Abstract
Mycoplasmas are cell wall-less bacteria considered among the smallest and simplest prokaryotes known, and yet several species including Mycoplasma pneumoniae have a remarkably complex cellular organization highlighted by the presence of a differentiated terminal organelle, a membrane-bound cell extension distinguished by an electron-dense core. Adhesin proteins localize specifically to the terminal organelle, which is also the leading end in gliding motility. Duplication of the terminal organelle is thought to precede cell division, but neither the mechanism of its duplication nor its role in this process is understood. Here we used fluorescent protein fusions and time-lapse digital imaging to study terminal organelle formation in detail in growing cultures of M. pneumoniae. Individual cells ceased gliding as a new terminal organelle formed adjacent to an existing structure, which then migrated away from the transiently stationary nascent structure. Multiple terminal organelles often formed before cytokinesis was observed. The separation of terminal organelles was impaired in a nonmotile mutant, indicating a requirement for gliding in normal cell division. Examination of cells expressing two different fluorescent protein fusions concurrently established their relative order of appearance, and changes in the fluorescence pattern over time suggested that nascent terminal organelles originated de novo rather than from an existing structure. In summary, spatial and temporal analysis of terminal organelle formation has yielded insights into the nature of M. pneumoniae cell division and the role of gliding motility in that process.
Keywords: cell division, gliding motility, adherence, fluorescent protein fusion
Mycoplasma pneumoniae causes chronic infections of the human respiratory tract, including bronchitis and primary atypical or “walking” pneumonia, accounting for up to 30% of all community-acquired pneumonia, particularly among older children and young adults. M. pneumoniae infections can result in chronic or permanent lung damage, and a growing body of evidence supports a correlation with the onset, exacerbation, and recurrence of asthma. Furthermore, extrapulmonary sequelae are not uncommon, reflecting both invasive and immunopathological components to M. pneumoniae disease (1).
In addition to its significant impact on public health, M. pneumoniae is intriguing from a biological perspective. Mycoplasmas have no cell wall and are among the smallest known cells, with M. pneumoniae having a cell volume only ≈5% of that of Escherichia coli. Likewise, at 816 kb the M. pneumoniae genome is among the smallest known for a cell capable of a free-living existence, lacking genes for cell wall production, de novo synthesis of nucleotides and amino acids, and two-component or other common bacterial transcriptional regulators (2, 3). Nevertheless, a remarkable level of structural complexity underlies what are otherwise considered minimal cells (4). Thus, experimental evidence indicated the presence of cytoskeletal structure and function in M. pneumoniae well before cytoskeleton-like elements were described in walled bacteria (5, 6). Furthermore, M. pneumoniae cells possess a complex, differentiated polar extension of the cell body that mediates both adherence to host cells (cytadherence) and gliding motility (7, 8). Adhesin proteins, including P1 and P30, as well as cytadherence-associated proteins of undefined function, such as P41 and P65 (9), localize to this structure. The terminal organelle is defined by an electron-dense core that is also a component of the Triton X-100-insoluble, cytoskeletal fraction (5, 6). Loss of certain cytadherence-accessory proteins results in failure to assemble a core or to exhibit the polarity characteristic of wild-type M. pneumoniae cells (10, 11). By conventional electron microscopy of thin sections the electron-dense core appears as two parallel flattened rods (12, 13) but recent analysis by electron cryotomography has revealed a complex, multisubunit composition to this structure (14, 15). Nevertheless, little is known regarding its molecular architecture or functional mechanisms.
By light microscopy M. pneumoniae cell division appears to begin with formation of a second terminal organelle adjacent to the first and the migration of one structure toward the opposite cell pole (16). Limits of resolution and the small size of the mycoplasma cell restrict the conclusions that might be drawn by light microscopy, but cell images by electron microscopy (17) as well as data correlating DNA content and the number and location of terminal organelles in fixed cells (18) are consistent with this model. Here we used fluorescent protein fusions with terminal organelle proteins P30, P41, and P65 and time-lapse digital imaging to observe directly the formation and maturation of the terminal organelle in individual cells during M. pneumoniae growth. Gliding ceased as a new terminal organelle formed adjacent to an existing structure at a cell pole, and P41 appeared to precede P30 and P65 in terminal organelle development. Migration of an existing terminal organelle was responsible for separation from the nascent structure, a process which was impaired in a nonmotile mutant, indicating a requirement for gliding function for normal cell division. Finally, incorporation of P30, P41, and P65 into nascent terminal organelles appeared to result from new protein synthesis rather than from an existing organelle, as has been suggested by electron microscopy images (13).
