Abstract
The mRNA nuclear export receptor Mex67/Mtr2 is recruited to mRNAs through RNA-binding adaptors, including components of the THO/TREX complex that couple transcription to mRNA export. Here we show that the ubiquitin-associated (UBA) domain of Mex67 is not only required for proper nuclear export of mRNA but also contributes to recruitment of Mex67 to transcribing genes. Our results reveal that the UBA domain of Mex67 directly interacts with polyubiquitin chains and with Hpr1, a component of the THO/TREX complex, which is regulated by ubiquitylation in a transcription-dependent manner. This interaction transiently protects Hpr1 from ubiquitin/proteasome-mediated degradation and thereby coordinates recruitment of the mRNA export machinery with transcription and early messenger ribonucleoproteins assembly.
Keywords: nuclear export, THO complex
Concomitantly to their transcription, nascent transcripts are loaded with mRNA-binding proteins implicated in the processing and packaging of mRNA into stable and export competent messenger ribonucleoproteins (mRNPs). The production of mature mRNPs involves 5′ capping, splicing, and 3′ end cleavage/polyadenylation. All these cotranscriptional but biochemically distinct reactions are tightly coupled and coordinated by the RNA polymerase II C-terminal domain (CTD), which acts as a recruitment platform for the different processing machineries (1–5).
Fully mature and correctly packaged yeast mRNPs are released from the transcription site and transported into the cytoplasm by the heterodimeric export receptor Mex67/Mtr2 (or TAP/p15 in metazoan), which promotes their translocation through the nuclear pore complexes (NPC) by direct interactions with FG-nucleoporins (6). Mtr2 promotes the interaction of Mex67 with the NPC (7). The adaptor protein Yra1/REF contributes to mRNA export by facilitating the binding of Mex67 to the mRNP (8, 9). Yra1/REF and its partner Sub2 (UAP56 in metazoan) become associated with nascent transcripts during transcription elongation (10, 11). Yra1 and Sub2 copurify with THO, a tetrameric complex associated with the transcription machinery, to form the TREX complex proposed to link mRNA transcription and export (12). Constituents of THO include Hpr1, Tho2, Mft1, and Thp2 (12, 13), and evidence supports the notion that early recruitment of Sub2 is promoted through its direct interaction with Hpr1. Sub2 may then in turn facilitate the loading of Yra1 to mRNA (11, 12, 14, 15).
Loss of any of the four THO components impairs transcription elongation, genome stability, and mRNA export (12, 13). The current view is that THO primarily contributes to efficient mRNP assembly by promoting correct loading of mRNA-binding proteins. In THO mutants, the production of improperly packaged mRNP complexes results in the formation of DNA–RNA hybrids (R-loops), which interfere with RNA polymerase II processivity and transcription elongation and consequently increase transcription-dependent recombination events (16, 17). Furthermore, the absence of THO components induces retention and eventually degradation of malformed transcripts, i.e., heat-shock HSP104 mRNAs, at or close to the transcription site by nuclear surveillance mechanisms (18).
Previous studies have indicated that the ubiquitin pathway is involved in regulation of nuclear transport of both poly(A)+RNA and proteins (19). In particular, Tom1 and Rsp5, two ubiquitin ligases from the HECT family, have been shown to play a role in nuclear export of poly(A)+RNA in Saccharomyces cerevisiae (20–23). We recently reported that Hpr1, a component of the THO complex, is polyubiquitylated both in vitro and in vivo by Rsp5 before its degradation by the 26S proteasome. Hpr1 turnover, which is more active at 37°C, appeared linked to on-going RNA-polymerase II-dependent transcription, whereas the other members of the THO complex, such as Mft1 or Thp2, were not affected in similar conditions (24). Hpr1 thus represents a key factor whose stability controls the integrity and activity of the whole THO complex. Paradoxically, HPR1 deletion, but also Hpr1 stabilization by inactivation of Rsp5, correlated with a poly(A)+ RNA nuclear export defect, suggesting that tight control of both expression and active ubiquitin-dependent turnover of Hpr1 is required for proper transport function.
