Abstract
The RIG-G gene, originally isolated from an acute promyelocytic leukemia cell line NB4, codes for a 60-kDa cytoplasmic protein that is induced by all-trans retinoic acid (ATRA) treatment along with the induction of morphological differentiation of NB4 cells. Here, we provide evidence that ectopic expression of Rig-G in U937 cells can lead to a significant accumulation of cells at G1/S transition. Growth arrest seems to occur by modulating several major cell cycle regulatory players. Interestingly, Rig-G alters JAB1 cellular distribution through interacting with this protein and increases the intracellular level of p27 by preventing it from the JAB-1-dependent and ubiquitin/proteasome-mediated degradation. Furthermore, we demonstrate a role of Rig-G for c-myc down-regulation that results in an up-regulation of p21, tightly associated with cell cycle arrest. In addition, our studies reveal that Rig-G is a direct target of STAT1, a key transcription factor in regulating IFN responses, and may be one of the first experimentally proven molecular mediators for the antiproliferative effect of IFN-α. Considering that IFN-α and ATRA synergistically inhibit growth along the intracellular pathways triggered by the two compounds in many cell types, we suggest that Rig-G may also represent one of the key molecular nodes of signaling cross-talk between ATRA and IFN-α.
Keywords: cell growth inhibition, retinoic acid, Rig-G, STAT1
Interferons (IFNs) are a group of cytokines having potent antiviral, growth-inhibitory, and immunomodulatory activities. The binding of IFNs to their cell surface receptors results in the rapid autophosphorylation of the IFN receptors-associated JAKs (Janus activated kinases), which in turn phosphorylate STAT (signal transducer and activator of transcription) proteins and activate the JAK-STAT signaling pathway. Phosphorylated STATs generally form homo- or heterodimers that enter the nucleus and bind specific DNA sites in the promoters of IFN-stimulated genes to initiate the transcription of these genes, thereby regulating various downstream cascades (1). Seven members of the STAT family have been identified in mammals: STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6. It has been known that STAT proteins play a key role in controlling cell cycle progression and apoptosis. In particular, STAT1 works as a tumor suppressor involved in growth arrest, and STAT3 and STAT5 are implicated in promoting cell proliferation and cellular transformation (2, 3). So far, the antiproliferative ability of IFNs has been largely confirmed in the treatment of different disorders, such as hairy-cell leukemia, chronic myeloid leukemia, and various solid tumors (1). However, the molecular mechanisms of growth-inhibitory action of IFNs are far from clear.
In our previous work, we identified the RIG-G gene by PCR-differential display due to its dramatically up-regulated transcriptional expression in acute promyelocytic leukemia cell line NB4 after treatment with all-trans retinoic acid (ATRA) for 72 h (4). We showed that the RIG-G gene located on chromosome 10q24 and contained 2 exons encoding for a 60-kDa protein with 490 amino acids. The fact that ATRA-induced RIG-G mRNA up-regulation occurred at a relatively later stage and could be completely blocked by protein synthesis inhibitor cycloheximide, indicated that RIG-G was not an ATRA-induced primary response gene. Nevertheless, we found that RIG-G shared a high homology with several IFN-stimulated genes, representing a member of the human IFN-stimulated gene family (4, 5). A synergistic induction of RIG-G mRNA in NB4 cells by the treatment with ATRA and IFNs implied a possible role of Rig-G in cross-talk between these two signaling pathways. Here, we report our recent work concerning the transcriptional regulation and the biological function of Rig-G. Our data suggest that Rig-G acts as a key molecular mediator of the antiproliferative activity of IFN-related pathways through, among others, up-regulation of cell cycle inhibitors p21 and p27.
Results
Essential Role of STAT1 Protein in Rig-G Expression.
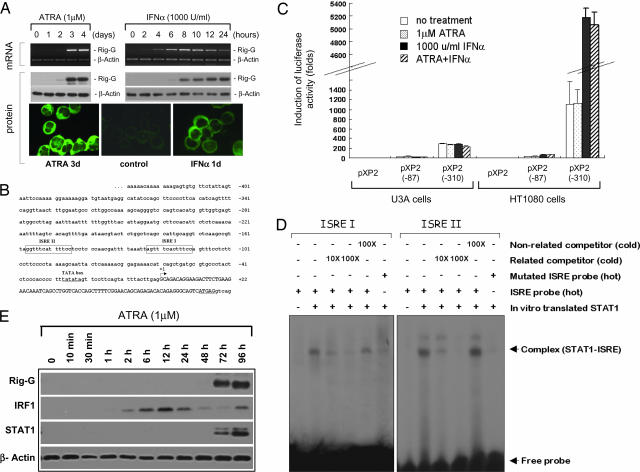
In this work, using the polyclonal antibody against Rig-G, we first detected the induction of Rig-G at the protein level. Western blot analysis showed a significant up-regulation of Rig-G in ATRA-treated NB4 cells for 72 h, which was perfectly consistent with the kinetics of RIG-G mRNA expression (Fig. 1A). A diffuse cytoplasmic distribution of Rig-G protein was determined by indirect immunofluorescence staining (Fig. 1A). Because Rig-G was a member of IFN-inducible (IFI) gene family, we then examined the effect of IFN-α and -γ on the expression of Rig-G. As a result, we found that the induction of Rig-G by IFN-α occurred in NB4 cells at as early as 4–6 h after treatment (Fig. 1A), whereas IFN-γ alone had no effect on Rig-G expression (data not shown). The similar induction of Rig-G by IFN-α was also found in U937 cells (data not shown), indicating the implication of Rig-G in IFN-α signaling pathways.