Results
Visualization of Terminal Organelle Development in Growing M. pneumoniae Cultures.
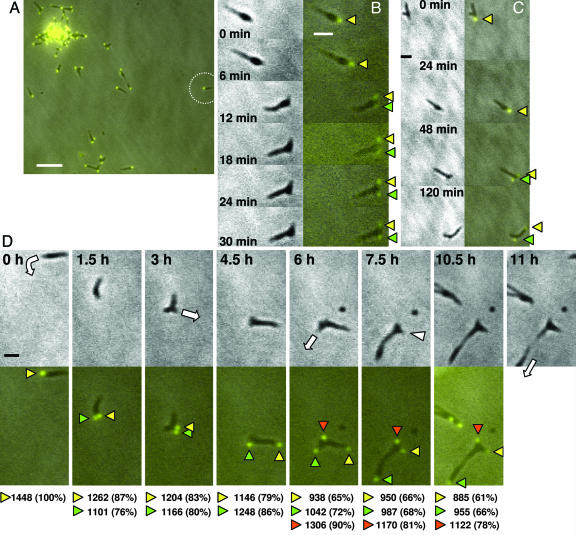
Protein P30 is a terminal organelle component required for cytadherence and gliding motility (19, 20). A recombinant P30 fusion with yellow fluorescent protein (YFP) introduced by transposon delivery localizes to the terminal organelle and restores cytadherence and gliding to a mutant lacking P30 (20); transformants with the recombinant transposon in an intergenic site exhibit a phenotype essentially indistinguishable from wild-type M. pneumoniae producing P30-YFP. As P30-YFP also yielded the strongest signal of the fluorescent protein fusions examined here (data not shown), we focused initially on this fusion. We monitored mycoplasma cells having a single P30-YFP focus for the formation of nascent terminal organelles, seen as the appearance of additional fluorescent foci over time. Ten phase contrast/fluorescence time-lapse digital movies were generated and analyzed, three with observation periods of 8–12 h and image capture at 0.5–1.5-h intervals, and seven of at least 2 h and image capture at 6- to 20-min intervals. Each had ≥20 cells per field initially, most in clusters or stationary with >1 P30-YFP focus (Fig. 1A). Of the 296 cells examined, 69 had 1 P30-YFP focus; nearly 90% of these were gliding, similar to published values (20). A new P30-YFP focus appeared adjacent to the single existing focus and coincident with cessation of gliding in 38% of the motile cells during the observation period (Fig. 1 B and C). However, even with image capture at 6-min intervals it was not possible to establish more precisely the sequence of these events, as the impact of photobleaching on detection, enhanced by the small cell size, restricted the number of fluorescence images possible per field. The remaining motile cells glided beyond the field of view, collided with other cells, or failed to form a new terminal organelle during the observation period. In Fig. 1B cell gliding ceased and a second P30-YFP focus appeared adjacent to the first between the 6- and 12-min time points. Over the next 18 min the original and new foci appeared to decrease and increase in fluorescence intensity, respectively. The same pattern was seen in Fig. 1C, where longer observation made it possible to see the initial focus (yellow arrowhead) move away from the new focus (green arrowhead), which remained relatively fixed. In Fig. 1D, a single cell (circled in Fig. 1A) was monitored over 11 h. Again the appearance of a second P30-YFP focus coincided with gliding cessation, although the interval between observations does not permit more precise sequencing of these events. Nascent foci were initially less intense than existing foci and required up to 3 h from initial detection to achieve maximum fluorescence (Fig. 1D, 0–3 h). During this time cells were not motionless, but gliding of whole cells was not observed. Original and nascent foci separated, in most cases the result of the former moving away from the latter (Fig. 1 C and D). Cytokinesis, when observed, generally required an additional 3 h minimum, but usually a third and occasionally a fourth new P30-YFP focus formed before daughter cell separation was observed (Fig. 1D, 6–10.5 h and data not shown).
Fig. 1.
