Abstract
An N-ethyl-N-nitrosourea mutagenesis screen in mice was performed to isolate regulators of circulating platelet number. We report here recessive thrombocytopenia and kidney disease in plt1 mice, which is the result of a severe but partial loss-of-function mutation in the gene encoding glycoprotein-N-acetylgalactosamine-3-β-galactosyltransferase (C1GalT1), an enzyme essential for the synthesis of extended mucin-type O-glycans. Platelet half-life and basic hemostatic parameters were unaffected in plt1/plt1 mice, and the thrombocytopenia and kidney disease were not attenuated on a lymphocyte-deficient rag1-null background. gpIbα and podocalyxin were found to be major underglycosylated proteins in plt1/plt1 platelets and the kidney, respectively, implying that these are key targets for C1GalT1, appropriate glycosylation of which is essential for platelet production and kidney function. Compromised C1GalT1 activity has been associated with immune-mediated diseases in humans, most notably Tn syndrome and IgA nephropathy. The disease in plt1/plt1 mice suggests that, in addition to immune-mediated effects, intrinsic C1Gal-T1 deficiency in megakaryocytes and the kidney may contribute to pathology.
Keywords: core 1 galactosyltransferase, N-ethyl-N-nitrosourea mutagenesis, nephropathy, platelet
To explore the molecular regulation of platelet production in health and disease, we have used large-scale mutagenesis to create mouse models of thrombocytopenia. N-ethyl-N-nitrosourea (ENU) is a potent, random mutagen of the mouse germ line that induces heritable DNA changes (1), and breeding schemes can be established that allow mice with dominant and recessive phenotypes to be identified and the causative mutations to be genetically mapped and positionally cloned. While positional cloning has traditionally been a bottleneck in genetic screens, the recent elucidation of the mouse and human genome sequences has dramatically simplified mutation identification. Thus, genetic screens are feasible in the mouse and are a powerful tool in the dissection of diverse biological systems (2–4). We have demonstrated the utility of ENU mutagenesis in the analysis of platelet regulation with the isolation of unique mutations in the gene encoding c-Myb that result in thrombocytosis (5).
Here we describe recessively inherited thrombocytopenia and kidney disease in plt1 mice, which have an ENU-induced point mutation in the gene encoding glycoprotein-N-acetylgalactosamine-3-β-galactosyltransferase [core1-β1,3-galactosyltransferase, C1GalT1 (EC 2.4.1.122)]. C1GalT1 generates the core1 disaccharide O-glycan Galβ1–3GalNAcα1-Ser/Thr, also known as the T antigen, by addition of Gal to GalNAcα1-Ser/Thr (the Tn antigen), which is initially generated by the action of N-acetylgalactosaminyltransferases on specific Ser or Thr residues of target proteins. The core1 disaccharide is the precursor for the synthesis of many of the extended mucin-type O-glycans found on mammalian glycoproteins (6). Compromised C1GalT1 activity has been associated with disease in humans, most notably Tn syndrome, a rare hematological disorder characterized by reduced numbers of blood cells (7), and IgA nephropathy, a common primary glomerulonephritis (8). Embryonic lethality in knockout mice lacking C1GalT1 (9) has prevented the development of effective animal models of C1GalT1 deficiency in adult physiology and disease. plt1 mice, in which C1GalT1 is expressed with very low residual enzymatic activity, reveal indispensable roles of C1GalT1 in thrombopoiesis and kidney homeostasis.
Results
The plt1 Mouse.
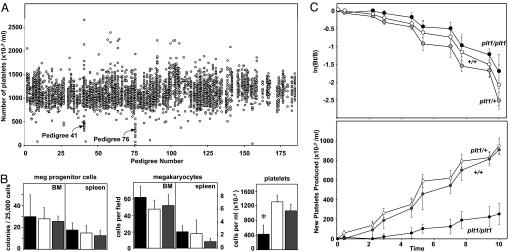
A mutagenesis screen was performed in which C57BL/6 mice were treated with ENU and bred to generate pedigrees of third-generation (G3) mice. In two G3 pedigrees multiple individuals displayed low platelet counts (pedigrees 41 and 76; Fig. 1A), consistent with segregation of ENU-induced mutations causing thrombocytopenia. Pedigree 41 has not been pursued, but offspring of matings between affected pedigree 76 mice and WT C57BL/6 mice all had normal platelet counts, whereas intercrosses yielded thrombocytopenic mice at an approximate frequency of one in four, consistent with segregation of a recessively acting mutation in pedigree 76, which we termed plt1.
Fig. 1.
Recessive thrombocytopenia in plt1 mice. (A) Platelet counts from pedigrees of G3 mice segregating ENU-induced mutations. Each data point represents the platelet count from an individual mouse with data from mice of the same pedigree grouped vertically. Two pedigrees in which thrombocytopenia segregated were identified, pedigrees 41 and 76, the latter being subsequently referred to as plt1. (B) Homozygous plt1/plt1 mice exhibit thrombocytopenia without deficiencies in megakaryocyte or megakaryocyte progenitor cell numbers in the bone marrow and spleen. Means ± standard deviations are shown for numbers of megakaryocyte progenitor cells, n = 2–5; megakaryocytes, n = 6; and platelets, n = 15–22 mice per genotype. ∗, P < 0.01 for comparison of data from plt1/plt1 mice with that from plt1/+ and +/+ mice. (C) Reduced rate of platelet production in plt1/plt1 mice. (Upper) The decline in the numbers of biotinylated platelets after pulse labeling in vivo is shown with data expressed as the logarithm of the ratio of the number of labeled platelets at a given time over the initial number of labeled platelets (ln[Bt/B]). (Lower) Rate of appearance of unlabeled platelets after pulse labeling in vivo, a measure of new platelet production. Data from plt1/plt1 mice are shown in black, with that from plt1/+ and +/+ mice shown in white and gray, respectively.