Fig. 1.
Transcriptional regulation of Rig-G expression. (A) NB4 cells were treated by 1 μM ATRA or 1,000 units/ml IFN-α for the indicated times. Rig-G expression was detected by RT-PCR, Western blot analysis, and immunofluorescence staining. The expression of β-actin was used as internal control for the amount of mRNA and proteins. (B) Partial 5′ end genomic sequences of the RIG-G gene. Two putative ISRE I and II are boxed. The TATA box is underlined. The transcriptional start site +1 is indicated by an arrow. The capital letters represent the first exon of the Rig-G gene. (C) U3A (STAT−/−) or HT1080 (STAT+/+) cells were, respectively, transfected with pXP2 vector and RIG-G promoter-luciferase constructs pXP2 (−87) and pXP2 (−310). Twenty-four hours after transfection, all of the cells were treated without or with 1,000 units/ml IFN-α and/or 1 μM ATRA as indicated. Luciferase activities were determined 24 h after the treatment. The error bars represent the SDs of the average of three independent experiments. (D) EMSA analysis of the binding of STAT1 protein to the RIG-G ISRE I and II. (E) Time courses of IRF1 and STAT1 induction by ATRA in NB4 cells were analyzed by Western blotting.
We thus analyzed the sequence of 5′-flanking region of the RIG-G gene in an attempt to uncover the transcriptional regulation of RIG-G. Although no typical retinoic acid response elements were revealed, two well conserved ISREs (IFN-stimulated response elements, hereafter ISRE I and ISRE II) were found upstream of a TATA box (Fig. 1B). For a detailed functional analysis of these elements, two RIG-G promoter-reporter constructs were prepared. After transient transfection into HT1080 cells, the pXP2 (−310)-luciferase construct containing intact ISRE I and II showed a significant baseline expression that could be further enhanced up to 4-fold by IFN-α, whereas the pXP2 (−87)-luciferase construct lost all of these activities because of deletion of ISRE I and II (Fig. 1C). These results strongly supported a transcriptional basis for induction of Rig-G by IFN-α. We noted no obvious effect of ATRA on RIG-G promoter, which was in agreement with the absence of retinoic acid response elements and the indirect up-regulation of Rig-G by ATRA in NB4 cells (4). In addition, we found that the activity of RIG-G promoter as well as its IFN-α inducibility could be completely abrogated in STAT1-deficient U3A cells (Fig. 1C), indicating that the STAT1 protein was a prerequisite to the expression of Rig-G.
Furthermore, in vitro translated STAT1 proteins were used to test for their binding to two probes respectively corresponding to ISRE I and II (Fig. 1B). By EMSA, both ISRE I and II showed their effective binding abilities with STAT1 protein. The specificity of these bindings was examined by adding cold cognate or nonrelated probes for competitive inhibition and by using mutated ISRE I and II oligonucleotides (Fig. 1D). These results indicated that both ISRE I and II were functional, and RIG-G was indeed a primary target gene of IFN-α. Because STAT1 protein is involved in growth arrest in response to many cytokines and growth factors, RIG-G gene may represent one of the key transcription targets of STAT1 to exert its growth inhibitory function.
In contrast with IFN-α-induced primary up-regulation of Rig-G, we found that, although ATRA could not directly induce the Rig-G expression in NB4 cells, the agent was able to up-regulate some crucial transcription factors associated with IFN-α pathway. Here, we showed that ATRA could rapidly up-regulate interferon regulatory factor 1 (IRF1) expression, which was followed by induction of STAT1 with relatively slower kinetics, suggesting that these proteins could act as mediators of ATRA signaling cascade and contribute to Rig-G induction by ATRA (Fig. 1E).
Effect of Exogenous Rig-G Protein on U937 Cell Proliferation.
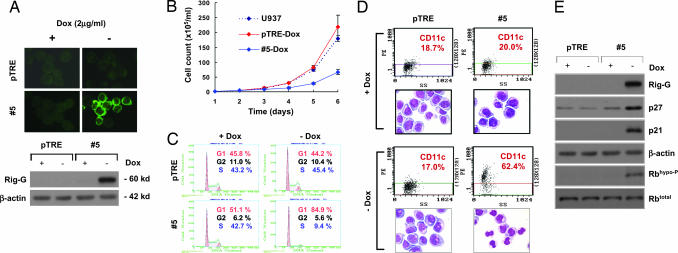
To investigate the biological function of Rig-G in both IFN-α and ATRA signaling pathways, the U937 sublines #5 stably expressing Rig-G were established by using the Tet-Off transfection system (BD Biosciences). In the absence of doxycycline (Dox), the induced expression of Rig-G in sublines #5 was respectively confirmed by immunofluorescence staining and Western blot analysis (Fig. 2A). No Rig-G expression was detected in the cells transfected by empty vector pTRE.