Time-lapse phase contrast/fluorescence microscopy of terminal organelle appearance, development, and separation during M. pneumoniae growth. Mutant II-3 plus recombinant P30-YFP has a wild-type phenotype (20); shown are phase contrast and fluorescence images of II-3 plus P30-YFP taken at various intervals over observations of 30 min to 11 h. (A) A representative field for analysis of terminal organelle development. The circle indicates an individual cell monitored in D over 11 h. (Scale bar, 5 μm.) (B) Images were captured over a 30-min period at 6-min intervals. (Left) Phase contrast images. (Right) Merged P30-YFP fluorescence/phase contrast images. Yellow arrowheads indicate the original focus, and green arrowheads indicate the nascent focus. (Scale bar, 1 μm.) For further detail, see Movie 1, which is published as supporting information on the PNAS web site. (C) Selected frames over a 2-h observation. The arrowheads and scale are the same as in B. For further detail, see Movie 2, which is published as supporting information on the PNAS web site. (D) Selected frames over an 11-h observation. (Upper) Phase contrast images. (Lower) Merged P30-YFP fluorescence/phase contrast images. White arrows indicate the direction of cell gliding; white arrowheads indicate cell retraction. Yellow, green, and orange arrowheads indicate the first, second, and third terminal organelles, respectively. The values below the images indicate relative fluorescence units, with the values in parentheses being standardized to the single focus at 0 h. (Scale bar, 1 μm.) For further detail, see Movie 3, which is published as supporting information on the PNAS web site.
Gliding Motility Is Required for Normal Cell Division.
Although the appearance of nascent P30-YFP foci coincided with cessation of gliding, subsequent terminal organelle separation and cytokinesis appeared to result from transient resumption of gliding, almost without exception with an older fluorescent focus moving away from a new focus. Only rarely was the converse true or were both foci seen to separate simultaneously. In Fig. 1D the original focus (but not the entire cell) glided to the right (3.0 and 4.5 h; yellow arrowheads) whereas the nascent focus (green arrowheads) remained relatively stationary except for perhaps some rotation. Between 4.5 and 6.0 h a third focus formed (orange arrowheads), and the second focus migrated downward in the field, eventually leading to cytokinesis. This was accompanied by the movement of the initial focus back toward the cell body (7.5 h; Fig. 1D, white arrowhead), as though detached from the surface to allow cell retraction and partitioning.
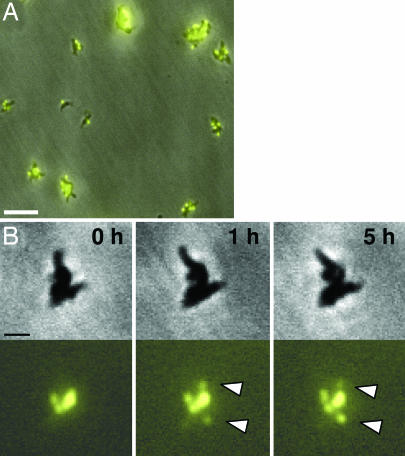
The association between M. pneumoniae gliding motility and cell division was examined further to determine whether separation of P30-YFP foci was impaired in the nonmotile mutant III-4, which forms a terminal organelle and localizes P30 to that structure but lacks cytadherence-accessory proteins B and C (10, 11, 21, 22). This mutant formed P30-YFP foci which were not observed to separate over periods of up to 12 h (Fig. 2 and data not shown), consistent with a requirement for gliding competence for normal cell division. We cannot rule out the possibility that appearance of new foci simply reflects movement of existing foci into the depth of field in these large cell clusters; nevertheless no separation of fluorescent foci was observed in this nonmotile mutant.
Fig. 2.
Time-lapse phase contrast/fluorescence microscopy of terminal organelle development without separation during growth of nonmotile M. pneumoniae mutant III-4. (A) Representative field for examination of nonmotile mutant III-4 plus P30-YFP. (Scale bar, 5 μm.) (B) Nonmotile mutant III-4 with selected frames over a 5-h observation during growth in a chamber slide. (Upper) Phase contrast images. (Lower) P30-YFP fluorescence images, with white arrowheads indicating new foci evident during the observation period. (Scale bar, 1 μm.)
Examination of Assembly Sequence by Using Fluorescent Protein Fusions.