Megakaryocytopoiesis and Platelet Turnover in plt1/plt1 Mice.
Platelet counts in plt1/plt1 mice were 40% of those observed in WT mice and were accompanied by increased platelet volume (Fig. 1B and Table 1). The hematocrit and numbers of circulating white blood cells were normal in plt1/plt1 mice (Table 1). There was no reduction in megakaryocyte numbers in plt1/plt1 mutant bone marrow or spleen compared with WT or plt1/+ mice, and numbers of megakaryocyte progenitor cells and progenitor cells for other myeloid lineages were unaffected (Fig. 1B and data not shown). Megakaryocytes from plt1/plt1 mice demonstrated a DNA ploidy distribution similar to WT or mildly shifted to higher values, and electron microscopic examination of megakaryocytes and platelets from plt1/plt1 mice suggested no major obvious ultrastructural abnormalities (data not shown).
Table 1.
Hematological profile of plt1 mutant mice
| Peripheral blood | Genotype |
|||||
|---|---|---|---|---|---|---|
| +/+,n = 18 | plt1/+,n = 20 | plt1/plt1,n = 15 | rag1−/− +/+,n = 5 | rag1−/−plt1/+,n = 9 | rag1−/−plt1/plt1, n = 5 | |
| Platelets, ×10−6 per ml | 1,265 ± 242 | 1,305 ± 188 | 522 ± 255* | 1,363 ± 253 | 1,394 ± 189 | 461 ± 320‡ |
| Platelet volume, fl | 8.4 ± 0.5 | 8.5 ± 0.3 | 10.0 ± 1.5* | 7.6 ± 0.7 | 7.4 ± 0.5 | 9.2 ± 0.9 |
| Hematocrit, % | 55.4 ± 1.7 | 56.3 ± 1.9 | 55.2 ± 4.2 | 52.7 ± 2.2 | 55.4 ± 2.2 | 54.9 ± 2.6 |
| White blood cells, ×10−3 per ml | 9.0 ± 1.5 | 9.4 ± 1.9 | 11.1 ± 2.3 | 1.8 ± 0.5† | 1.5 ± 0.4† | 1.2 ± 0.9† |
| Neutrophils | 0.9 ± 0.2 | 0.8 ± 0.2 | 0.8 ± 0.3 | 0.6 ± 0.3 | 0.6 ± 0.2 | 0.4 ± 0.5 |
| Lymphocytes | 7.7 ± 1.5 | 8.1 ± 1.7 | 9.6 ± 2.1 | 0.8 ± 0.1† | 0.7 ± 0.3† | 0.5 ± 0.3† |
| Monocytes | 0.05 ± 0.03 | 0.05 ± 0.03 | 0.04 ± 0.02 | 0.05 ± 0.02 | 0.04 ± 0.02 | 0.02 ± 0.02 |
| Eosinophils | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.2 ± 0.1 | 0.1 ± 0.03 | 0.1 ± 0.1 |
Means ± standard deviations are shown. P < 0.05 for comparison of plt1/plt1 with +/+ mice (∗), rag−/− data with that of rag+/+ mice of a given plt1 genotype (†), and rag1−/− plt1/plt1 with rag1−/− +/+mice (‡) (Student's t test with P values adjusted for multiple testing).
In vivo labeling studies revealed that the half-life of platelets in plt1/plt1 mice was similar to control mice. However, the generation of unlabeled platelets after pulse labeling occurred at a slower rate (Fig. 1C). Thus, the thrombocytopenia in plt1/plt1 mice is not caused by impaired megakaryocyte production or accelerated clearance of platelets, but appears caused by compromised generation of platelets from megakaryocytes.
plt1/plt1 Mice Develop Kidney Disease.
Young adult plt1/plt1 mice were smaller than control mice (plt1/plt1 males, 15.4 ± 3.1 g and females, 14.4 ± 0.7 versus 24.0 ± 1.7 and 18.1 ± 0.7, respectively, for WT). All major organs in plt1/plt1 mice were histologically normal with the exception of the kidney, which displayed lesions affecting clusters of glomeruli and their corresponding proximal tubules; the latter showed fatty degeneration and contained protein casts. The glomerular-tubular architecture was distorted, and often there were infiltrating inflammatory cells in affected foci. The Bowman's capsule, normally a single layer of cells, was often two to four cells thick in plt1/plt1 mice, but there were no deposits in the glomerular vessels. plt1/plt1 mice displayed abnormally high levels of urinary protein from an early age, which mass spectrometric analysis revealed to be predominantly serum albumin (data not shown), consistent with compromised renal function.