Fig. 2.
Effects of Rig-G-inducible ectopic expression on U937 cells fate. (A) The U937 sublines pTRE and #5 were cultured, respectively, in the presence or absence of Dox (2 μg/ml). The expression of Rig-G protein was detected by both immunofluorescence staining (Upper) and immunoblotting (Lower). (B) The viability of the indicated cells was assessed by trypan blue dye exclusion method. Each value represents the mean ± SD of three independent experiments. (C) Flow cytometric analysis of cell cycle distribution by propidium iodide staining. The experiments were repeated at least three times. (D) The expression of CD11c of the indicated cells was examined as described in Materials and Methods. Cell morphology was determined by Wright–Giemsa staining. All of the experiments were repeated at least three times. (E) The expression of cell cycle regulatory proteins p21, p27, and Rb in the indicated cells was analyzed by Western blotting.
Although Dox itself exerted an inhibitory effect on cell growth, we found that, after removal of Dox, the growth of cells was dramatically inhibited concomitantly to the augmentation of Rig-G exogenous expression (Fig. 2B). Analysis of cell cycle distribution further showed that cells expressing Rig-G significantly accumulated at the G1/S transition phase (Fig. 2C). In addition, an increase of CD11c expression as well as some cells with morphologic features of partial differentiation (smaller cell size, reduced nucleus-cytoplasm ratio, notched nucleus, and coarse chromatin) were also observed in Rig-G-expressing #5 cells (Fig. 2D). These data suggested that Rig-G played an important role in the cell cycle inhibition and could also facilitate cell differentiation.
To better understand the molecular mechanism by which Rig-G induced G1 phase arrest, the expression of several G1/S-transition-related genes was then screened before and after Rig-G induction. We found a dramatic increase of p21 and p27 proteins in Rig-G-expressing cells (Fig. 2E), indicating that the cell growth-suppressive actions of Rig-G were correlated with, and probably mediated by, the modulation of these regulatory proteins.
Both p21 and p27 are cyclin-dependent kinase (CDK) inhibitors that decrease G1 cyclin–CDK complex kinase activity (6). We then examined the effect of Rig-G expression on the phosphorylation of tumor suppressor Rb, a critical substrate in G1 of the cdk-associated kinase. The accumulation of hypophosphorylated Rb was found in Rig-G-expressing #5 cells, although the total Rb protein level was unchanged (Fig. 2E). This observation was in line with the Rig-G-induced up-regulation of p21 and p27 levels, resulting in a maintenance of Rb in a hypophosphorylated state.
Rig-G Increases p27 Protein Level Through Interaction with JAB1.
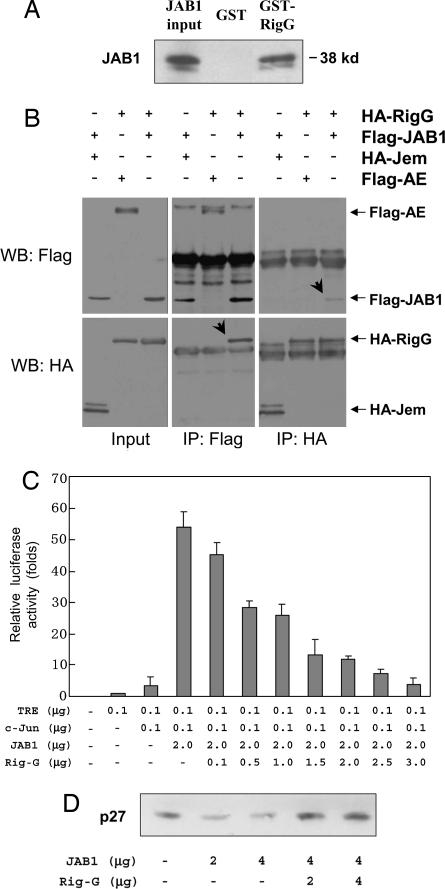
To address the question of how Rig-G could regulate these G1/S transition negative regulators, we further searched for Rig-G-interacting proteins by means of the yeast two-hybrid assay. The coding region of Rig-G fused to the DNA-binding domain of GAL4 was used as a bait to screen a human bone marrow cDNA library. After tests for nutritional selection and β-galactosidase activity, one clone containing a cDNA insert of the human JAB1 gene showed evidence for Rig-G interaction. The specificity of the interaction between Rig-G and JAB1 was then confirmed by GST pull-down assay (Fig. 3A). The purified recombinant GST-tagged Rig-G showed a strong binding with 35S-labeled in vitro synthesized JAB1 protein, whereas the GST alone showed no binding. In addition, the coimmunoprecipitation experiments were also performed to verify the interaction of these two partners in COS-7 cells cotransfected with HA-tagged Rig-G and Flag-tagged JAB1 (Fig. 3B). Specific coprecipitation of Rig-G by JAB1 could be detected by Western blot analysis with anti-HA antibody when anti-Flag antibody was used for immunoprecipitation. Vice versa, the coprecipitation of JAB1 by Rig-G could be obtained while interchanging the two antibodies. Two unrelated proteins respectively tagged with HA and Flag were used for control and showed no binding with Rig-G or JAB1. Finally, the interaction between endogenous JAB1 and Rig-G was further confirmed by using protein extracts from ATRA-treated NB4 cells (data not shown). Collectively, these results strongly indicated that JAB1 is one of the Rig-G-interacting proteins.