The loss of certain M. pneumoniae terminal organelle proteins results in accelerated turnover of others and is thought to reflect sequential requirements in the assembly process (9). Cataloging the downstream consequences of loss of essential binding partners has yielded a model for terminal organelle assembly whereby some components are predicted to be incorporated early and others, including P30 and P65, are incorporated later (9, 18, 23). To begin to test that model, we examined fluorescence patterns in growing mycoplasma cultures producing either P41-YFP or P65-YFP and P30-cyan fluorescent protein (CFP) for their relative timing of appearance. Proteins P65 and P41 are novel cytoskeletal elements of unknown function expressed from the same transcriptional unit in M. pneumoniae and localize to the distal end and base, respectively, of the terminal organelle (24, 25). Transformants producing P65-YFP alone exhibited a pattern of new terminal organelle development comparable to that of P30-YFP, and in cells producing both fusions, P30-CFP and P65-YFP were observed in pairs in >95% of the cells examined, suggesting near-concurrent incorporation into the terminal organelle (data not shown).
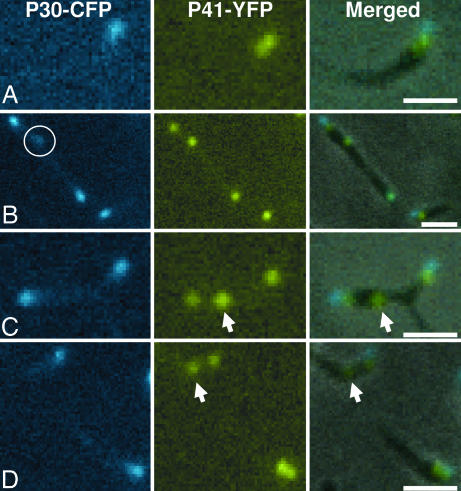
Results for P41-YFP alone differed from those for P30-YFP and P65-YFP in that 26% of gliding cells had two P41-YFP foci in tandem (Fig. 3) compared with <2% for P30-YFP and P65-YFP, suggesting that P41 may precede P30 in the assembly sequence. P30-CFP foci at the tip of terminal organelles were almost exclusively observed paired with P41-YFP at the base of those structures (Fig. 4), but in some cells the P30-CFP was faint and diffuse (Fig. 4B, circle), and nearly 30% of P41-YFP foci were unpaired with P30-CFP (Fig. 4 C and D, arrows), a frequency similar to that of gliding cells having two P41-YFP foci. Taken together, these observations are consistent with a model where P41 is incorporated into the developing terminal organelle before P30 or P65; time-lapse images were consistent with appearance of P41 before P30, but the toxicity and rapid photobleaching of CFP limited image capture (data not shown). Thus, we were unable, for example, to determine whether new P41 foci invariably matured into terminal organelles on the basis of subsequent acquisition of a P30 focus. However, those new P41 foci were observed in phase contrast images to gain adherence and gliding function (data not shown).
Fig. 3.
Time-lapse phase contrast/fluorescence microscopy demonstrating the appearance of P41-YFP before cessation of gliding. Selected frames over a 24-min observation during growth in a chamber slide. Yellow arrowheads indicate the leading P41-YFP focus, and green arrowheads indicate the trailing P41-YFP focus in a single gliding cell. (Scale bar, 1 μm.) To see the trailing focus unpaired with P30-CFP, see Movie 4, which is published as supporting information on the PNAS web site.
Fig. 4.
Phase contrast/fluorescence microscopy of M. pneumoniae cells producing P30-CFP and P41-YFP. (Left) P30-CFP fluorescence images. (Center) P41-YFP fluorescence images. (Right) Merged phase contrast and P41-YFP plus P30-CFP fluorescence images. (A and B) Cells with paired P30-CFP and P41-YFP foci. The circle in B indicates faint and diffuse P30-CFP focus. (C and D) Cells with unpaired P41-YFP foci (arrows). (Scale bars, 1 μm.)
Quantitation of Fluorescence Suggests That New Foci Develop de Novo.