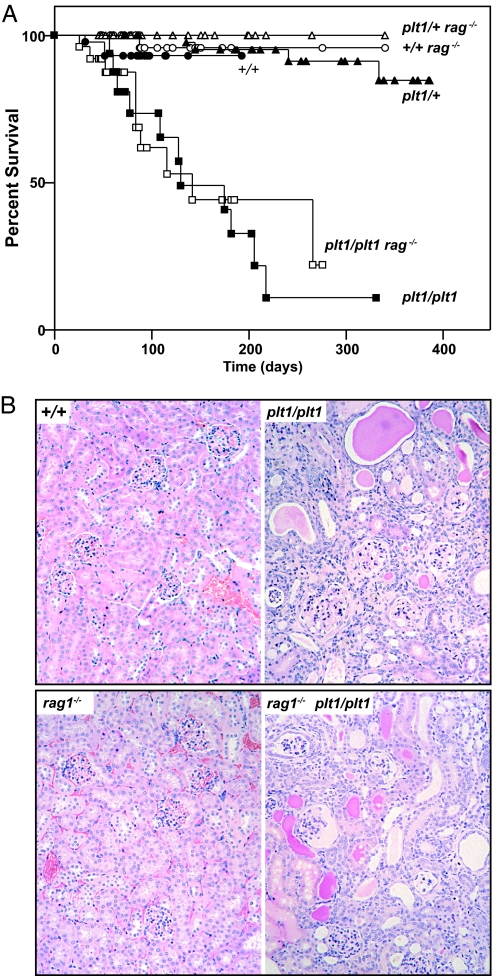
plt1/plt1 mice became ill from ≈10 weeks of age, and 90% were moribund by 200 days (Fig. 2A). The kidneys of sick mice exhibited cortical atrophy, glomerular loss, inspissated ducts, and inflammatory infiltration (Fig. 2B), and serum from sick plt1/plt1 mice contained excessive levels of urea (plt1/plt1 169 ± 33 mmol/liter versus WT 9 ± 2) and creatinine (plt1/plt1 200 ± 14 mmol/liter versus WT 47 ± 4).
Fig. 2.
plt1/plt1 mice develop kidney disease. (A) Mortality in plt1/plt1 mice. (B) Histological sections of kidneys from a sick plt1/plt1 or plt1/plt1 rag1−/− mouse and healthy controls illustrating the extensive inflammatory infiltration, loss of glomeruli, and tubular disorganization in the plt1/plt1 kidneys.
To determine whether immune mechanisms contributed to disease, plt1/plt1 mice were crossed with rag1−/− mice, which lack mature B and T cells. plt1/plt1 rag1−/− mice exhibited thrombocytopenia and high platelet volume (Table 1) and proteinuria and renal lesions characteristic of plt1/plt1 rag1+/+ mice and succumbed to disease with similar kinetics (Fig. 2).
plt1 Is a Mutation in the Gene Encoding Core1-β1,3-Galactosyltransferase.
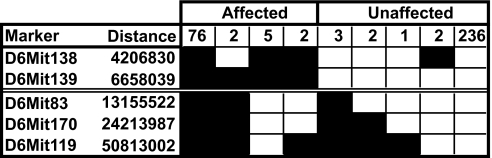
Genetic mapping revealed that the mutation causing the plt1 phenotype was located in a region of ≈6.5 Mb on chromosome 6 between markers D6Mit139 and D6Mit83 (Fig. 3), a region that contained a number of putative and known genes. cDNA was extracted from plt1/plt1 and WT organs, and the protein-encoding region of each of 12 candidate genes was amplified and sequenced. Sequences derived from plt1/plt1 mice were identical with those from WT mice with the single exception of the gene encoding C1GalT1. In this case, a mutation in which nucleotide T1108 was altered to A, leading to substitution of Tyr for Asn at amino acid 321 was observed. The mutation was confirmed in cDNA from two plt1/plt1 and two WT mice and all four exons from genomic DNA of each of four plt1/plt1 and WT mice.
Fig. 3.
Mapping of the Plt1 mutation. A cohort of 329 (C57BL/6-BALB/c) × C57BL/6 N2 mice was used to map the Plt1 mutation. The genotype of key markers on chromosome 6 is shown (filled boxes, loci that are homozygous for C57BL/6 homozygous allele; open boxes, loci that are heterozygous), demonstrating that the mutation lies in a 6.5-Mb interval between D6Mit139 and D6Mit83.
