Abstract
During cell entry, reovirus particles with a diameter of 70–80 nm must penetrate the cellular membrane to access the cytoplasm. The mechanism of penetration, without benefit of membrane fusion, is not well characterized for any such nonenveloped animal virus. Lysis of RBCs is an in vitro assay for the membrane perforation activity of reovirus; however, the mechanism of lysis has been unknown. In this report, osmotic-protection experiments using PEGs of different sizes revealed that reovirus-induced lysis of RBCs occurs osmotically, after formation of small size-selective lesions or “pores.” Consistent results were obtained by monitoring leakage of fluorophore-tagged dextrans from the interior of resealed RBC ghosts. Gradient fractionations showed that whole virus particles, as well as the myristoylated fragment μ1N that is released from particles, are recruited to RBC membranes in association with pore formation. We propose that formation of small pores is a discrete, intermediate step in the reovirus membrane-penetration pathway, which may be shared by other nonenveloped animal viruses.
Keywords: cell entry, hemolysis, membrane penetration, reoviridae
Crossing the cellular membrane is a key step in the infectious life cycle of all viruses. Although the understanding of enveloped-virus entry via membrane fusion has seen many advances (1–3), the entry mechanisms of nonenveloped animal viruses remain elusive. Mammalian orthoreovirus (reovirus) is a large nonenveloped virus with a 10-segmented dsRNA genome. During entry, it must penetrate the cellular membrane to deliver icosahedral particles of 70–80 nm in diameter to the cytoplasm, where particle-encased polymerases synthesize the viral mRNAs (4–6). The manner in which this large “payload” (7) is delivered across the membrane, without benefit of membrane fusion, remains largely unknown.
Enveloped viruses mediate membrane fusion by virally encoded fusion proteins, which undergo conformational rearrangements that are often promoted by low pH or receptor binding and lead to exposure of membrane-interacting sequences. After insertion into the target membrane, further rearrangements bring the lipid bilayers into proximity, favoring fusion. The paradigm of promoted conformational rearrangements is reflected in the mechanisms of nonenveloped viruses as well. For example, the poliovirus capsid is promoted by receptor binding to undergo conformational rearrangements exposing the N terminus of capsid protein VP1 and the myristoylated autocleavage peptide VP4; these newly exposed sequences are then thought to form a transmembrane pore, through which the genomic RNA may be extruded into the cytoplasm (8). Similarly, adenovirus capsids can be promoted by low pH to undergo rearrangements that expose the membranolytic protein VI, which is thought to mediate endosome rupture (9).
During reovirus infection, the outermost capsid protein σ3 is digested by intestinal or endosomal proteases (10–15), leaving capsid protein μ1 exposed on the particle surface (16). The resulting infectious subvirion particles (ISVPs) can also be generated in vitro by protease digestion (17, 18). ISVPs are metastable and can be promoted to undergo conformational rearrangements leading to the ISVP* particle (19). ISVP→ISVP* conversion is correlated with membrane-perforation activities both in vivo and in vitro (19, 20). Other hallmarks of this conversion include μ1 rearrangement to a protease-sensitive conformer and elution of the adhesion protein σ1 (18, 19, 21, 22). In addition, a myristoylated N-terminal fragment of μ1, μ1N, is generated by autocleavage and released from particles (23–26). Several lines of evidence suggest that μ1 directly engages in membrane perforation (12, 19, 20, 24, 25, 27–31), but little is yet known about this mechanism.
The membrane-perforation activity of ISVP*s can be assayed in vitro by lysis of RBCs (12, 19, 32, 33). In this study we demonstrated that hemolysis occurs osmotically, after formation of small size-selective lesions, or “pores,” in the RBC membrane. Both ISVP* particles and the myristoylated fragment μ1N were recruited to the membranes, implicating one or both of them in pore formation. We propose that the capability of reovirus to form small pores in target membranes represents a discrete, intermediate step in the membrane-penetration pathway during cell entry.
Results
Reovirus-Induced Hemolysis Is Inhibited by Osmotic Protection.
PEGs inhibit hemolysis but not μ1 conformational change.
We previously thought that reovirus-mediated hemolysis (12, 19, 32, 33) reflects gross disruption of the RBC membrane. An alternative hypothesis is that viral components form pores, leading to an influx of water down the osmotic gradient and consequent cell swelling and lysis. To test this hypothesis, hemolysis reactions were performed in the presence of PEGs (Fig. 1), which serve as osmotic protectants and have been used in numerous systems to demonstrate pore formation in RBC membranes (34–36).