Fig. 3.
Molecular interactions between Rig-G and JAB1 and effects of Rig-G protein on JAB1 function. (A) GST and GST-Rig-G fusion proteins were used to pull down in vitro translated 35S-labeled JAB1 proteins. The bound JAB1 proteins were detected by SDS/PAGE. (B) COS-7 cells were cotransfected as indicated with the plasmids respectively expressing HA-tagged Rig-G, Flag-tagged JAB1, HA-tagged Jem, and Flag-tagged AE proteins. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody followed by Western blot analysis (WB) with anti-HA antibody or immunoprecipitated with anti-HA antibody followed by WB analysis with anti-Flag antibody. Jem and AE are two nonrelated proteins used for control. (C) COS-7 cells were cotransfected with 0.1 μg of AP-1-driven reporter plasmid TRE-luciferase and the indicated expression plasmids pRSV-c-Jun, pcDNA-JAB1, and pcDNA-Rig-G. The total amount of transfected DNA was standardized by the addition of empty vector. The error bars represent the SD of the average of three independent experiments. (D) NIH 3T3 cells were cotransfected with expression plasmids of JAB1 and Rig-G as indicated. The p27 protein level was analyzed by Western blotting.
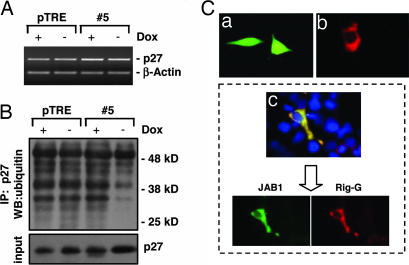
Because JAB1 is a protein involved in AP-1 activation and p27 degradation (7, 8), we successively examined whether Rig-G expression could affect these functions of JAB1. Using luciferase reporter gene containing AP-1 response elements, we found that the JAB1-enhanced AP-1 transactivation could be decreased in a dose-dependent manner by coexpressed Rig-G (Fig. 3C). It has been reported that the cytoplasmic shuttling and subsequent degradation of p27 is mediated by JAB1 (9, 10). Here, we found that although ectopic expression of JAB1 accelerated the down-regulation of p27 as expected, overexpression of Rig-G could reverse the decreased level of p27 in a dose-dependent manner (Fig. 3D). This result was in complete accord with the up-regulation of p27 level in #5 subline expressing Rig-G (Fig. 2E). However, the mRNA expression of p27 remains stable regardless of Rig-G expression (Fig. 4A). Therefore, it is most likely that Rig-G increase p27 level through posttranslational regulation mechanisms. The evidence that Rig-G could impair the ubiquitination of p27 (Fig. 4B) strongly supported the idea that Rig-G may have an inhibitory effect on JAB1-mediated p27 degradation.
Fig. 4.
Effects of Rig-G protein on JAB1-mediated p27 degradation. (A) RT-PCR analysis of p27 mRNA expression in U937 sublines pTRE and #5. The cells were cultured respectively in the presence or absence of Dox (2 μg/ml). (B) The pTRE and #5 cells were incubated with the proteasome inhibitor MG132 (20 μM) for 5 h, lysed, and subjected to immunoprecipitation (IP) with anti-p27 antibodies. The resulting precipitates were detected by Western blot analysis (WB) with antiubiquitin antibodies. (C) NIH 3T3 cells were respectively transfected with pcDNA-JAB1 alone (Ca), transfected with pcDNA-Rig-G alone (Cb), or cotransfected with both pcDNA-JAB1 and pcDNA-Rig-G (Cc). The intracellular distribution of JAB1 and Rig-G was analyzed by immunofluorescence staining.
According to the literature, JAB1 protein is localized in both cytoplasm and nucleus (Fig. 4Ca). Because Rig-G was a cytoplasmic protein (Figs. 1A and 4Cb), we examined whether the intracellular distribution of JAB1 was altered because of its association with Rig-G. By immunofluorescence staining, we noted that when Rig-G and JAB1 were coexpressed in NIH 3T3 cells, the nuclear JAB1 was markedly diminished. A colocalization of JAB1 and Rig-G in the cytoplasm was shown by superimposition of the staining for these two proteins (Fig. 4Cc). These findings suggested that the interference of Rig-G on the biological functions of JAB1 may result from JAB1 sequestration in the cytoplasm.
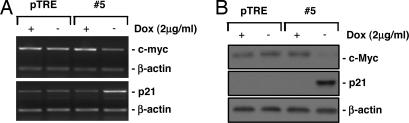
Rig-G Up-Regulates p21 Through Inhibition of c-Myc Expression.