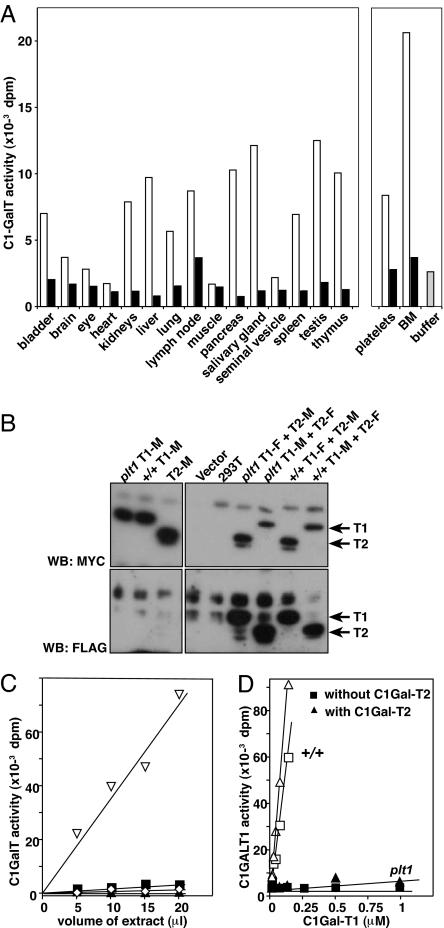
C1GalT1 enzymatic activity was detected in extracts from a number of WT tissues. In contrast, little activity above background levels was evident in extracts from plt1/plt1 organs (Fig. 4A). Titration studies with liver extracts estimated plt1/plt1 C1GalT1 activity at <5% of WT, and this activity was reduced to background levels upon the addition of EDTA (Fig. 4C). Because C1GalT1 is a Mn-dependent enzyme, this finding is consistent with the plt1 mutation causing a profound, but not complete, loss of C1GalT1 activity.
Fig. 4.
Reduced core1-β1,3-galactosyltransferase activity in plt1/plt1 mice. (A) Core1-β1,3-galactosyltransferase activity normalized to 50 μg total protein in organ extracts from plt1/plt1 (filled bars) and +/+ (open bars) mice. (B) Expression of FLAG (F) or Myc (M) epitope-tagged recombinant C1GalT1 (T1) and C1GalT2 (T2) in 293T cells. Proteins were precipitated from cell extracts using anti-Myc antibodies for Western blotting (WB) with either anti-Myc (Upper) or anti-FLAG (Lower) antibodies. (C) Comparison of core1-β1,3-galactosyltransferase activity in titrations of liver extracts from plt1/plt1 and +/+ mice. ▿, +/+ liver extract; ■, plt1/plt1 liver extract; ◇, +/+ extract plus EDTA; ▴, plt1/plt1 extract plus EDTA. (D) Core1-β1,3-galactosyltransferase activity of purified recombinant WT (+/+) and plt1 C1GalT1 in the presence and absence of C1GalT2. The concentration of C1GalT1 was titrated in the presence of a constant amount of C1GalT2 (five times the highest concentration of C1GalT1 or plt1 C1GalT1).
A C1GalT1-related protein, Cosmc or C1GalT2, interacts with C1GalT1 and has been proposed to provide a chaperone function (10) or have enzyme activity itself (11). Recombinant extracellular domains known to have enzymatic activity (12) were used to show that both FLAG and MYC epitope-tagged forms of recombinant plt1 C1GalT1 were expressed in 293T cells at similar levels to WT in both the presence and absence of recombinant C1GalT2. Moreover, like WT protein, recombinant plt1 C1GalT1 protein proved capable of forming a complex with C1GalT2 (Fig. 4B). Although C1GalT2 had no detectable activity when expressed alone, the enzyme activity of recombinant WT C1GalT1 was evident in the absence of C1GalT2 and further enhanced by its addition. In contrast, the residual activity of plt1 C1GalT1 completely depended on the presence of C1GalT2 (Fig. 4D).
Anomalous Protein Glycosylation in plt1/plt1 Mice.
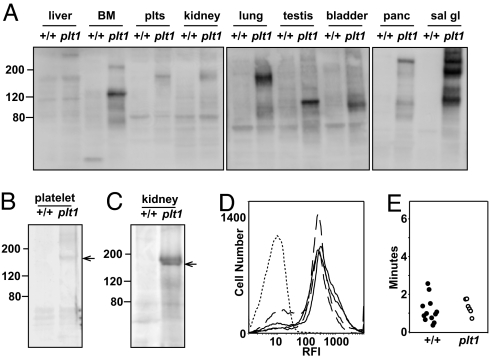
Reduced activity of C1GalT1 results in exposure of the Tn antigen on proteins normally targeted for modification by this enzyme, and this antigen is recognized by the lectin Helix pomatia agglutinin (HPA) (9). Consistent with deficient C1GalT1, the Tn antigen was detected in a range of tissue extracts from plt1/plt1 but not WT mice (Fig. 5A). Proteins displaying the Tn antigen in extracts of plt1/plt1-affected tissues, platelets, and kidney were affinity-purified with HPA-agarose (Fig. 5 B and C) and identified by mass spectrometry. The major HPA-reactive protein in plt1/plt1 platelets was glycoprotein Ib-α (gpIbα), and a mixture of aminopeptidase N and podocalyxin comprised the major protein band purified from kidney.
Fig. 5.
Anomalous protein glycosylation in plt1/plt1 mice. (A) Western blot of protein extracts from tissues of plt1/plt1 (plt1) and WT (+/+) mice probed with the Tn-specific lectin HPA. (B and C) Purification of Tn-bearing proteins from plt1/plt1 platelet (B) and kidney (C) extracts by using HPA-agarose affinity chromatography. (D) gp1bα expression on purified platelets from WT (solid line) and plt1/plt1 (broken line) mice. Platelets were incubated with an antibody to gp1bα or control isotype antibody (dotted line) and analyzed by flow cytometry. (E) The time to cessation of bleeding after resection of the terminal region of the tail in WT (●) and plt1/plt1 (○) mice is shown. BM, bone marrow; sal gl, salivary gland; plts, platelets; panc, pancreas.
