription of drosomycin (Fig. 5B) as well as the ectopic melanization phenotype of a Tl3 mutant (Fig. 5C), whose Toll receptor contains a dominant gain-of-function mutation (5, 30, 31). Therefore, we concluded that Caspar does not affect the Toll pathway.
Fig. 5.
Caspar does not affect the Toll pathway. (A and B) Northern blot analyses of drosomycin expression in adult female flies with the indicated genotypes at indicated times after B. subtilis infection (A) or under uninfected conditions (B). rRNA was used as a loading control. (C) Melanization frequencies (%) in adult female flies with indicated genotypes. Error bars indicate standard deviations among three independent experiments. (D and E) Fat bodies isolated from adult female flies with the indicated genotypes and treatments were analyzed by anti-DIF immunostaining (green, Anti-DIF). DNA was visualized by Hoechst 33258 (blue) in merged images (Merge).
To further validate that Caspar is not involved in the Toll pathway, we also analyzed the subcellular localization of DIF, which migrates into the nucleus upon activation of the Toll signaling pathway and activates downstream genes such as drosomycin (8, 32). Consistent with the results described above, the nuclear localization of DIF was not affected by Caspar overexpression (Fig. 5D) or by casparP1 mutation (Fig. 5E), in stark contrast with the behavior of Relish (Fig. 4).
Caspar Suppresses the Imd Pathway.
It is well known that the Imd pathway regulates Relish nuclear translocation and diptericin induction, whereas the Toll pathway controls DIF nuclear localization and drosomycin expression (1, 2). Because Caspar specifically affected the localization of Relish and diptericin gene expression, we inferred Caspar to be involved in the Imd pathway.
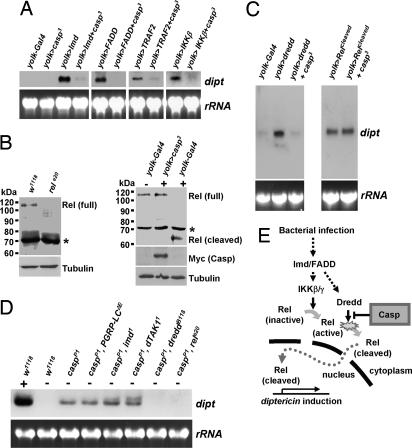
To substantiate the involvement of Caspar in the Imd pathway, we performed genetic interaction assays using transgenic flies expressing the components of the pathway. As previously reported (6), overexpression of Imd robustly induced diptericin gene expression (Fig. 6A). However, coexpression of Caspar dramatically inhibited the Imd-dependent induction of diptericin (Fig. 6A), showing that Caspar is capable of suppressing the Imd-dependent immune responses. Similarly, Caspar strongly suppressed diptericin expression induced by other components in the Imd pathway including FADD, TRAF2, and IKKβ (Fig. 6A), implying that Caspar-dependent inhibition of the Imd pathway occurs downstream of these molecules.
Fig. 6.
Caspar suppresses the Imd pathway by inhibiting Dredd-dependent cleavage of Relish. (A) Northern blot analyses of diptericin in uninfected adult female flies with indicated genotypes. rRNA was used as a loading control. (B) Immunoblot analyses of w1118, relish-null (relE20), yolk–Gal4, and yolk>caspar3 adult female flies using anti-N-terminal Relish antibody. The flies were infected (+) or kept uninfected (−) at 4 h before sampling. Asterisks indicate nonspecific bands. Transgenic Caspar was detected by anti-myc antibody. Tubulin was used as a loading control. (C) Uninfected adult female flies with indicated genotypes were subjected to Northern blot analyses of diptericin. rRNA was used as a loading control. (D) Northern blot analyses of diptericin in adult flies with indicated genotypes after E. coli infection (+) or no infection (−). (E) A schematic model of in vivo roles of Caspar in Drosophila.
Caspar Inhibits the Dredd-Dependent Cleavage of Relish.