The large array of cellular processes involving ubiquitin modification is likely mediated through recognition of ubiquitin moieties by effectors containing ubiquitin-binding domains. Several families of ubiquitin-interacting motifs have been recently identified including UBA (ubiquitin associated), the most frequent ubiquitin-binding motif, UIM (ubiquitin-interacting motif), CUE (coupling of ubiquitin conjugation to endoplasmic reticulum), NZF (Npl4 Zn finger), and UEV (ubiquitin E2 enzyme variant) (reviewed in ref. 25). Structural and molecular features of UBA–ubiquitin interactions and ubiquitin linkage selectivity have been analyzed in a restricted number of UBA-containing proteins, exemplified by yeast Rad23 or its human ortholog hHR23A, leading to the general assumption that UBA domains preferentially interact with Lys-48-linked polyubiquitylated proteins (26–32). In contrast to this current view, a recent study on 30 distinct UBA motifs revealed that, in vitro, a UBA motif may or may not be selective for ubiquitin linkage but may also display no measurable affinity for ubiquitin (33). In addition, the idea emerges that the intramolecular environment of a UBA domain may influence the specificity of the UBA domain (33), but ubiquitin-binding domains described to date have not been reported to display any particular specificity for a ubiquitylated protein. Interestingly, the yeast mRNA export receptor Mex67 as well as its metazoan counterpart TAP harbor a UBA domain in their C terminus that participates in the interaction with FG nucleoporins at nuclear pores, whereas their N-terminal domain binds mRNP by RNA-binding adaptors (8, 9, 34–37). The UBA domain of Mex67 consists of the characteristic triple helix bundle plus an additional fourth helix. Mex67 is so far the only nuclear transport factor containing an ubiquitin-associating motif, but its role in connecting mRNA nuclear export to regulation by ubiquitin modification remains to be determined.
Here we show that Mex67 is recruited to transcribing genes, and that its UBA domain contributes to its recruitment. Lack of recruitment correlates with a defect in mRNA export. Our data show that the UBA domain of Mex67 not only recognizes ubiquitin and polyubiquitylated proteins but also physically interacts with Hpr1. In addition, an excess of UBA-Mex67 or the absence of UBA (Mex67-ΔUBA), respectively, induces a decrease or increase in the rate of degradation of Hpr1, consistent with a role for the UBA-Mex67 domain in transient protection of ubiquitylated Hpr1 from degradation by the 26S proteasome. The Mex67–Hpr1 interaction may contribute to the appropriate coordination of the different steps of mRNP biogenesis
Results and Discussion
To precisely characterize the function of the UBA domain of Mex67 (UBA-Mex67) in mRNA nuclear export, the subcellular distribution of mRNA was analyzed upon deletion or overexpression of the UBA-Mex67 domain. Strains were constructed that lacked the chromosomal MEX67 gene and expressed HA-tagged wild-type Mex67 (Mex67–3HA) or Mex67ΔUBA (mex67ΔUBA-3HA) from a plasmid. Western blotting with anti-HA antibodies confirmed that the wild-type (encoding amino acids 1–599) and mutant proteins (encoding amino acids 1–542) were expressed to comparable levels (not shown). In agreement with results reported in ref. 7, deletion of the UBA-Mex67 domain rendered cells thermosensitive for growth at 37°C (not shown). It should be noted that the Mex67ΔUBA protein used in this study lacked the C-terminal 75 amino acids, whereas Mex67ΔUBA-3HA lacks only the last 57 amino acids, strictly corresponding to the UBA domain. Consistent with its growth phenotype, the mex67ΔUBA-3HA strain displayed a weak mRNA export defect at 23°C [8% of the cells with nuclear accumulation of poly(A)+RNA], whereas poly(A)+RNA accumulated in the nucleus of 74% of the cells after a 1-h shift to the restrictive temperature. In contrast, only 1% of the Mex67–3HA cells presented nuclear accumulation of poly(A)+RNA under the same experimental conditions (Fig. 1A). Similarly, yeast cells overexpressing the Lex-UBA chimeric protein clearly accumulated poly(A)+RNA in their nucleus after a 2-h shift to 37°C compared with 23°C or to control cells (Fig. 1C). GAL1 mRNA, whose transcription was induced for 90 min in galactose at 23°C, mainly accumulated within a nuclear dot at or close to the site of transcription and was poorly detected in the cytoplasm of mex67ΔUBA-3HA cells, whereas this mRNA was distributed throughout Mex67–3HA cells with a detectable but much weaker dot staining, indicating a role for the UBA-Mex67 domain in the release and export of this specific transcript (Fig. 1B). These results indicate that the UBA domain is required for Mex67 to ensure proper nuclear export function. The more pronounced effect of UBA deletion on both cell growth and nuclear export at 37°C and the capacity of Mex67ΔUBA to bind nucleoporins in vitro (7) suggest the UBA domain may play a role distinct from its nuclear pore complex-targeting function.