The p21 gene is a target for diverse signals that control G1/S progression as well as cell differentiation (6). The induction of p21 by Rig-G was more likely through transcriptional regulation, or p21 mRNA stability, because p21 transcripts increased in Rig-G-expressing cells in accordance with its protein level (Fig. 5). It has been demonstrated that the promoter of p21 gene contains a variety of cis-acting elements for transcription factors, including p53, Sp1/2/3, STATs, C/EBP, Smads, Vitamin D receptor, RAR, Miz-1, etc. (11, 12). The different extracellular signals can regulate transcription of p21 via distinct transcription factors or combinations of transcription factors. To explore the mechanism by which Rig-G up-regulated p21, we examined the expression of a series of transcription factors involved in p21 transcriptional regulation in the cells before and after Rig-G induction. No obvious changes were observed except a down-regulation of c-Myc protein in Rig-G-expressing cells #5 (Fig. 5).
Fig. 5.
Effects of inducible Rig-G protein on c-Myc and p21 expression. The U937 sublines pTRE and #5 were respectively cultured in the presence or absence of Dox. The expression of c-Myc and p21 genes was detected by RT-PCR (A) and Western blot analysis (B).
The biological activities of c-Myc are characterized by induction of proliferation and inhibition of differentiation. Some reports showed that c-Myc could repress the expression of p21 in hematopoietic cells through association with the transcription factor Miz-1 (13–15). A Myc/Miz-1 switch model for p21 gene expression regulation has been proposed in U937 cells. It was shown that Miz-1 could bind the transcriptional start site of p21 gene and repress p21 expression when interacting with c-Myc, thereby promoting cell growth and inhibiting differentiation (13). This Miz-1-related model was also proposed to explain the c-Myc-mediated p15 repression (16). Recently, another compatible mechanism has also been reported, that c-Myc can down-regulate p21 through formation of an inactive complex with SP1-Smad on its promoter (17–19). Although there may be other mechanisms that c-Myc could use to repress p21, there is no doubt that Rig-G can induce p21 expression through c-myc down-regulation. However, the mechanism by which Rig-G exerts its inhibitory effect on c-Myc expression must be further addressed.
Discussion
In this work, we show that RIG-G, a gene whose expression is triggered by both IFN-α and ATRA, plays an important role in leukemia cell growth inhibition. Indeed, a survey of the literature indicates that the induction of Rig-G by ATRA or IFN-α occurs not merely in acute promyelocytic leukemia but also in many types of solid tumors, including head and neck squamous carcinoma cells, non-small-cell lung cancer H460 and A549 cells, cervical carcinoma HeLa cells, and epithelium-like WISH cells (5, 20), suggesting that the significance of Rig-G-induced cell growth inhibition is well beyond the acute promyelocytic leukemia model.
On the basis of bioinformatics analysis, RIG-G belongs to the IFI gene family and is also called IFI60 (5). It is worth pointing out that the expression of other members in this family, such as IFI54, IFI56, and IFI58, can also be induced with comparable kinetics by IFN-α or ATRA in NB4 cells (data not shown). Of note, these genes are tightly clustered at 10q23–24 and share similar cis elements in the promoters, indicating that the human IFI gene family members, including RIG-G, most likely evolve from one common gene ancestor because of gene duplication, and their expression may comply with the same transcriptional mechanisms.
It has been reported that the STAT1 gene contains a retinoic acid response element in its promoter, suggesting that the STAT1 may be a direct target of ATRA (21). However, this is doubtful in NB4 cells because of the slower kinetics of STAT1 up-regulation by ATRA as shown in Fig. 1E. According to our data and literature, several conceivable situations in NB4 cells are proposed. First, ATRA-induced STAT1 induction depends on the rapid up-regulation of IRF1 (22, 23), a primary target of ATRA. This idea is supported by the presence of ISRE and GAS elements in the promoter of STAT1 gene, which is molecularly essential for STAT1 response to IRF1 (24). Second, the fact that ATRA can induce IFN-α synthesis and secretion in NB4 cells (22, 25) together with the observed time course of STAT1 and Rig-G expression collectively support the idea that ATRA-induced STAT1 indeed may be mediated by IFN-α. Nonetheless, we observed that in NB4 cells, ATRA was unable to induce tyrosine phosphorylation of STAT1 (data not shown), which is a crucial step in the IFN-α-mediated classical JAK-STAT pathway (1). Therefore, it was postulated that ATRA and IFN-α induce Rig-G expression through distinct mechanisms (IRF1 and STAT1 activation, sequentially).