GpIbα is a component of a platelet membrane protein complex important in hemostasis and GpIbα−/− mice exhibit thrombocytopenia and prolonged bleeding times (13). Anomalous glycosylation of gpIbα in plt1/plt1 mice did not appear to compromise cell-surface expression (Fig. 5D), and isolation of Tn-antigen-bearing gpIbα by HPA chromatography from plt1/plt1 platelet extracts resulted in the copurification of gpIbβ, gpIX, and gpV (data not shown), suggesting that altered glycosylation of gpIbα does not prevent adhesion receptor assembly. Tests of blood clotting were also normal in plt1/plt1 mice with no abnormality in bleeding times (Fig. 5E), in prothrombin time (0.95 ± 0.04 in plt1/plt1 mice versus 0.95 ± 0.08 for +/+ mice, n = 5–6 mice per genotype), in activated partial thromboplastin time (28.5 ± 3.3 s in plt1/plt1 mice versus 33.7 ± 4.2 for +/+ mice, n = 5–6 mice per genotype), or in collagen-induced platelet aggregation (data not shown). Thus, reduced C1GalT1-mediated glycosylation of gp1bα appears to significantly affect platelet production without a major impact on basic hemostatic parameters.
Discussion
C1GalT1 is a widely expressed type II transmembrane protein. Purification studies suggested that C1GalT1 could form dimers and that both dimeric and monomeric forms were enzymatically active (14). A second transmembrane protein, C1GalT2 or Cosmc, which displays 26% sequence homology to C1GalT1, has also been described. Initial studies suggested that C1GalT2 acted as a molecular chaperone with evidence that C1GalT1 and C1GalT2 interacted physically, and that C1GalT1 was rapidly degraded in the absence of C1GalT2 (10). However, an independent report suggested that C1GalT2 might itself have core1-β1,3-galactosyltransferase activity (11). We have established here that in in vitro assays using purified recombinant soluble proteins C1GalT2 displayed no detectable enzymatic activity. C1GalT1 was active, but the combination of the two proteins resulted in significantly greater activity. In contrast to WT C1GalT1, the plt1 mutant form of recombinant C1GalT1, which has substitution of Tyr for Asn at amino acid 321, was inactive in vitro. However, the mixture of mutant C1GalT1 and WT C1GalT2 was active, albeit at a level <5% of that of the WT complex. The simplest interpretation of these data are that the most potent core1-β1,3galactosyltransferse is a heterodimer or heterooligomeric configuration of C1GalT1 and C1GalT2. The plt1 mutation appears not to disrupt grossly the interaction between C1GalT1 and C1GalT2 but significantly reduces the enzymatic activity of the complex.
C1GalT1 is essential for developmental angiogenesis with fatal brain hemorrhage and disorganized microvascular networks in midgestation embryos lacking C1GalT1 (9). The plt1 mouse provides insights into the indispensable physiological roles of C1GalT1 in the adult, with key functions revealed in thrombopoiesis and kidney integrity. Anomalously glycosylated proteins were observed in tissues other than platelets and kidney, although no major phenotype was evident. Because the plt1 mutant form of C1GalT1 is partially functional, this residual activity may be sufficient in these tissues. The major anomalously glycosylated protein in plt1/plt1 platelets was gpIbα. GpIbα, in complex with gpIbβ, gpIX, and gpV, comprises an adhesion receptor on the platelet surface. GpIbα provides the binding surface for a number of ligands, including von Willebrand factor and thrombin, and key functions of the receptor are to facilitate thrombus formation by mediating adhesion between platelets and vessel walls at the site of injury and to augment thrombin-mediated platelet activation (15). A key role for gpIbα in megakaryocyte maturation has emerged from analysis of mice lacking this protein. GpIbα−/− mice exhibit thrombocytopenia, large platelets, and dysmorphic megakaryocytes (13, 16). Macrothrombocytopenia in both gp1bα−/− and plt1/plt1 mice implies that hypoglycosylation of gpIbα contributes significantly to plt1 thrombocytopenia. Because plt1/plt1 platelets exhibit a normal circulating half-life and mice exhibit thrombocytopenia in the absence of a reduction in megakaryocyte numbers, C1GalT1-mediated glycosylation of gpIbα appears particularly critical in the process of megakaryocyte maturation and/or platelet release. In contrast, unlike GpIbα−/− mice, basic clotting parameters and bleeding times were not affected in plt1/plt1 mice, suggesting that the contribution of gpIbα glycosylation to these hemostatic functions is less critical.
plt1/plt1 mice succumb to a fatal kidney disease characterized by glomerulonephritis and proteinuria. Lectin binding studies revealed that a major Tn-bearing protein in the kidney was podocalyxin. Podocalyxin is a sialoprotein prominent at the apical surface of renal podocyte foot processes (17). Mice lacking podocalyxin (podx1−/−) die soon after birth with defective kidney development and anuria (18) consistent with podocalyxin acting as an antiadhesin on the podocyte surface to ensure filtration slits remain patent. The importance of glycosylation in podocalyxin function is highlighted by the removal of sialic acid, which has been shown to affect cell adhesion and junctional permeability (19). Because plt1/plt1 mice are able to produce urine, hypoglycosylated podocalyxin appears sufficient for at least partial renal function. However, the kidney disease that develops in these mice suggests that C1GalT1-mediated glycosylation of podocalyxin may be essential for maintenance of normal kidney structure and function.