Fig. 1.
The UBA domain of Mex67 contributes to its mRNA export activity. (A) Subcellular localization of poly(A)+ RNA was analyzed by FISH by using oligodT Cy3 as probe in Mex67–3HA or in mex67ΔUBA-3HA shuffle strains grown overnight at 23°C and then shifted to 37°C for 1 h. (B) GAL1 mRNA was analyzed by FISH by using a specific probe in Mex67–3HA or mex67ΔUBA-3HA shuffle strains after a 30-min induction in galactose at 23°C. (C) Cells expressing Lex or Lex-UBA Mex67 under the control of a galactose-inducible promoter were grown overnight in galactose containing medium at 23°C and then shifted to 37°C for 2 h before FISH analysis by using oligodT Cy3 as probe.
Mex67/Mtr2 is recruited to mature mRNPs through RNA-binding adaptors that interact with its N-terminal domain. To determine whether the UBA-Mex67 domain may function in an earlier more upstream event of mRNA biogenesis, the ability of Mex67 to be recruited to actively transcribed genes was analyzed by ChIP of the galactose-inducible GAL10 gene (Fig. 2A) and the constitutively expressed PMA1 gene in cells grown, respectively, in galactose and glucose (Fig. 2B). Our results show that Mex67 becomes associated with the PMA1 gene and with GAL10 when cells are grown in galactose, indicating that recruitment of Mex67 is transcription-dependent (Fig. 2). Mex67 was enriched in the middle of both PMA1 and GAL10 genes, showing an association profile similar to that observed earlier for the THO complex component Hpr1 and the mRNA adaptor Yra1 (Fig. 2 A and B; ref. 11). Interestingly, absence of the UBA domain resulted in the clear decrease of cotranscriptional recruitment of Mex67 all along GAL10 and PMA1 genes (Fig. 2 A and B). However, neither CTD nor Yra1 recruitment was significantly affected in the mutant, indicating that the reduced recruitment of Mex67-ΔUBA was not a consequence of a transcriptional defect, and that the cotranscriptional recruitment of Mex67–3HA was mainly because of its UBA domain (Fig. 2 A and B). Despite a normal recruitment of both CTD and Yra1 in mex67ΔUBA-3HA cells, we observed a decrease of Hpr1 on GAL10 gene in these cells, again suggesting that UBA-Mex67 could interfere with the function of some factors involved in the coupling between transcription and mRNA export (Fig. 2B and see below).
Fig. 2.
The UBA domain of Mex67 contributes to its cotranscriptional recruitment. (A) ChIP experiments on GAL10 gene were performed with extracts prepared from Mex67–3HA or mex67ΔUBA-3HA strains shifted to galactose for 2 h by using the indicated antibodies. (B) ChIP experiments on the PMA1 gene were performed with extracts prepared from Mex67–3HA or mex67ΔUBA-3HA strains by using antibodies against HA or CTD. Each immunoprecipitation was repeated at least from three different extracts. Error bars correspond to standard deviations.