At the end point of the Imd pathway, Dredd-dependent cleavage of Relish is essential for the nuclear localization of Relish (19). Therefore, we suspected that Caspar suppresses the Dredd-dependent processing of Relish to prevent its nuclear migration. To verify this possibility, we examined Relish using immunoblot analyses. In uninfected wild-type flies, a single 110-kDa band corresponding to a full-length Relish was detected (18) (Fig. 6B). Upon bacterial infection, as previously reported (18, 19), the 110-kDa band disappeared and instead a 68-kDa band corresponding to a cleaved form of Relish was newly detected (Fig. 6B). However, in the flies overexpressing Caspar, the infection-dependent cleavage of Relish was completely blocked (Fig. 6B). These results strongly suggested that Caspar suppresses the nuclear translocation of Relish by inhibiting the Dredd-mediated cleavage.
Interestingly, overexpression of Dredd alone has been reported to be sufficient for inducing antibacterial gene transcription in the absence of upstream infection signals (14) (Fig. 6C). To determine whether Caspar is indeed able to suppress Dredd activity, we examined diptericin expression in the flies coexpressing Dredd and Caspar. Remarkably, diptericin expression induced by Dredd overexpression was completely suppressed by Caspar (Fig. 6C). However, surprisingly, Caspar failed to suppress the diptericin expression induced by overexpression of the cleaved form of Relish (Fig. 6C), which localizes to the nucleus and induces antibacterial gene transcription independent of Dredd and other upstream signals (19, 20). These results demonstrated that Caspar specifically inhibits the Imd pathway by blocking Dredd-dependent proteolytic activation of Relish.
Finally, to investigate whether endogenous Caspar negatively regulates the Imd pathway, we generated double-mutant lines carrying both the casparP1 mutation and a mutation affecting the Imd pathway, and we monitored their diptericin expression. Intriguingly, although the mutation of PGRP-LC, imd, or TAK1 did not significantly affect diptericin gene induction caused by casparP1 mutation, the mutation of dredd or relish completely abolished it (Fig. 6D), showing that endogenous Caspar controls the activity of Dredd and Relish to suppress the Imd pathway. These genetic data further confirmed our conclusion that Caspar specifically inhibits Dredd-dependent cleavage of Relish (Fig. 6E).
Discussion
Through a screen for Drosophila mutants with hyperactivated innate immunity, we have identified an immune suppressor, Caspar. Because mutation of caspar increased the resistance to infection by Gram-negative bacteria (Fig. 1), we expected Caspar to be involved in an antibacterial defense reaction that depends on the Imd pathway. Indeed, casparP1 mutation induced a modest expression of the antibacterial peptide diptericin even in uninfected conditions (Fig. 2). Conversely, Caspar-overexpressing flies exhibited a strong suppression of diptericin induction, highly increasing the flies' susceptibility to bacterial infection (Fig. 3). These phenotypes were strikingly similar to those observed in the loss-of-function mutants for antibacterial immune signaling components (1, 2, 6, 17).
To dissect the molecular mechanism underlying the Caspar-dependent inhibition of Drosophila immunity, we examined the functional relationship between Caspar and the Imd pathway. Surprisingly, Caspar specifically suppressed nuclear localization of the Relish transcription factor (Fig. 4), and this suppression was exerted by inhibition of the Dredd-dependent cleavage of Relish (Fig. 6). These data also excluded the possibility that the diptericin induction by casparP1 mutation indirectly occurs by ectopic melanization (33). Because regulation of the Dredd-dependent cleavage of Relish is regarded as a critical step in the Imd pathway, further biochemical characterization of how Caspar inhibits the cleavage process will yield valuable insight into understanding the regulation of antibacterial signaling pathways in Drosophila.