In this study, we find that the antiproliferative effect of Rig-G is correlated with increased p21 and p27, two negative regulators of the cell cycle progression from G1 into S phase. The induction of p21 by Rig-G has been shown to be at the transcriptional level. It is most likely that the c-Myc-dependent mechanisms act with regard to the p21 up-regulation (13, 14, 17). By contrast, we provide evidence that the Rig-G-induced up-regulation of p27 occurs at posttranslational level by disturbing the JAB1 function. JAB1 was first identified as a Jun activating binding protein (8), which can stabilize the binding of c-Jun or Jun D-containing AP-1 complex to their consensus DNA sites and increase the transcription of AP-1-dependent target genes. Besides, it has been also reported that JAB1 can physically interact with p27 in the nucleus and stimulate its nuclear export and subsequent proteolysis via the ubiquitin/26S proteasome pathway (9, 10). In our case, the interaction between Rig-G and JAB1 in the cytosol could partially prevent JAB1 from entering the nucleus and allow the nuclear accumulation of p27 and rescue of p27 from the degradation machinery. Similarly, the formation of Rig-G/JAB1 complex in the cytosol can also account for this inhibitory effect of Rig-G on JAB1-enhanced AP-1 transactivities, leading to cell proliferation inhibition. Taken together, our data suggest that Rig-G may work as a negative regulator of JAB1 function. Over the last several years, JAB1 also has been known as COP9 signalosome subunit 5 (CSN5), which is a component of the COP9 signalosome regulatory complex (26–28). Some reports show that JAB1/CSN5 subunit possesses a metalloprotease catalytic activity that is required for the COP9 holocomplex-directed deneddylation of the Cullins, a subunit of SCF (Skp1-Cullin-F-box protein) ubiquitin E3 ligases (29, 30). Although JAB1/CSN5 can exist in a monomeric form and act independently of COP9 holocomplex (7, 28), it should be interesting to explore in the future whether Rig-G has effects on the function of COP9 holocomplex through interaction with JAB1/CSN5.
The hallmark of tumors consist of a rapid, uncontrolled cell proliferation, accompanied by differentiation block and accumulation of genetic alterations. Induction of G1/S arrest and cell cycle exit can sensitize cells to differentiation and keep the cells from becoming tumorous. In our study, the potential of Rig-G to promote cell differentiation was also found, indicating that up-regulation of p21 and p27 might also have an effect on leukemia cell maturation. This is reminiscent of the observation that ectopic overexpression of p21 and/or p27 in U937 cells can induce the expression of the monocyte/macrophage-specific differentiation markers in the absence of hormone (31). It has been suggested that several cell cycle regulatory proteins may have dual functions (growth inhibition and/or differentiation), such as pRb family proteins, p21 and p27. The molecular mechanisms whereby the interplay between differentiation and cell cycle operates remain to be determined.
Materials and Methods
Cell Culture.
The cells NB4 [gift from M. Lanotte (32)] and U937 were cultured in RPMI medium 1640 supplemented with 10% FBS, 2 mM l-glutamine, and antibiotics. The adherent cells COS-7, NIH 3T3, U3A, HT1080, and 293T were cultured in DMEM supplemented with 10% calf serum, 2 mM l-glutamine, and antibiotics. The U3A and HT1080 cells were provided by G. R. Stark (33). Cell cultures were performed at 37°C in humidified air with 5% CO2.
Establishment of U937T-Rig-G Stable Transformants.
RIG-G cDNA (1.5 kb) was constructed in an expression vector pTRE by using the Tet-Off system (BD Biosciences). The U937T cells [gift from G. Grosveld (34)] were U937 stably transfected with a tet-VP16 fusion gene under the control of a tetracycline-inducible promoter and maintained in the presence of 2 μg/ml Dox and 0.5 μg/ml puromycin (Sigma). Electroporations were done by using a Gene-Pulser (Bio-Rad) at 280 V and 960 μF. Positive clones were selected by hygromycin B (Sigma).
Analysis of Cell Differentiation and Cell Cycle.
The expression of CD11c was detected by direct immunofluorescence with PE-conjugated anti-human CD11c monoclonal antibody (Beckman Coulter). A minimum of 104 cells were analyzed by using flow cytometry (EPICS XL; Beckman Coulter). For cell cycle analysis, cells were fixed overnight in 70% cold ethanol. After washing in PBS and treating with RNase, cells were stained with 25 μg/ml propidium iodide (Sigma). A minimum of 104 cells were then evaluated for cell cycle distribution by a flow cytometry.
Western Blot Analysis.
Twenty-microgram protein extracts were loaded on 10% SDS-polyacrylamide gels, subjected to electrophoresis, and blotted onto Hybond-C Extra membranes (Amersham Pharmacia). After blocking with 5% nonfat milk, the membrane was successively incubated with the indicated primary antibody and HRP-conjugated secondary antibody (Santa Cruz Biotechnology). The immunocomplex was visualized by using an ECL kit (Cell Signaling). Both rabbit polyclonal anti-Rig-G sera and mouse monoclonal anti-Rig-G ascites were raised against the GST-Rig-G fusion protein generated in Escherichia coli (Shanghai Genomics). Other primary antibodies included mouse anti-JAB1 from Genetex; rabbit anti-c-Myc, mouse anti-STAT1, and anti-p21 from Cell Signaling; mouse anti-RB, rabbit anti-p27, anti-ubiquitin, and anti-IRF1 from Santa Cruz Biotechnology; and mouse anti-β-actin from Sigma.