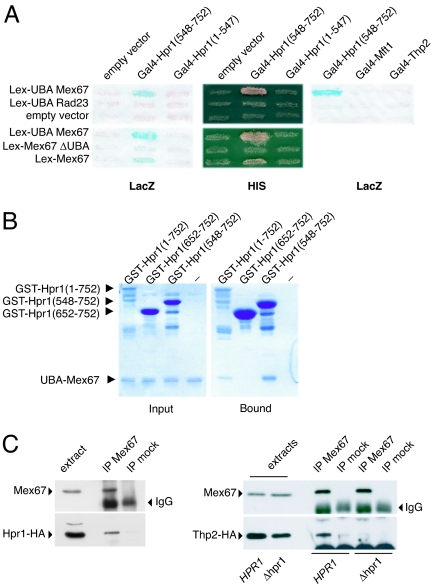
To dissect the mechanisms responsible for the roles of UBA-Mex67, partners of this domain were searched by using a two-hybrid screen. This strategy allowed the identification of a 203-aa C-terminal fragment of Hpr1 (amino acids 548–752) that interacted with UBA-Mex67, as well as with full-length Mex67. No interaction was observed with Mex67 lacking its UBA domain (Fig. 3A). The specificity of the interaction between Hpr1 and Mex67 was confirmed by using UBA-Rad23 as well as an Hpr1 molecule deleted of its C-terminal fragment [Hpr1 (1–547)]. Interestingly, no significant interaction was measured between UBA-Mex67 and Mft1 or Thp2, suggesting that the interaction between Mex67 and Hpr1 does not involve the other members of the THO complex (Fig. 3A). In agreement with the two-hybrid results, recombinant purified His-tagged UBA-Mex67 could interact in vitro with GST-Hpr1. This interaction occurred through the C-terminal fragment of Hpr1 [GST-Hpr1 (548–752)], corresponding to the two-hybrid fragment, whereas a smaller C-terminal fragment [Hpr1 (652–752)] was not sufficient to significantly bind UBA-Mex67 in vitro (Fig. 3B). Finally, coimmunoprecipitation was equally observed between Hpr1-HA and Mex67 and Thp2-HA and Mex67, indicating that Mex67 can interact with Hpr1 in the context of the THO complex in intact cells when expressed at physiological levels (Fig. 3C). However, no coimmunoprecipitation between Mex67 and Thp2-HA could be observed in absence of Hpr1 (Fig. 3C), thus demonstrating that this interaction is also mediated by Hpr1 in vivo. Together, these results clearly show that UBA-Mex67 promotes binding of Mex67 to Hpr1, whereas an unrelated UBA domain cannot provide this function.
Fig. 3.
The UBA domain of Mex67 interacts with Hpr1. (A) Two-hybrid analysis of the Lex-Mex67 baits vs. Gal4-Hpr1-derived preys. (B) Purified recombinant UBA-Mex67 and GST-Hpr1 fusion proteins were mixed (input) and complexes were analyzed after purification on glutathione-Sepharose beads (bound). (C) In vivo Mex67/THO complex interaction was analyzed by coimmunoprecipitating HA-tagged Hpr1 with anti-Mex67 antibodies or alternatively HA-tagged Thp2 with anti-Mex67 antibodies in Thp2-HA (HPR1) or Δhpr1/Thp2-HA (Δhpr1) strains. Preimmune serum (IP mock) was used as negative control.
Based on a few well-studied examples such as UBA-Rad23, UBA folds are believed to interact with and either expose or protect polyubiquitylated substrates to/from the 26S proteasome (38–40). However, a recent study of 30 distinct UBA domains revealed that ≈30% do not bind mono- or polyubiquitin chains (33). The ability of UBA-Mex67 to interact with ubiquitin moieties was therefore carefully analyzed both in vivo and in vitro. GST pull-down experiments by using the GST-Mex67/Mtr2 dimer revealed that the mRNA export receptor was able to bind polyubiquitylated cellular proteins accumulated upon treatment with the proteasome inhibitor MG132 (Fig. 4A). In vitro titration experiments indicate that GST-Mex67/Mtr2 clearly interacts with ubiquitin, with a higher affinity for tetraubiquitin (Kd = 5.9 +/− 0.7 μM) than for monoubiquitin (Kd = 22.7 +/− 4.2 μM), values comparable to those reported for other proteins harboring a UBA domain (33, 39). Importantly, deletion of the UBA domain abolished the ability of Mex67 to bind ubiquitin both in vivo and in vitro (Kd > 400 μM; Fig. 4A and Table 1). These results therefore demonstrate that Mex67 binds polyubiquitylated cellular proteins through its UBA domain.
Fig. 4.