Reduced C1GalT1 activity has been associated with the human diseases Tn syndrome (7) and IgA nephropathy (8). Several studies have suggested that Tn syndrome arises from an acquired C1GalT1 deficiency that leads to exposure of the Tn antigen on multiple lineages of blood cells, caused at least in some cases by mutations affecting C1GalT2 (20). Mild hemolysis, thrombocytopenia, and/or leucopenia is thought to ensue via reactivity of the Tn antigen with naturally occurring anti-Tn antibodies (7, 21). Consistent with our findings in plt1/plt1 mice, gpIb has been shown to be a major Tn-bearing protein in at least one Tn-syndrome patient (22). Because thrombocytopenia in plt1/plt1 mice appears not to be caused by immune-mediated clearance, the phenotype of the plt1/plt1 mice suggest that an intrinsic action of gp1bα is required for appropriate megakaryocyte maturation and platelet release and that thrombocytopenia of Tn syndrome might potentially also have a nonimmune, megakaryocyte-intrinsic component.
IgA nephropathy is a common glomerulonephritis characterized by elevated circulating IgA, which exhibits abnormal, galactose-deficient O-linked glycans in the hinge region of the IgA protein (8). Reduced IgA clearance and/or the formation of immune complexes result in IgA deposition and pathology in the kidney. Because the O-linked glycosylation sites in the hinge region of human IgA are not conserved in the mouse, this mechanism is unlikely to underlie kidney disease in plt1/plt1 mice. IgA levels in plt1/plt1 mice were not elevated (data not shown), and the development of renal disease in rag1−/−plt1/plt1 mice excludes an Ig-mediated pathogenesis. Although reduced C1GalT1 activity has been shown clearly in B cells from IgA nephropathy patients (22), the phenotype of plt1/plt1 mice suggests that reduced C1GalT1 activity in the kidney may cause renal pathology independent of immune effects and the potential contribution of intrinsic deficiency in C1GalT1 to renal disease may be worthy of closer examination in IgA nephropathy and other kidney disorders. In addition, mutations in several key kidney podocyte proteins have been implicated in congenital human diseases, including nephrin in congenital nephritic syndrome, podocin in steroid-resistant nephritic syndrome, and α-actinin-4 in focal segmental glomerular sclerosis (23). Because our data suggest that defective glycosylation of podocaxylin may cause kidney disease, plt1 mice may provide a valuable model for nephropathy in which podocyte function is compromised.
Materials and Methods
ENU Mutagenesis.
Male C57BL/6 mice were treated with three doses of 100 mg/kg ENU at weekly intervals as described (24) and mated with C57BL/6 females to yield G1 progeny. G1 males were mated with C57BL/6 females, and matings between G2 females and their G1 fathers were established to produce pedigrees of G3 mice. Blood was collected into EDTA, and the number of platelets was determined with an Advia 120 analyzer (Bayer, Tarrytown, NY). A total of 3,523 mice from 150 G3 pedigrees were analyzed.
Genetic Mapping and Mutation Detection.
Male C57BL/6 plt1/plt1 mice were mated to WT BALB/c females to yield plt1/+ (C57BL/6×BALB/c)F1 progeny. F1 female mice were backcrossed to male C57BL/6 plt1/plt1 mice to produce an N2 generation. At 7 weeks of age, proteinuria and platelet number and size were measured, and using a set of simple sequence-length polymorphisms across the genome (25) we determined whether a marker was homozygous C57BL/6 or heterozygous C57BL/6-BALB/c. For markers not linked to the plt1 mutation no correlation between genotype and phenotype was expected, whereas for markers closely linked to the mutation, affected mice should be homozygous C57BL/6 and unaffected animals should be heterozygous. The only region of the genome to follow this pattern was at the centromeric end of chromosome 6. Standard protocols were used to sequence cDNA and exon and intron-exon boundaries of 12 genes within the interval (obtained from the Ensembl mouse genome browser annotaion of National Center for Biotechnology Information m33 released Feb. 2005): Acn9, tachykinin 1 (Tac1), asparagine synthetase (Asns), core 1 UDP-galactose:N-acetylgalactosamine-α-R β 1,3-galactosyltransferase (C1galt1), NM_145374, replication protein A3 (Rpa3), glucocorticoid-induced transcript 1 (Glcci1), islet cell autoantigen 1, neurexophilin 1 precursor (Nxph1), NADH:ubiquinone oxidoreductase MLRQ subunit (Ndufa4), PHD finger protein 14 (Phf14), and NM_027992.
Hematological Assays.
Blood cell counts were determined by using manual or automated counting techniques. Clonal cultures of hemopoietic cells (2.5 × 104 bone marrow or 5 × 104 spleen cells) were performed as described (26) with a mixture of 100 ng/ml stem cell factor, 10 ng/ml IL-3, and 2 units/ml erythropoietin as the stimulus. Megakaryocyte counts were performed from H&E-stained sections of sternum and spleen. Megakaryocyte ploidy was determined as described (27).