Yeast Two-Hybrid Screening.
The Matchmaker 3 two-hybrid system (Clontech) was used according to the manufacturer's protocol. The coding cDNA of RIG-G was subcloned into vector pGBK-T7 by the sites of BamHI and SalI. The resulting plasmid was used as bait to screen a human bone marrow cDNA AD library (Clontech) constructed in pACT2 vector. Yeast transformants were selected on Trp−Leu− medium, and the positive colonies were further tested for β-gal activity by colony-lift filter assay.
GST Pull-Down Assay.
The plasmid pGST-Rig-G was constructed by inserting the 1.5-kb RIG-G cDNA into the BamHI and XhoI sites of pGEX-5X-2 vector (Amersham Pharmacia). The plasmid pcDNA3-JAB1 was a gift from M. Naumann (35). A GST pull-down procedure was performed according to the standard protocol.
Coimmunoprecipitation.
The HA-tagged Rig-G plasmid was subcloned in a eukaryotic expression vector (36). The pCMV4-Flag-JAB1 expression plasmid was a gift from M. Naumann (35). These two expression plasmids were cotransfected into COS-7 cells by using SuperFect (Qiagen). The protein extracts were prepared in the buffer containing 150 mM NaCl, 50 mM Tris·HCl (pH 8.0), and 0.5% Nonidet P-40 and then mixed with protein A-agarose (Santa Cruz Biotechnology) and mouse anti-HA (Roche) or anti-Flag (Sigma) antibodies at 4°C overnight with rotation. The precipitated proteins were eluted by boiling beads in SDS-loading buffer and analyzed by Western blotting.
Luciferase Reporter Assay.
Two plasmids, pXP2 (−310) and pXP2 (−87) were, respectively, constructed by inserting the genomic fragment from −310 bp to +51 bp of 5′-flanking region of RIG-G gene and the fragment from −87 bp to +51 bp (Fig. 1B) into pXP2 vector (American Type Culture Collection), then transfected into U3A or HT1080 cells by using SuperFect (Qiagen). pRL-SV40 vector (Promega) was cotransfected into the cells as an internal control for normalizing the transfection efficiency. The report plasmid pTRE-luciferase containing AP-1 responsive element was provided by M. Naumann (35).
RT-PCR.
Total RNA was extracted with TRIzol reagent (Invitrogen). After reverse transcription, cDNA were amplified with the gene-specific primers by using a Bio-Rad iCycler thermal cycler at the following conditions: 94°C for 5 min; 25–31 cycles of 94°C for 30 s, 52–58°C for 45 s, and 70°C for 50 s; and 70°C for 10 min. The following gene-specific primers were used to detect the indicated gene transcripts: RIG-G, 5′-ACCTCGAGACACAGAGGGCAGTC-3′ and 5′-ACGGATCCGCCTTGTAGCAGCACCCAATC-3′; c-Myc, 5′-CTTGCAGCTGCTTAGACGCT-3′ and 5′-GTGCTGATGTGTGGAGACGT-3′; p27, 5′-ACGTGCGAGTGTCTAACGGGAGC-3′ and 5′-GTCCATTCCATCTTCAGAGCGA-3′; p21, 5′-GTCAGTTCCTTGTGGAGCCGGA-3′ and 5′-GAATTCAGGTCTGAGTGTCCAGGA-3′. The amplification of β-actin gene was used as an internal control for cDNA loading with the primers 5′-CATCCTCACCCTGAAGTACCCC-3′ and 5′-AGCCTGGATGCAACGTACATG-3′.
EMSA.
The in vitro translated STAT1 proteins were preincubated with or without unlabeled competitor probes in binding buffer (Promega) at room temperature for 10 min and at 4°C for 30 min. Labeled probes was then added and further incubated at room temperature for 30 min. DNA–protein complexes were resolved on 4% polyacrylamide gels equilibrated in 0.5× TBE under 300 V. The probes were 32P-labeled by using the T4 polynucleotide kinase (Promega) and their sequences were as follows: ISRE I, 5′-AATTAGTTTCACTTTCCAGT-3′; ISRE II, 5′-GTTAGGTTTCATTTTCCTCC-3′; mutated ISRE I, 5′-AATTAGTGGTCCTTTCCAGT-3′; mutated ISRE II, 5′-GTTAGGTTTCATTTTCCTCC-3′.
Acknowledgments
We thank all members of the Shanghai Institute of Hematology for their support. This work was supported in part by National Key Basic Research Project 973 (2002CB512805); Chinese National High Tech Program 863; National Natural Science Foundation of China Grants 30570778, 30670882, and 30070416; the “Shu Guang” Program of Shanghai Municipal Commission for Education; and the Samuel Waxman Cancer Research Foundation.
Abbreviations
- ATRA
all-trans retinoic acid
- Dox
doxycycline
- IFI
interferon-inducible
- IRF1
interferon regulatory factor 1
- ISRE
IFN-stimulated response element.
Footnotes
The authors declare no conflict of interest.