The UBA domain of Mex67 interacts with polyubiquitin chains and prevents proteasome-mediated degradation of Hpr1. (A) Pull-down assays using the indicated GST fusion recombinant proteins and extracts from Δerg6 cells +/− MG132. (B) The stability of Hpr1-HA was analyzed in cells (YFS 1748) transformed with a pLex10-UBA-Mex67 or a pLex10-UBA-Rad23 plasmid and collected before (0) or after a shift at 37°C for 15 min in the absence (−CX) or presence (+CX) of cycloheximide (Left) or alternatively in Mex67–3HA or in mex67ΔUBA-3HA shuffle strains shifted to 37°C in the presence of cycloheximide for different incubation times (Right). (C) Ni-Purified 6His-ubiquitin conjugated forms of Hpr1-HA from cim3.1 cells transformed with pYEp96–6His-Ub and a p426ADH-Lex-UBA-Mex67 or a p426ADH-Lex-UBA-Rad23 plasmid and shifted to 37°C during 4 h. The purified material was examined by Western blotting with an anti-HA antibody.
Table 1.
Dissociation constants of Mex67-derived GST fusion proteins and Rad23 for mono- and tetraubiquitin
| Proteins | Kd for monoubiquitin, μM | Kd for tetraubiquitin, μM |
|---|---|---|
| Rad23-6His | 8.7 ± 1.4 | 1.7 ± 0.23 |
| GST-Mex67/Mtr2-6His | 22.7 ± 4.2 | 5.9 ± 0.7 |
| GST-Mex67ΔUBA/Mtr2-6His | >500 | 400–500 |
| Mtr2-6His | No binding | No binding |
The association with ubiquitin was measured in vitro by monitoring the fluorescence enhancement of fluorescently labeled ubiquitin.
We recently reported that Rsp5, a WW domain-containing ubiquitin ligase involved in the control of mRNA export (22, 24) polyubiquitylates Hpr1 before its degradation by the proteasome in a temperature- and transcription-dependent fashion. The lysine-rich C-terminal domain of Hpr1 is required not only for proper ubiquitylation and degradation of Hpr1 (24) but also for interaction with UBA-Mex67 (Fig. 3 A and B), suggesting that ubiquitylation of Hpr1 may influence its binding to UBA-Mex67, which might in turn affect Hpr1 degradation. To examine the potential effect of the UBA-Mex67 domain on Hpr1 turnover, Hpr1 levels were analyzed when the UBA domain was either absent or present in excess. The data show that loss of UBA-Mex67 resulted in faster degradation of Hpr1 after a shift to 37°C, whereas overexpression of Lex-UBA-Mex67 was able to protect Hpr1 from degradation at 37°C when compared with overexpression of the Lex-UBA-Rad23 fusion protein (Fig. 4B). Interfering with the interaction between Hpr1 and Mex67 thus clearly affects the degradation of Hpr1. To distinguish whether the interaction of Hpr1 with UBA-Mex67 prevents Hpr1 degradation by inhibiting Hpr1 ubiquitylation or rather by protecting the polyubiquitin chain from the proteasome, polyubiquitin-conjugated species of Hpr1 were analyzed upon overexpression of UBA-Mex67 or UBA-Rad23. For this purpose, His-6-tagged ubiquitin and UBA domains were overexpressed in cim3.1 temperature-sensitive mutants, impaired in proteasomal activity and grown at the restrictive temperature (24). Affinity purification followed by Western blot analysis showed that polyubiquitylated species of Hpr1 accumulated upon overexpression of UBA-Mex67 compared with UBA-Rad23 (Fig. 4C). In agreement with these results, we found that, in vitro, the affinity of UBA-Mex67 for ubiquitin was strongly increased upon binding to Hpr1 (M.H. N.I., C.G., F.S., G.D., et al., unpublished observations). The specificity of the interaction between UBA-Mex67 and Hpr1 thus allows UBA-Mex67 to interfere with the ubiquitin/proteasome-mediated degradation of Hpr1 by transiently protecting ubiquitylated Hpr1 from the 26S proteasome.