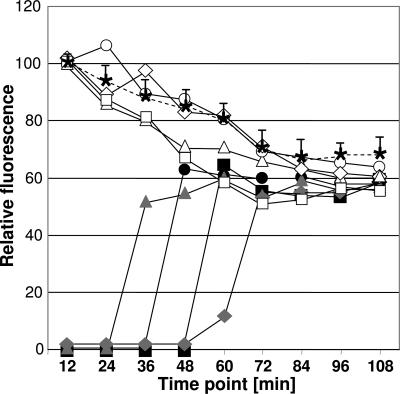
Electron microscopic images of the M. pneumoniae terminal organelle splitting at the distal end (13) suggest that its duplication may occur by a semiconservative process. By this scenario one would predict a rapid decrease in fluorescence intensity of an existing P30-YFP focus when a new focus appeared. Preliminary analysis of relative fluorescence intensities of existing and nascent P30-YFP foci over time suggested that no such decrease occurred (Fig. 1D). However, for a more systematic examination we compared P30-YFP fluorescence intensity over time for existing foci in cells that were not dividing, with that in cells forming a second focus during the observation period (Fig. 5). Each group exhibited the same gradual decline in fluorescence of the existing P30-YFP focus, suggesting that P30 accumulating at a nascent terminal organelle did not originate from a preexisting terminal organelle but from new protein synthesis. Interestingly, the maximum intensity of new P30-YFP foci appearing during each observation was typically lower than that of existing foci at 0 h, suggesting that some photobleaching of P30-YFP occurred before localization, providing a parameter by which it might be possible to establish the timeframe from synthesis to focus formation. Finally, to test the requirement for new protein synthesis directly in the appearance of new P30-YFP foci during mycoplasma growth, we examined wild-type cells over time in the presence of chloramphenicol (30 μg/ml). Inhibition of protein synthesis blocked formation of new fluorescent foci, whereas a normal pattern of terminal organelle appearance returned with removal of the chloramphenicol (data not shown).
Fig. 5.
Quantitation of P30-YFP fluorescence intensity over time. Fluorescence was measured in 12-min intervals for 10 cells that formed new P30-YFP foci during the observation period and 10 cells that did not. The average fluorescence for the latter group (asterisks on the dashed line, with positive standard deviations indicated) is compared with fluorescence of existing (open symbols) and new (solid symbols) P30-YFP foci in four individual, representative cells that began with a single fluorescent focus that was duplicated at approximately the same time point during the observation period and for which no collisions or other contact with neighboring cells was observed.
Discussion
Early studies by light microscopy and microcinematography suggested that the terminal organelle of M. pneumoniae duplicates before cell division. However, the exceptionally small size of M. pneumoniae cells, the limits of resolution, and therefore the inability to identify terminal organelles definitively, made a conclusive determination impossible. More recently Seto et al. correlated the number and position of terminal organelles with genome content (18), but their technique required cell fixation, hence it was not possible to document the sequence of events that led to those images or to rule out the potential introduction of fixation artifacts. In the current study we used fluorescent protein fusions and digital image analysis to examine assembly and function of the terminal organelle over time in growing cultures, confirming that M. pneumoniae terminal organelle duplication and separation precede cell division. Moreover, our findings add considerable detail regarding development and gain of function for nascent terminal organelles and demonstrate that, contrary to previous thinking (18), multiple new terminal organelles often form before cell division is observed.
The coordination of gliding motility and cell division was noted previously (8, 16, 26); our findings support and extend the current understanding of that relationship. Gliding ceased as nascent P30 foci emerged, as expected. However, the frequent presence of nascent P41-YFP foci in gliding cells suggests that assembly of the new terminal organelle begins earlier than previously recognized. Furthermore, subsequent separation of terminal organelles was a function of resumption of gliding as opposed to migration of the nascent structure along the cell body to the opposite pole by some undefined mechanism, as has been considered (26). Almost without exception (>98% of the cells examined), an existing terminal organelle initiated gliding and separation whereas the newest structure remained transiently fixed. For example in Fig. 1D, between 3 and 4.5 h the second focus (green arrowhead) remained fixed whereas the first focus (yellow arrowhead) glided to the right, and between 6 and 7.5 h the second focus acquired gliding capacity and moved downward in the field whereas the third focus (orange arrowhead, now the newest), remained fixed. Thus, P30 is required for both adherence and gliding motility (20, 27), but acquisition of the capacity to adhere occurred rapidly, whereas the initiation of gliding by a nascent terminal organelle was delayed, hence the localization of P30 to a nascent terminal organelle was not sufficient for gliding competence. In addition, at times terminal organelles appeared to release from the glass to allow retraction of the cell body (Fig. 1D, white arrowhead). We saw no change in the appearance of a fluorescent focus for P30, P41 or P65 during retraction, and the molecular mechanism responsible for release from the glass is not known. Regardless, both gliding and adherence appear to be dynamic functions during mycoplasma development and cell division. Finally, analysis of the nonmotile mutant III-4 demonstrated that loss of gliding function impairs normal separation of developing terminal organelles, expanding on recent findings that cell extension during gliding is responsible for the characteristic filamentous morphology of M. pneumoniae (20). Like mutant II-3 in that study, mutant III-4 here grows both on agar and in liquid culture, hence the inability of daughter cells to separate normally is not lethal. However, the large cell clusters characteristic of both mutants in liquid culture (unpublished observation) likely reflect a failure to separate rather than a propensity to aggregate.