Notably, Caspar contains multiple ubiquitin-related domains, namely ubiquitin-associated (UAS) domain and ubiquitin-like domain, which are conserved in hFAF1 (Fig. 10). Because hFAF1 regulates protein degradation via these ubiquitin-related domains (34), it is likely that Caspar performs similar biochemical activities. Moreover, mutations of some components in the ubiquitin–proteasome pathway, such as SkpA, dCullin, and Slimb (an F-box protein), have been shown to suppress the Imd pathway by controlling Relish activity (20). Therefore, it would be interesting to investigate whether the ubiquitin-related domains of Caspar are involved in suppression of the Dredd-dependent cleavage of Relish.
casparP1 mutants also showed interesting semilethal phenotypes, which might have resulted from deleterious side effects caused by abnormally activated immune responses in uninfected and quiescent situations. However, because the lethality was only slightly suppressed by a relish-null mutation (Fig. 12, which is published as supporting information on the PNAS web site), abnormal activation of the Imd pathway does not seem to be the sole cause of the semilethality in casparP1 mutants. Therefore, it would not be surprising to find that Caspar may have additional physiological roles that are independent of antibacterial immunity.
Because the loss of caspar is beneficial for efficient antibacterial immune reactions (Fig. 1), Caspar down-regulation could actually be advantageous for the survival of an infected organism. Intriguingly, we found decreased caspar transcript levels after bacterial infection (Fig. 13, which is published as supporting information on the PNAS web site), implying that, when the host is infected, the activity of Caspar is suppressed in vivo. More importantly, the reduced transcription level of caspar was maintained even 24 h after initial infection when antibacterial immune responses had almost completely silenced (5, 6, 35) (Fig. 13). From these results, we boldly suggest that Caspar is a feedback-immune regulator that enables an organism to be more sensitive to immune defense; initial infection induces a long-term silencing of caspar transcription to reinforce antibacterial immunity against additional bacterial infection that may follow.
Because Caspar and the NF-κB-dependent immune signaling pathways are highly conserved (1, 2), it is possible that the in vivo functions of Caspar revealed in this study are conserved in mammals as well. Furthermore, because previous studies on the negative regulation of the NF-κB pathway by FAF1 have been limited to biochemical and cell culture-based experiments (23), our current genetic studies using Drosophila will provide new insight into understanding the in vivo functions of mammalian FAF1 as well as negative regulation of mammalian innate immunity.
Methods
Fly Strains.
Stocks were raised on a standard cornmeal–yeast agar medium at 25°C. Each EP line carries a P-element containing Gal4-binding sites and a basal promoter oriented to direct expression of the genomic sequences downstream of its insertion site (36). For the screening, we scored homozygous phenotypes of 15,000 independent nonlethal EP lines from the GenExel's EP fly collection without using any Gal4 drivers. w1118 was used as wild type because it is the parental line of all of the GenExel EP lines. UAS-Imd, EP(X)DTRAF2, UAS-IKKβ, UAS–dredd, UAS–relish, Tl3, PGRP–LCΔE, dreddB118, imd1, TAK11, key1, relE20, diptericin–lacZ, drosomycin–GFP, hs–Gal4, da–Gal4, and yolk–Gal4 fly lines are described elsewhere (5–7, 10–12, 14, 17, 26, 29, 37). EP(3)FADD was purchased from GenExel. Transgenes were expressed by using the Gal4/UAS binary system. To generate UAS–caspar flies, caspar EST cDNA (Berkeley Drosophila Genome Project accession no. LD03368) was cloned into the myc-tagged pUAST vector and microinjected into w1118 embryos.
Infection Experiments.
Bacterial infection was carried out by pricking third-instar larvae or 7-day-old adults with a thin needle dipped into a concentrated solution of the following bacterial strains: wild-type E. coli, ampicillin-resistant E. coli, Er. carotovora, or B. subtilis. Survival experiments were performed with 50 flies at 25°C for each fly line grown in the same conditions. Surviving flies were transferred to fresh vials, and counts were taken every day for 7 days. For Figs. 1 B and 2 C and D, the average of three replicate experiments is presented as a graph, and the standard deviation is indicated as an error bar (5, 16). P values were calculated by one-way ANOVA. Examination of bacterial growth in flies was performed as previously described (5). The average of three replicate experiments is presented as a bar graph, and the standard deviation is indicated by an error bar.