To determine whether the interaction between UBA-Mex67 and ubiquitylated Hpr1 not only affects Hpr1 turnover but also contributes to the cotranscriptional recruitment of Mex67 and mRNA export, these functions were analyzed in a mutant affected in Hpr1 ubiquitylation. The interaction between Rsp5 E3 ligase and Hpr1 most likely involves the recognition of the LPxY motif of Hpr1 (amino acids 335–338; Fig. 5A) by the second and third WW repeats of Rsp5 (22, 24). Indeed, mutation of tyrosine 338 to alanine (hpr1-Y338A) slowed down Hpr1 turnover at 37°C, confirming that ubiquitin-dependent degradation of Hpr1 is affected by a mutation in the Rsp5-binding motif LPxY (Fig. 5A). This partial block in Hpr1 ubiquitylation also resulted in a defect of GAL1 mRNA nuclear export, illustrated by a clear and reproducible accumulation of GAL1 mRNA within a marked nuclear dot and a lack of cytoplasmic staining after a 30-min shift to galactose at 30°C in a majority of hpr1-Y338A cells compared with WT cells (Fig. 5B). However, GAL1 transcripts were detected in the cytoplasm after longer time points, indicating that this partial defect most likely results from a delayed release from the transcription site rather than an export defect per se. Accordingly, this ubiquitylation defect led to a 40% decrease of the cotranscriptional recruitment of Mex67 to the GAL10 gene at 25°C. This effect did not result from a decreased transcription of the gene nor a defect in hpr1-Y338A recruitment or THO complex formation and recruitment, as measured by the association of CTD, Hpr1, and Thp2 respectively (Fig. 5C). Notably, the hpr1-Y338A mutation resulted in a slight increase of CTD and Hpr1 recruitment on GAL10, whereas recruitment of Thp2 remained unchanged, suggesting that Hpr1 ubiquitylation could influence the THO complex integrity to some extent. Together, these data are consistent with the RNA-independent early binding of Mex67 (41) and further support the view that ubiquitylation of Hpr1 facilitates cotranscriptional recruitment of Mex67 and mRNA export by mediating an interaction with the UBA domain of Mex67.
Fig. 5.
Ubiquitylation of Hpr1 is required for proper mRNA export and cotranscriptional recruitment of Mex67. (A) Mutation Y338A within the Rsp5-binding site of Hpr1 slowed down the turnover of Hpr1. (B) GAL1 mRNA localization was analyzed by FISH by using a specific probe in Hpr1wt or hpr1Y338A mutant strains after a 30-min induction in 2% galactose at 30°C. (C) The association of Mex67 with the 5′, middle, 3′, and 3′UTR regions of the GAL10 gene was analyzed by ChIP by using anti-Mex67, anti-CTD, anti-Hpr1, or anti-HA antibodies and extracts from Hpr1wt/Thp2-HA or hpr1Y338A/Thp2-HA cells induced with 2% galactose for 1 h at 25°C. Values correspond to the mean of three independent experiments.
Hpr1 is a key component of the THO complex, whose stability is likely to control the integrity and activity of the whole THO complex (12, 24). Because ubiquitylation of Hpr1 depends on active transcription, the UBA-Mex67-mediated recruitment of Mex67 and protection of Hpr1 are likely to occur during transcription itself and may contribute to increasing the local concentration of Mex67 in the vicinity of transcribed genes, and to coordinating THO complex metabolism and mRNA export. In other words, ubiquitin-mediated degradation does not appear to be the “raison d'être” of this modification but a consequence of the accomplished assembly/recruitment function. The mechanisms described here represent the first example of the role of polyubiquitylation in the coordination between transcription and nuclear export through the tight control of the transport machinery assembly, disassembly, and degradation. The involvement of at least another ubiquitin ligase Tom1 (20, 21), in the regulation of mRNA export, leads us to suspect that such a mechanism is not likely to be unique and could be used by eukaryotic cells to organize and control the chronology of concerted molecular events.
Experimental Procedures
Plasmids and Cloning.
See supporting information, which is published on the PNAS web site.
FISH Experiments.