Platelet Half-Life.
Mice were injected with 0.6 mg of N-hydroxysuccinimide-biotin. Blood samples were collected at regular intervals and incubated with fluorescein-conjugated anti-CD41 and streptavidin-conjugated phycoerythrin. The proportion of biotin-labeled platelets (CD41+ biotin+) was determined by flow cytometry. An initial platelet count was determined and assumed to remain constant over the course of the experiment. Platelet clearance was determined by plotting the logarithm of the ratio of the number of labeled platelets at a given time to the initial number of labeled platelets. The number of new platelets produced, a measure of the rate of platelet production, was determined by subtracting the initial number of unlabeled platelets from the number of unlabeled platelets at a given time.
Protein Analysis.
Epitope-tagged WT C1GalT1 and C1GalT2 and plt1 mutant C1GalT1 in pEF-BOS were transfected into 293T cells. Three days later, cells were lysed in 1% (vol/vol) Triton X-100 in 50 mM Tris·HCl (pH 7.4)/150 mM NaCl/1 mM EDTA with 10% (vol/vol) glycerol, 1 mM PMSF, and complete proteinase inhibitor mixture. Proteins were immunoprecipitated with anti-FLAG (M2; Sigma, St. Louis, MO) or anti-MYC (9E10; Santa Cruz Biotechnology, Santa Cruz, CA) antibodies, separated by SDS/PAGE, transferred to PVDF membrane, and Western blotted by using anti-FLAG or anti-MYC antibodies followed by detection with ECL reagents. FLAG-tagged soluble WT C1GalT1 and C1GalT2 and Plt1 mutant C1GalT1 that lacked the transmembrane domain were expressed 293T cells. FLAG-tagged proteins were purified from medium by using M2-agarose affinity gel columns and eluted with FLAG peptide. Monomeric protein was used in all enzyme assays. Core1-β1,3-galactosyltransferase activity was measured as described (14). Reactions contained 100 mM Mes (pH 6.8), 20 mM MnCl2, 1 mM GalNAcα1-O-phenyl, 0.4 mM UDP-Gal, 107 dpm of UDP-[3H]Gal, 2 mM ATP, and 5–20 μl of purified enzyme or tissue extracts in a volume of 55 μl. After incubation at 37°C for 1 h, reactions were stopped with 1 ml of cold water, loaded onto 360- mg Sep-Pak Plus C18 cartridges, and processed as described (14).
Lectin Blotting and Affinity Purification.
Lectin blotting was performed as described (9). For lectin affinity purification, cleared tissue extracts were incubated for 2 h at 4°C with HPA agarose. HPA-binding proteins were eluted in 0.1 M N-acetyl-d-galactosamine/0.1% Triton X-100/0.15 M NaCl/EDTA-free protease inhibitor mixture. Eluates were concentrated and analyzed by SDS/PAGE. Coomassie-stained protein bands were excised and digested in situ with trypsin (28). Peptides were separated by capillary chromatography and sequenced by using an on-line electrospray ionization ion-trap mass spectrometer (LCQ; Thermo-Finnigan, San Jose, CA) as described (29).
FACS Analysis.
Whole blood was collected into EDTA-coated tubes, erythrocytes were lysed in 168 mM ammonium chloride, and platelets were stained with FITC-conjugated anti-gpIbα (CD42b) antibodies (Emfret Analytics, Wuerzburg, Germany). Analyses were performed on a FACScan (Becton-Dickinson, Franklin Lakes, NJ).
Hemostatic Functions.
Prothrombin time (international normalized ratio) and activated partial thromboplastin time were measured by using platelet-poor plasma prepared from whole blood collected by inferior vena caval puncture with 0.105 M buffered sodium citrate as anticoagulant. Studies were conducted on the semiautomated STart 4 (Diagnostica STAGO/Roche Diagnostics, Indianapolis, IN) using manufacturer-supplied protocols. Whole blood platelet aggregometry was performed by using a model 570 whole blood aggregometer (Chrono-log Corp, Havertown, PA) operated in the impedance mode. Aggregation was induced by the addition of collagen (4 μg/ml). For bleeding time determination, mice were anesthetized, and then 3–5 mm of distal tail was amputated. The tail was immersed in saline warmed to 37°C, and the time to cessation of bleeding was recorded.
Acknowledgments
We thank Janelle Lochland, Jason Corbin, Ladina DiRago, Sandra Mifsud, Danielle L. Krebs, Lynne Hartley, and the staff of the Peter MacCallum Cancer Centre's Microscopy Core for valuable assistance and advice. Protein identification by mass spectrometry was carried out by the Joint Proteomics Facility of the Walter and Eliza Hall Institute for Medical Research and the Ludwig Institute of Cancer Research. This work was supported by the Australian National Health and Medical Research Council (Program 257500), the Cancer Council Victoria, and MuriGen Pty Ltd.
Abbreviations
- ENU
N-ethyl-N-nitrosourea
- HPA
Helix pomatia agglutinin
- gpIbα
glycoprotein Ib-α.
Footnotes
The authors declare no conflict of interest.