Histology and Molecular Analyses.
Immunostaining of fat bodies was carried out as previously described (10) by using anti-N-terminal Relish (18) or anti-DIF (32) antibody. X-gal staining of fat bodies was performed as previously described (10). Immunoblot analysis was performed as previously described (38) by using anti-N-terminal Relish antibody. RT-PCR analysis was performed as previously described (38). Primer sequences for RT-PCR of caspar mRNA are 5′-GACGCGAGTCCATCAGATTAG-3′ (3′ region, forward), 5′-CAGCTTGAGCGACTCCAATG-3′ (3′ region, reverse), 5′-ATGTCAGAGAACAAGGACGAG-3′ (5′ region, forward), and 5′-GATGTGCGGCTGATTTAGGTTTAG-3′ (5′ region, reverse). Northern blot experiments and quantification of the signals on three independent blots were conducted as previously described (5). More specifically, signals of diptericin or drosomycin expression were normalized with a corresponding value of rRNA signals. The levels of diptericin in wild-type flies 6 h after infection and the levels of drosomycin in wild-type flies 12 h after infection were each standardized as 100, and the results are presented as relative activity in percent, in two separate graphs. For Fig. 2 A and B, the averages of three independent experiments are presented as a bar graph, and the standard deviations are indicated by error bars. Melanization frequencies were calculated in proportion of the number of flies with melanization mass to the total number of flies (n > 100) from three independent experiments under the same conditions. For Fig. 5C, the averages of three independent experiments are presented as a bar graph, and the standard deviations are indicated by error bars.
Supplementary Material
Acknowledgments
We thank Drs. B. Lemaitre (CNRS), Y. Engstrom (Stockholm University), J. Royet (Universite Louis Pasteur), W. J. Lee (Ewha Woman's University), D. Hultmark (Umeå University), K. Anderson (Sloan–Kettering Institute), M. Meister (Universite Paul Sabatier), S. Stoven (Umeå University), and the Bloomington Stock Center for providing fly stocks and antibodies.
Abbreviations
- Imd
immune deficiency
- DIF
Dorsal-related immunity factor
- FAF1
Fas-associating factor 1
- hFAF1
human FAF1
- UAS
ubiquitin-associated.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Brennan CA, Anderson KV. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann JA. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 3.Khush RS, Lemaitre B. Trends Genet. 2000;16:442–449. doi: 10.1016/s0168-9525(00)02095-3. [DOI] [PubMed] [Google Scholar]
- 4.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. Proc Natl Acad Sci USA. 1995;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 6.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA. Dev Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 7.Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, Hultmark D. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 8.Rutschmann S, Jung AC, Hetru C, Reichhart JM, Hoffmann JA, Ferrandon D. Immunity. 2000;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- 9.Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 10.Cha GH, Cho KS, Lee JH, Kim M, Kim E, Park J, Lee SB, Chung J. Mol Cell Biol. 2003;23:7982–7991. doi: 10.1128/MCB.23.22.7982-7991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y, Wu LP, Anderson KV. Genes Dev. 2001;15:104–110. doi: 10.1101/gad.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutschmann S, Jung AC, Zhou R, Silverman N, Hoffmann JA, Ferrandon D. Nat Immunol. 2000;1:342–347. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- 13.Vidal S, Khush RS, Leulier F, Tzou P, Nakamura M, Lemaitre B. Genes Dev. 2001;15:1900–1912. doi: 10.1101/gad.203301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leulier F, Vidal S, Saigo K, Ueda R, Lemaitre B. Curr Biol. 2002;12:996–1000. doi: 10.1016/s0960-9822(02)00873-4. [DOI] [PubMed] [Google Scholar]