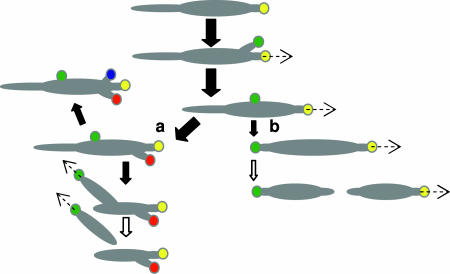
On occasion we observed duplication and separation of terminal organelles in wild-type mycoplasmas followed directly by cytokinesis, consistent with the existing model for M. pneumoniae cell division (18). However, for most dividing cells examined additional terminal organelles developed before new daughter cells emerged, representing a major shift in our understanding of M. pneumoniae cell division (Fig. 6) and indicating that terminal organelle duplication and cytokinesis are not tightly coordinated under these culture conditions. The extent to which terminal organelle duplication is coordinated with DNA replication remains unclear; we did not examine DNA content here, and the studies by Seto et al. (18) correlating DNA content with number and location of terminal organelles were limited to M. pneumoniae cells having only one or two terminal organelles.
Fig. 6.
Model for M. pneumoniae terminal organelle duplication and growth cycle. The yellow circle represents the initial terminal organelle, and green, red, and blue circles represent subsequent organelles, appearing in that order. The dashed arrows reflect movement of the indicated terminal organelle, and open arrows indicate cytokinesis. Solid arrows indicate steps in the cell cycle, with arrow size reflecting relative frequency. In most cases, multiple duplications of the terminal organelle occur before daughter cells emerge (a), although rarely some cells do undergo a single duplication of the terminal organelle followed by cytokinesis (b), according to the previous model for cell division in M. pneumoniae (18).
Images of wild-type M. pneumoniae by electron microscopy occasionally reveal cells with a bifurcated terminal organelle (13), suggesting that formation of a nascent structure may begin with the splitting of the existing terminal organelle. However, in the current study the fluorescence intensity patterns in cells forming a second focus during the observation period did not support this model but suggested de novo incorporation into nascent terminal organelles for both P30 (Fig. 5) and P65 (data not shown) at the tip of the terminal organelle and P41 (data not shown) at the base of this structure. Although it remains to be determined whether the same applies for proteins known to be specifically associated with the electron-dense core, the inhibition of nascent terminal organelle formation in cells under chloramphenicol arrest is consistent with a requirement for new protein synthesis. This conclusion is likewise consistent with electron cryotomography images (14, 15) in which the dimensions of the two rods of the core are not identical, and therefore fail to support a semiconservative process. Taken together these observations expand our current understanding of terminal organelle development but also raise questions regarding regulation of synthesis and assembly of this structure and the coordination of the process with cell division, particularly given the dearth of typical transcriptional regulators in M. pneumoniae (2).
Cataloging the downstream consequences of loss of essential binding partners in terminal organelle assembly has yielded a model whereby some components are predicted to be incorporated early and others, including P30 and P65, are incorporated later (9, 18, 23). The ability to follow the relative order of appearance of new fluorescent foci in cells producing two different protein fusions makes it possible to test that model, albeit within the limitations of time-lapse imaging. In pair-wise comparisons P30 and P65 fluorescent foci developed concurrently, consistent with the prediction that both are required relatively late in terminal organelle assembly. In contrast, P41 foci appeared before P30, suggesting that this cytoskeletal protein is required earlier in the assembly sequence. However, it was not possible to follow terminal organelle development for many frames with CFP fusions, which exhibited rapid photobleaching and toxicity.