DF5 strain transformed with p426GAL1 plasmid encoding the fusion protein Lex-UBA-Mex67 or the empty vector were grown in YEP/2% raffinose/0.02% glucose medium at 23°C. The expression of Lex-UBA protein was induced by addition of 2% galactose overnight at 23°C. At OD600 = 0.8 cells were shifted to 37°C for 2 h. These cells were then analyzed by FISH by using Cy3 labeled oligo dT (50) performed as described (22, 42). Poly(A)+ mRNA in situ hybridization in Mex67–3HA and mex67-ΔUBA-3HA strains was performed by using Cy3-labeled oligo dT (50) on cells grown at 23°C or heated for 1 h at 37°C. Results were quantified by counting, in a blind experiment, the number of cells accumulating poly(A)+ mRNA in the nucleus within a total of 100 cells per each condition. GAL1 mRNA in situ hybridization in Mex67–3HA and mex67-ΔUBA-3HA or in Hpr1 WT and hpr1Y338A strains was performed on cells treated for 90 min with galactose at 25°C or 30 min with galactose at 30°C, respectively, using six Cy3-internally labeled 50-mer oligonucleotide probes.
ChIP Analysis.
See supporting information.
Purification of 6His-Tagged Ubiquitin-Hpr1 Conjugates.
cim3.1 ts cells expressing Hpr1-HA were transformed with a plasmid encoding 6His-ubiquitin under the CUP1 promoter (43) and a plasmid-encoding Lex-UBA-Mex67 or Lex-UBA-Rad23 from the ADH promoter. Cells were grown on selective media supplemented with 0.1 mM of CuSO4 and shifted to 37°C for 4 h. Purification was performed essentially as described (24) from 50 OD600 of cells (2.3 mg of total proteins) and modified Hpr1 was detected by using anti-HA antibody (Babco, Evanston, IL).
Coimmunoprecipitation Experiments.
Yeast cells expressing Hpr1-HA or Thp2-HA were grown up to an OD600 = 1.2. Cells were harvested and lysed at 4°C with glass beads in ice-cold immunoprecipitation (IP) buffer. The lysate was centrifuged for 30 min at 13,000 × g. The supernatant was incubated with protein G-Sepharose beads (Amersham Pharmacia, Uppsala, Sweden) and anti-Mex67 or preimmune antibodies for 2 h at 4°C. Beads were then washed with IP buffer and bound proteins were eluted by heating samples at 95°C for 5 min in Laemmli sample buffer before Western blot analysis by using anti-Mex67 (24) or anti-HA antibodies (Babco).
Fluorescence Titration Experiments.
Binding of Mex67, Mex67ΔUBA, and Rad23 to ubiquitin and tetraubiquitin was monitored by steady-state fluorescence spectroscopy as described (27). Fluorescence titrations were performed in 20 mM Tris·HCl (pH: 7.2)/150 mM NaCl/2 mM EDTA/1% glycerol, at 25°C. Fluorescently labeled ubiquitin and tetraubiquitin were prepared and purified as described (27). A fixed concentration of fluorescently labeled ubiquitin or tetraubiquitin (1 μM) was titrated with increasing concentrations of Mex67, Mex67ΔUBA, Rad23, and Mtr2 proteins. Fluorescence was measured at 460 nm upon excitation at 340 nm. Upon binding to either Rad23 or Mex67, the fluorescence of mono- or tetraubiquitin increased by a factor of 2–3. The titration curves were fitted according to a quadratic equation by using Grafit Software (Erithacus Software, Surrey, UK) (44).
Supplementary Material
Acknowledgments
We thank E. Hurt (University of Heidelberg, Heidelberg, Germany) for plasmids, A. Jacquier (Institut Pasteur, Paris, France) for the yeast library, and M. Rosbash (Brandeis University, Waltham, MA) for the GAL1-specific oligonucleotide probes. We also thank R. Haguenauer-Tsapis, D. Libri, and M. Morris for critical reading of the paper and D. Lumbroso, A. De Georges, and A. Vitaliano for technical help. This study was funded by grants from the Association de Recherche Contre le Cancer, the Ministère de la Recherche), the Swiss National Science Foundation (SNF; Grant 102235, to F.S.), the SNF program “Frontiers in Genetics” (to F.S.), and the Canton de Genève. C.G. is supported by the Centre National de la Recherche Scientifique, and M.H. is supported by the Ministère Délégué à la Recherche.
Abbreviations
- UBA
ubiquitin-associated
- CTD
RNA polymerase II C-terminal domain
- mRNP
messenger ribonucleoprotein.