- 15.Naitza S, Rosse C, Kappler C, Georgel P, Belvin M, Gubb D, Camonis J, Hoffmann JA, Reichhart JM. Immunity. 2002;17:575–581. doi: 10.1016/s1074-7613(02)00454-5. [DOI] [PubMed] [Google Scholar]
- 16.Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoven S, Ando I, Kadalayil L, Engstrom Y, Hultmark D. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, Maniatis T, Hultmark D. Proc Natl Acad Sci USA. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khush RS, Cornwell WD, Uram JN, Lemaitre B. Curr Biol. 2002;12:1728–1737. doi: 10.1016/s0960-9822(02)01214-9. [DOI] [PubMed] [Google Scholar]
- 21.Chu K, Niu X, Williams LT. Proc Natl Acad Sci USA. 1995;92:11894–11898. doi: 10.1073/pnas.92.25.11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu SW, Lee SJ, Park MY, Jun JI, Jung YK, Kim E. J Biol Chem. 2003;278:24003–24010. doi: 10.1074/jbc.M302200200. [DOI] [PubMed] [Google Scholar]
- 23.Park MY, Jang HD, Lee SY, Lee KJ, Kim E. J Biol Chem. 2004;279:2544–2549. doi: 10.1074/jbc.M304565200. [DOI] [PubMed] [Google Scholar]
- 24.Braun A, Hoffmann JA, Meister M. Proc Natl Acad Sci USA. 1998;95:14337–14342. doi: 10.1073/pnas.95.24.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peter A, Schottler P, Werner M, Beinert N, Dowe G, Burkert P, Mourkioti F, Dentzer L, He Y, Deak P, et al. EMBO Rep. 2002;3:34–38. doi: 10.1093/embo-reports/kvf012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reichhart JM, Meister M, Dimarcq JL, Zachary D, Hoffmann D, Ruiz C, Richards G, Hoffmann JA. EMBO J. 1992;11:1469–1477. doi: 10.1002/j.1460-2075.1992.tb05191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manfruelli P, Reichhart JM, Steward R, Hoffmann JA, Lemaitre B. EMBO J. 1999;18:3380–3391. doi: 10.1093/emboj/18.12.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fehlbaum P, Bulet P, Michaut L, Lagueux M, Broekaert WF, Hetru C, Hoffmann JA. J Biol Chem. 1994;269:33159–33163. [PubMed] [Google Scholar]
- 29.Ferrandon D, Jung AC, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, Reichhart J, Hoffmann JA. EMBO J. 1998;17:1217–1227. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu P, Pan PC, Govind S. Development (Cambridge, U.K.) 1998;125:1909–1920. doi: 10.1242/dev.125.10.1909. [DOI] [PubMed] [Google Scholar]
- 31.Lemaitre B, Meister M, Govind S, Georgel P, Steward R, Reichhart JM, Hoffmann JA. EMBO J. 1995;14:536–545. doi: 10.1002/j.1460-2075.1995.tb07029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ip YT, Reach M, Engstrom Y, Kadalayil L, Cai H, Gonzalez-Crespo S, Tatei K, Levine M. Cell. 1993;75:753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- 33.Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, Belvin M, Jiang H, Hoffmann JA, Reichhart JM. EMBO J. 2002;21:6330–6337. doi: 10.1093/emboj/cdf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song EJ, Yim SH, Kim E, Kim NS, Lee KJ. Mol Cell Biol. 2005;25:2511–2524. doi: 10.1128/MCB.25.6.2511-2524.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JH, Cho KS, Lee J, Yoo J, Chung J. Gene. 2001;271:233–238. doi: 10.1016/s0378-1119(01)00515-7. [DOI] [PubMed] [Google Scholar]
- 36.Tseng AS, Hariharan IK. Genetics. 2002;162:229–243. doi: 10.1093/genetics/162.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 38.Lee JH, Koh H, Kim M, Park J, Lee SY, Lee S, Chung J. Cell Death Differ. 2006;13:1110–1122. doi: 10.1038/sj.cdd.4401790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.