In conclusion, this study demonstrates the feasibility of spatial and temporal analysis of macromolecular assembly by using fluorescent protein fusion technology in mycoplasmas, extraordinarily small cell wall-less bacteria. Furthermore, our findings elucidate further the relationship between the assembly of the terminal organelle, adherence, and gliding motility in mycoplasma cell division. In addition, our results expand the current understanding of the mechanism of terminal organelle duplication and provide evidence for the order of assembly of this complex structure by an otherwise minimal microorganism.
Materials and Methods
Generation of M. pneumoniae Transformants Expressing Recombinant Protein Fusions.
Characterization of recombinant P30-YFP in wild-type M. pneumoniae was described previously (20); its introduction into M. pneumoniae mutant III-4 in transposon vector pMT85, which delivers recombinant alleles to the mycoplasma chromosome in a random-like manner, was by established protocols (20). YFP fusions to M. pneumoniae proteins P65 and P41 were constructed by using plasmid pEYFP (Clontech, Mountain View, CA) as described by Kenri et al. (25) with minor primer modifications to facilitate cloning of the PCR amplification products into BamH1 and EcoR1 sites in pMT85. M. pneumoniae transformants were evaluated for expression of the recombinant fusion proteins by Western immunoblotting and fluorescence microscopy; transformants for which transposition occurred at intergenic loci, as determined by sequencing across the insertion site and comparison with the genome sequence (2, 22), were evaluated further. P30-CFP was constructed as for P30-YFP but using plasmid pECFP (Clontech). For generation of recombinant transposons encoding two fusions in tandem, the gene for P30-CFP was excised by using XbaI and cloned into the corresponding site within the pMT85 derivatives encoding P41-YFP or P65-YFP described above. Constructs were transformed into wild-type M. pneumoniae, and multiple transformants were evaluated for the expression of each recombinant protein by Western immunoblotting and fluorescence microscopy.
Fluorescence Microscopy.
For analysis of terminal organelle assembly in actively dividing M. pneumoniae, frozen stocks (wild-type or the P30 mutant plus P30-YFP) were thawed, diluted 1:40 in SP-4 medium containing 3% gelatin, and incubated overnight in chamber slides (20). Digital phase-contrast and fluorescence images were captured with an exposure time of 1.0 sec and frame intervals ranging from 6 to 60 min on a DM IRB epifluorescence inverted microscope (Leica, Wetzlar, Germany) and merged. Fluorescence emission of P30-YFP, P41-YFP, and P65-YFP foci was quantitated for individual cells by using the profiling module of Openlab v3.0–4.0 software (Improvision, Lexington, MA). Emission intensities were calculated as the mean level of fluorescence signal emitted relative to the mean background noise of the immediately adjacent cell-free area. Fluorescence intensities were quantified for single pixels (65 nm2) resolving nascent terminal organelles assembling as near as 130 nm from the parental structure. Colocalization of P30-CFP with P41-YFP or P65-YFP in actively dividing cells was examined by using an exposure of 1.0 sec, with fluorescence excitation of both signals in rapid succession by alternating between dichroic YFP and CFP filter cubes (Chroma, Rockingham, VT).
Additional details can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank R. Herrmann (University of Heidelberg, Heidelberg, Germany) for providing pMT85 before publication, M. Balish for reading this manuscript, and M. Farmer for technical assistance. This work was supported by Public Health Service Research Grants AI22362 and AI49194 from the National Institute of Allergy and Infectious Diseases (to D.C.K.).
Abbreviations
- CFP
cyan fluorescent protein
- HMW
high molecular weight
- YFP
yellow fluorescent protein.
Footnotes
The authors declare no conflict of interest.