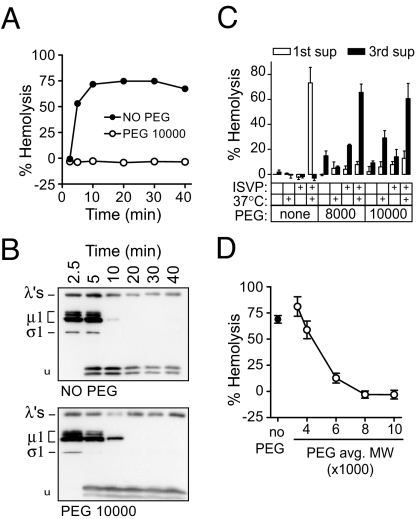
Fig. 1.
Inhibition of hemolysis by osmotic protection. (A) Hemolysis time courses were performed with or without 30 mM 10K-PEG. (B) Aliquots of supernatants from each time point in A were tested for protease-sensitivity of μ1. Arrowhead, trypsin-stable μ1 fragment. (C) Hemolysis reactions containing no PEG, 8K-PEG, or 10K-PEG were washed and resuspended in buffer with no PEG. (D) Hemolysis reactions were performed with PEGs of a range of sizes. Means ± SD of three experiments are shown.
Purified virions were first primed for hemolysis (19, 20) by protease digestion to yield ISVPs (18), then mixed with bovine RBCs and hemolysis buffer, with or without 30 mM PEG. Reactions were incubated at 37°C to promote ISVP→ISVP* conversion and the associated μ1 rearrangement and hemolysis (12, 19). After chilling on ice and centrifugation to remove unlysed RBCs, hemoglobin release into the supernatants was measured by absorbance at 405 nm (12, 33). PEG with a mean molecular weight of 10,000 (10K-PEG) was found to inhibit hemolysis completely (Fig. 1A), even though μ1 rearrangement as assayed by trypsin sensitivity (12, 19) was not affected (Fig. 1B). Essentially identical results were obtained with 8K-PEG (data not shown). These results indicate that 8K- and 10K-PEGs do not interfere with ISVP→ISVP* conversion but inhibit hemolysis at a later step.
Washing out PEG restores hemolysis.
When hemolysis reactions performed in the presence of 8K- or 10K-PEG were washed and resuspended in cold hemolysis buffer without PEG, lysis occurred without further incubation (Fig. 1C). The background levels of lysis in control reactions incubated on ice, or without virus particles, reflected nonspecific damage by PEG to the RBC membranes; however, nearly complete hemolysis was restored only in reactions containing virus particles that had been incubated in conditions that promote ISVP→ISVP* conversion. These findings suggest that ISVP→ISVP* conversion and membrane perforation occur in the presence of PEG, but the cells are osmotically protected from lysis. If the protectant is removed, then lysis occurs rapidly. Furthermore, maintenance of pores throughout the wash steps suggests that they are relatively stable and long-lived structures.
Osmotic protection depends on PEG size.
Whereas the largest PEGs tested (8K and 10K) completely inhibited hemolysis, the smallest PEGs (3.4K and 4K) had little or no effect, and 6K-PEG conferred an intermediate level of protection (Fig. 1D). These results are consistent with the hypothesis that small lesions or pores, of a discrete size or a narrow range of sizes, are being formed by viral components. The results suggest that if PEGs are small enough to pass through the pores, their concentration equilibrates rapidly across the membrane by diffusion, and they confer no osmotic protection; in contrast, PEGs large enough to be excluded retain water on the outside, preventing lysis. The intermediate effect of 6K-PEG may reflect the polydispersity of this polymer preparation and/or a distribution of pore sizes. Most of the size selectivity of protection in these experiments occurred with PEGs between 4,000 and 6,000 molecular weight. The hydrodynamic radii of these molecules have been estimated to be ≈2 nm and 3 nm, respectively (37, 38), suggesting a median pore diameter of 4–6 nm.
Reovirus Induces Size-Selective Leakage of Dextrans from Resealed Ghosts.
To assay for pores by a complementary approach, leakage of fluorescently labeled dextrans from resealed RBC ghosts (RGs) was examined by flow cytometry (Figs. 2 and 3). Because RGs do not contain enough solutes to create an osmotic gradient, and therefore are not expected to lyse osmotically, we hypothesized that virus-induced pores would allow small dextrans to leak out while large ones would remain trapped inside. Accordingly, RGs were loaded with carboxyfluorescein (CF) or fluorescein-conjugated dextran with mean molecular weights of 3,000, 10,000, 40,000, and 70,000.