Footnotes
The authors declare no conflict of interest.
References
- 1.Jensen TH, Dower K, Libri D, Rosbash M. Mol Cell. 2003;11:1129–1138. doi: 10.1016/s1097-2765(03)00191-6. [DOI] [PubMed] [Google Scholar]
- 2.Vinciguerra P, Stutz F. Curr Opin Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Ares M, Jr, Proudfoot NJ. Cell. 2005;120:163–166. doi: 10.1016/j.cell.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Bentley DL. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Aguilera A. Curr Opin Cell Biol. 2005;17:242–250. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Reed R, Hurt E. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- 7.Strasser K, Bassler J, Hurt E. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strasser K, Hurt E. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei EP, Krebber H, Silver PA. Genes Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Mol Cell Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 13.Chavez S, Beilharz T, Rondon AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A. EMBO J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strasser K, Hurt E. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- 15.Abruzzi KC, Lacadie S, Rosbash M. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huertas P, Aguilera A. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Mason PB, Struhl K. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH. Mol Cell Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azad AK, Tani T, Shiki N, Tsuneyoshi S, Urushiyama S, Ohshima Y. Mol Biol Cell. 1997;8:825–841. doi: 10.1091/mbc.8.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Utsugi T, Hirata A, Sekiguchi Y, Sasaki T, Toh-e A, Kikuchi Y. Gene. 1999;234:285–295. doi: 10.1016/s0378-1119(99)00197-3. [DOI] [PubMed] [Google Scholar]
- 21.Duncan K, Umen JG, Guthrie C. Curr Biol. 2000;10:687–696. doi: 10.1016/s0960-9822(00)00527-3. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez MS, Gwizdek C, Haguenauer-Tsapis R, Dargemont C. Traffic. 2003;4:566–575. doi: 10.1034/j.1600-0854.2003.00115.x. [DOI] [PubMed] [Google Scholar]
- 23.Neumann S, Petfalski E, Brugger B, Grosshans H, Wieland F, Tollervey D, Hurt E. EMBO Rep. 2003;4:1156–1162. doi: 10.1038/sj.embor.7400026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gwizdek C, Hobeika M, Kus B, Ossareh-Nazari B, Dargemont C, Rodriguez MS. J Biol Chem. 2005;280:13401–13405. doi: 10.1074/jbc.C500040200. [DOI] [PubMed] [Google Scholar]
- 25.Hicke L, Schubert HL, Hill CP. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 26.Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 27.Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. J Mol Biol. 2001;313:955–963. doi: 10.1006/jmbi.2001.5105. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Shinde U, Ortolan TG, Madura K. EMBO Rep. 2001;2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 30.Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Proc Natl Acad Sci USA. 2002;99:745–750. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohno A, Jee J, Fujiwara K, Tenno T, Goda N, Tochio H, Kobayashi H, Hiroaki H, Shirakawa M. Structure (London) 2005;13:521–532. doi: 10.1016/j.str.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 33.Raasi S, Varadan R, Fushman D, Pickart CM. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 34.Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, et al. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suyama M, Doerks T, Braun IC, Sattler M, Izaurralde E, Bork P. EMBO Rep. 2000;1:53–58. doi: 10.1093/embo-reports/kvd009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grant RP, Neuhaus D, Stewart M. J Mol Biol. 2003;326:849–858. doi: 10.1016/s0022-2836(02)01474-2. [DOI] [PubMed] [Google Scholar]
- 38.Chen L, Madura K. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raasi S, Pickart CM. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- 40.Glockzin S, Ogi FX, Hengstermann A, Scheffner M, Blattner C. Mol Cell Biol. 2003;23:8960–8969. doi: 10.1128/MCB.23.24.8960-8969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dieppois G, Iglesias N, Stutz F. Mol Cell Biol. doi: 10.1128/MCB.00870-06. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R, Dargemont C. Nat Cell Biol. 2003;5:661–667. doi: 10.1038/ncb1003. [DOI] [PubMed] [Google Scholar]
- 44.Heitz F, Morris MC, Fesquet D, Cavadore JC, Doree M, Divita G. Biochemistry. 1997;36:4995–5003. doi: 10.1021/bi962349y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.