References
- 1.Waites KB, Talkington DF. Clin Microbiol Rev. 2004;17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li BC, Herrmann R. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dandekar T, Huynen M, Regula JT, Ueberle B, Zimmermann CU, Andrade MA, Doerks T, Sanchez-Pulido L, Snel B, Suyama M, et al. Nucleic Acids Res. 2000;28:3278–3288. doi: 10.1093/nar/28.17.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morowitz HJ. Isr J Med Sci. 1984;20:750–753. [PubMed] [Google Scholar]
- 5.Gobel U, Speth V, Bredt B. J Cell Biol. 1981;91:537–543. doi: 10.1083/jcb.91.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng KE, Pfister RM. J Bacteriol. 1980;144:390–399. doi: 10.1128/jb.144.1.390-399.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier AM, Clyde WA., Jr Am Rev Respir Dis. 1974;110:765–773. doi: 10.1164/arrd.1974.110.6P1.765. [DOI] [PubMed] [Google Scholar]
- 8.Radestock U, Bredt W. J Bacteriol. 1977;129:1495–1501. doi: 10.1128/jb.129.3.1495-1501.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krause DC, Balish MF. Mol Microbiol. 2004;51:917–924. doi: 10.1046/j.1365-2958.2003.03899.x. [DOI] [PubMed] [Google Scholar]
- 10.Seto S, Miyata M. J Bacteriol. 2003;185:1082–1091. doi: 10.1128/JB.185.3.1082-1091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baseman JB, Cole RM, Krause DC, Leith DK. J Bacteriol. 1982;151:1514–1522. doi: 10.1128/jb.151.3.1514-1522.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biberfeld G, Biberfeld P. J Bacteriol. 1970;102:855–861. doi: 10.1128/jb.102.3.855-861.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegermann J, Herrmann R, Mayer F. Naturwissenschaften. 2002;89:453–458. doi: 10.1007/s00114-002-0359-2. [DOI] [PubMed] [Google Scholar]
- 14.Henderson GP, Jensen GJ. Mol Microbiol. 2006;60:376–385. doi: 10.1111/j.1365-2958.2006.05113.x. [DOI] [PubMed] [Google Scholar]
- 15.Seybert A, Herrmann R, Franqakis AS. J Struct Biol. 2006 doi: 10.1016/j.jsb.2006.04.010. in press. [DOI] [PubMed] [Google Scholar]
- 16.Bredt W. Pathol Microbiol. 1968;32:321–326. doi: 10.1159/000162074. [DOI] [PubMed] [Google Scholar]
- 17.Boatman ES. In: The Mycoplasmas. Barile MF, Razin S, editors. Vol. 1. New York: Academic; 1979. pp. 63–102. [Google Scholar]
- 18.Seto S, Layh-Schmitt G, Kenri T, Miyata M. J Bacteriol. 2001;183:1621–1630. doi: 10.1128/JB.183.5.1621-1630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baseman JB, Morrison-Plummer J, Drouillard D, Puleo-Scheppke B, Tryon VV, Holt SC. Isr J Med Sci. 1987;23:474–479. [PubMed] [Google Scholar]
- 20.Hasselbring BM, Jordan JL, Krause DC. J Bacteriol. 2005;187:6281–6289. doi: 10.1128/JB.187.18.6281-6289.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krause DC, Leith DK, Wilson RM, Baseman JB. Infect Immun. 1982;35:809–817. doi: 10.1128/iai.35.3.809-817.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasselbring BM, Page CA, Sheppard ES, Krause DC. J Bacteriol. 2006;188:6335–6345. doi: 10.1128/JB.00698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krause DC, Balish MF. FEMS Microbiol Lett. 2001;198:1–7. doi: 10.1111/j.1574-6968.2001.tb10610.x. [DOI] [PubMed] [Google Scholar]
- 24.Krause DC, Proft T, Hedreyda CT, Hilbert H, Plagens H, Herrmann R. J Bacteriol. 1997;179:2668–2677. doi: 10.1128/jb.179.8.2668-2677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenri T, Seto S, Horino A, Sasaki Y, Sasaki T, Miyata M. J Bacteriol. 2004;186:6944–6955. doi: 10.1128/JB.186.20.6944-6955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodwell AW, Mitchell A. In: The Mycoplasmas. Barile MF, Razin S, editors. Vol. 1. New York: Academic; 1979. pp. 103–132. [Google Scholar]
- 27.Romero-Arroyo CE, Jordan J, Peacock SJ, Willby MJ, Farmer MA, Krause DC. J Bacteriol. 1999;181:1079–1087. doi: 10.1128/jb.181.4.1079-1087.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.