References
- 1.Hitotsumachi S, Carpenter DA, Russell WL. Proc Natl Acad Sci USA. 1985;82:6619–6621. doi: 10.1073/pnas.82.19.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hrabe de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, et al. Nat Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 3.Nelms KA, Goodnow CC. Immunity. 2001;15:409–418. doi: 10.1016/s1074-7613(01)00199-6. [DOI] [PubMed] [Google Scholar]
- 4.Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, et al. Nat Genet. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- 5.Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, Hyland CD, Mifsud SL, Di Rago L, Hilton AA, Willson TA, Roberts AW, et al. Proc Natl Acad Sci USA. 2004;101:6553–6558. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanisch FG. Biol Chem. 2001;382:143–149. doi: 10.1515/BC.2001.022. [DOI] [PubMed] [Google Scholar]
- 7.Berger EG. Biochim Biophys Acta. 1999;1455:255–268. doi: 10.1016/s0925-4439(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 8.Julian BA, Novak J. Curr Opin Nephrol Hypertens. 2004;13:171–179. doi: 10.1097/00041552-200403000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Xia L, Ju T, Westmuckett A, An G, Ivanciu L, McDaniel JM, Lupu F, Cummings RD, McEver RP. J Cell Biol. 2004;164:451–459. doi: 10.1083/jcb.200311112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ju T, Cummings RD. Proc Natl Acad Sci USA. 2002;99:16613–16618. doi: 10.1073/pnas.262438199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kudo T, Iwai T, Kubota T, Iwasaki H, Takayma Y, Hiruma T, Inaba N, Zhang Y, Gotoh M, Togayachi A, et al. J Biol Chem. 2002;277:47724–47731. doi: 10.1074/jbc.M205839200. [DOI] [PubMed] [Google Scholar]
- 12.Ju T, Brewer K, D'Souza A, Cummings RD, Canfield WM. J Biol Chem. 2002;277:178–186. doi: 10.1074/jbc.M109060200. [DOI] [PubMed] [Google Scholar]
- 13.Ware J, Russell S, Ruggeri ZM. Proc Natl Acad Sci USA. 2000;97:2803–2808. doi: 10.1073/pnas.050582097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju T, Cummings RD, Canfield WM. J Biol Chem. 2002;277:169–177. doi: 10.1074/jbc.M109056200. [DOI] [PubMed] [Google Scholar]
- 15.Lopez JA, Andrews RK, Afshar-Kharghan V, Berndt MC. Blood. 1998;91:4397–4418. [PubMed] [Google Scholar]
- 16.Poujol C, Ware J, Nieswandt B, Nurden AT, Nurden P. Exp Hematol. 2002;30:352–360. doi: 10.1016/s0301-472x(02)00774-9. [DOI] [PubMed] [Google Scholar]
- 17.Kerjaschki D, Sharkey DJ, Farquhar MG. J Cell Biol. 1984;98:1591–1596. doi: 10.1083/jcb.98.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyonnas R, Kershaw DB, Duhme C, Merkens H, Chelliah S, Graf T, McNagny KM. J Exp Med. 2001;194:13–27. doi: 10.1084/jem.194.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeda T, Go WY, Orlando RA, Farquhar MG. Mol Biol Cell. 2000;11:3219–3232. doi: 10.1091/mbc.11.9.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ju T, Cummings RD. Nature. 2005;437:1252. doi: 10.1038/4371252a. [DOI] [PubMed] [Google Scholar]
- 21.Felner KM, Dinter A, Cartron JP, Berger EG. Biochim Biophys Acta. 1998;1406:115–125. doi: 10.1016/s0925-4439(97)00090-2. [DOI] [PubMed] [Google Scholar]
- 22.Nurden AT, Dupuis D, Pidard D, Kieffer N, Kunicki TJ, Cartron JP. J Clin Invest. 1982;70:1281–1291. doi: 10.1172/JCI110727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khoshnoodi J, Tryggvason K. Curr Opin Genet Dev. 2001;11:322–327. doi: 10.1016/s0959-437x(00)00197-0. [DOI] [PubMed] [Google Scholar]
- 24.Bode VC. Genetics. 1984;108:457–470. doi: 10.1093/genetics/108.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Copeland NG, Jenkins NA, Gilbert DJ, Eppig JT, Maltais LJ, Miller JC, Dietrich WF, Weaver A, Lincoln SE, Steen RG, et al. Science. 1993;262:57–66. doi: 10.1126/science.8211130. [DOI] [PubMed] [Google Scholar]
- 26.Alexander WS, Roberts AW, Nicola NA, Li R, Metcalf D. Blood. 1996;87:2162–2170. [PubMed] [Google Scholar]
- 27.Jackson CW, Brown LK, Somerville BC, Lyles SA, Look AT. Blood. 1984;63:768–778. [PubMed] [Google Scholar]
- 28.Moritz RL, Eddes JS, Reid GE, Simpson RJ. Electrophoresis. 1996;17:907–917. doi: 10.1002/elps.1150170512. [DOI] [PubMed] [Google Scholar]
- 29.Simpson RJ, Connolly LM, Eddes JS, Pereira JJ, Moritz RL, Reid GE. Electrophoresis. 2000;21:1707–1732. doi: 10.1002/(SICI)1522-2683(20000501)21:9<1707::AID-ELPS1707>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]