References
- 1.Platanias LC. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 2.Bromberg J, Darnell JE., Jr Oncogene. 2000;19:2468–2473. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 3.Turkson J. Exp Opin Ther Targets. 2004;8:409–422. doi: 10.1517/14728222.8.5.409. [DOI] [PubMed] [Google Scholar]
- 4.Yu M, Tong JH, Mao M, Kan LX, Liu MM, Sun YW, Fu G, Jing YK, Yu L, Lepaslier D, et al. Proc Natl Acad Sci USA. 1997;94:7406–7411. doi: 10.1073/pnas.94.14.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Veer MJ, Sim H, Whisstock JC, Devenish RJ, Ralph SJ. Genomics. 1998;54:267–277. doi: 10.1006/geno.1998.5555. [DOI] [PubMed] [Google Scholar]
- 6.Sherr CJ, Roberts JM. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 7.Chamovitz DA, Segal D. EMBO Rep. 2001;2:96–101. doi: 10.1093/embo-reports/kve028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claret FX, Hibi M, Dhut S, Toda T, Karin M. Nature. 1996;383:453–457. doi: 10.1038/383453a0. [DOI] [PubMed] [Google Scholar]
- 9.Tomoda K, Kubota Y, Kato J. Nature. 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 10.Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, Tanaka T, Yoshida M, Yoneda-Kato N, Kato JY. J Biol Chem. 2002;277:2302–2310. doi: 10.1074/jbc.M104431200. [DOI] [PubMed] [Google Scholar]
- 11.Gartel AL, Tyner AL. Exp Cell Res. 1999;246:280–289. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- 12.Gartel AL, Radhakrishnan SK. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 13.Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, Eilers M, Leon J, Larsson LG. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- 14.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 15.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M. Mol Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 16.Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Moroy T, Bartek J, Massague J, Hanel F, Eilers M. Nat Cell Biol. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 17.Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, Tyner AL. Proc Natl Acad Sci USA. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng XH, Liang YY, Liang M, Zhai W, Lin X. Mol Cell. 2002;9:133–143. doi: 10.1016/s1097-2765(01)00430-0. [DOI] [PubMed] [Google Scholar]
- 19.Seoane J, Pouponnot C, Staller P, Schader M, Eilers M, Massague J. Nat Cell Biol. 2001;3:400–408. doi: 10.1038/35070086. [DOI] [PubMed] [Google Scholar]
- 20.Kim HJ, Lotan R. Cancer Res. 2004;64:2439–2448. doi: 10.1158/0008-5472.can-03-2643. [DOI] [PubMed] [Google Scholar]
- 21.Kolla V, Weihua X, Kalvakolanu DV. J Biol Chem. 1997;272:9742–9748. doi: 10.1074/jbc.272.15.9742. [DOI] [PubMed] [Google Scholar]
- 22.Matikainen S, Ronni T, Hurme M, Pine R, Julkunen I. Blood. 1996;88:114–123. [PubMed] [Google Scholar]
- 23.Lehtonen A, Matikainen S, Julkunen I. J Immunol. 1997;159:794–803. [PubMed] [Google Scholar]
- 24.Yan R, Qureshi S, Zhong Z, Wen Z, Darnell JE., Jr Nucleic Acids Res. 1995;23:459–463. doi: 10.1093/nar/23.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chelbi-Alix MK, Pelicano L. Leukemia. 1999;13:1167–1174. doi: 10.1038/sj.leu.2401469. [DOI] [PubMed] [Google Scholar]
- 26.Wei N, Chamovitz DA, Deng XW. Cell. 1994;78:117–124. doi: 10.1016/0092-8674(94)90578-9. [DOI] [PubMed] [Google Scholar]
- 27.Deng XW, Dubiel W, Wei N, Hofmann K, Mundt K. Trends Genet. 2000;16:289. doi: 10.1016/s0168-9525(00)02071-0. [DOI] [PubMed] [Google Scholar]
- 28.Richardson KS, Zundel W. Mol Cancer Res. 2005;3:645–653. doi: 10.1158/1541-7786.MCR-05-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 30.Wolf DA, Zhou C, Wee S. Nat Cell Biol. 2003;5:1029–1033. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- 31.Rots NY, Iavarone A, Bromleigh V, Freedman LP. Blood. 1999;93:2721–2729. [PubMed] [Google Scholar]
- 32.Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R. Blood. 1991;77:1080–1086. [PubMed] [Google Scholar]
- 33.McKendry R, John J, Flavell D, Muller M, Kerr IM, Stark GR. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boer J, Bonten-Surtel J, Grosveld G. Mol Cell Biol. 1998;18:1236–1247. doi: 10.1128/mcb.18.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naumann M, Bech-Otschir D, Huang X, Ferrell K, Dubiel W. J Biol Chem. 1999;274:35297–35300. doi: 10.1074/jbc.274.50.35297. [DOI] [PubMed] [Google Scholar]
- 36.Duprez E, Tong JH, Derre J, Chen SJ, Berger R, Chen Z, Lanotte M. Oncogene. 1997;14:1563–1570. doi: 10.1038/sj.onc.1200995. [DOI] [PubMed] [Google Scholar]