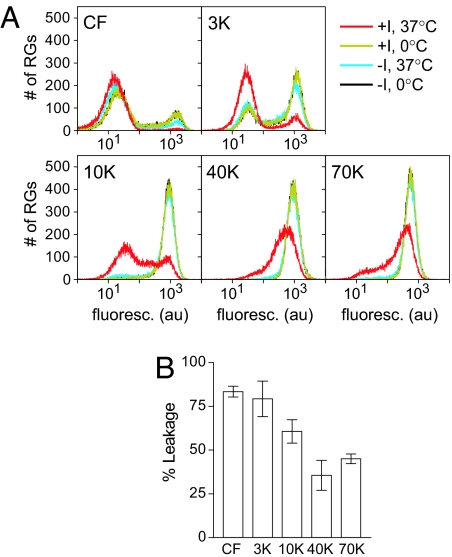
Fig. 2.
Leakage of fluorescent dextrans from RGs. (A) Hemolysis-like reactions with RGs containing CF, or fluorescein-labeled dextran of the indicated size, were analyzed by flow cytometry. (B) Percentage of leakage was as described in Materials and Methods. Means ± SD of three experiments are shown.
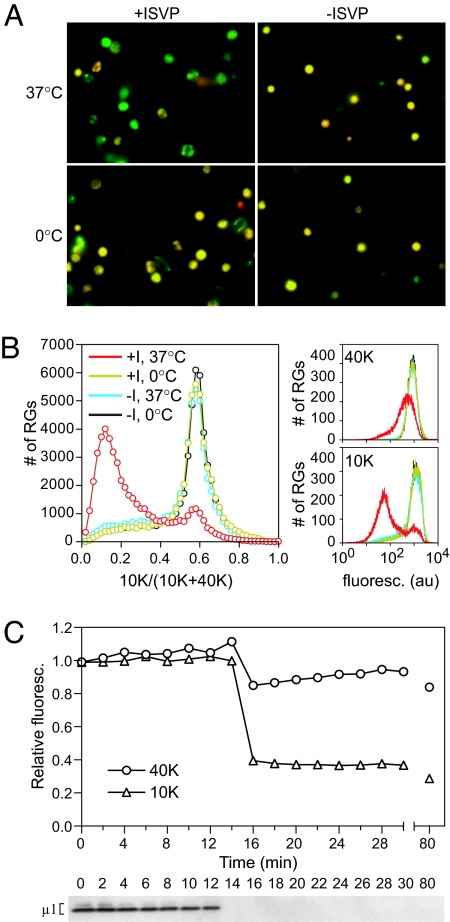
Fig. 3.
Leakage of fluorescent dextran from doubly labeled RGs. (A) RG leakage reactions with RGs containing both Texas red-10K-dextran (red) and fluorescein-40K-dextran (green) were imaged by fluorescent microscopy. (B) Reactions with RGs containing both Alexa Fluor 647–10K-dextran and fluorescein-40K-dextran were analyzed by flow cytometry. (Left) Frequency distribution of the ratio of signal from the 10K label to total signal. (Right) Frequency distribution of signal from each label. (C) Time courses were performed with RGs containing double label as in B. At each time point, an aliquot was analyzed by flow cytometry (Upper). The mean fluorescence intensity is shown for each time point, relative to that of a no-ISVP control at the same time point. Each time point was also assayed for protease sensitivity of μ1 (Lower).
Reovirus induces size-selective leakage of dextrans from singly labeled RGs.
RGs were mixed with ISVPs in hemolysis-like conditions and incubated at 37°C (Fig. 2A). Control reactions were included to assess the levels of initial loading and nonspecific leakage. After incubating on ice in the absence of virus particles, RGs loaded with either of the smallest labels (CF or 3K-dextran) occupied two distinct populations, reflecting a population of loaded cells and a population of RGs that were too leaky to confine these small labels. When incubated at 37°C without virus, some additional nonspecific leakage of CF or 3K-dextran was observed. Incubating with virus at 37°C, however, caused a further, major redistribution of cells from the labeled to the unlabeled population. RGs loaded with 10K-dextran, while exhibiting little or no nonspecific leakage, also underwent a major redistribution to a second population when incubated with virus at 37°C. In contrast, RGs loaded with either of the largest labels (40K- or 70K-dextran) showed only a modest shift in fluorescence signal, qualitatively different from that seen with labels 10K and smaller and consistent with these labels being largely retained within the RGs.
To quantify virus-specific leakage of each label in an equivalent way, the signal from unlabeled cells was first subtracted, and the remaining signal in the 37°C plus-virus sample was expressed relative to remaining signal in the 37°C minus-virus sample (Fig. 2B). The relationship between dextran size and percentage of leakage is reminiscent of the relationship between PEG size and percentage of hemolysis (see Fig. 1C) and again suggests that small pores of a defined size or narrow range of sizes are formed, allowing smaller dextrans to pass through but not larger ones.
Reovirus induces preferential leakage of smaller dextrans from doubly labeled RGs.
To confirm the size-selective behavior of the pores, RGs were doubly labeled with 10K- and 40K-dextran conjugated to different fluorophores, and hemolysis-like reactions were performed as described above for singly labeled RGs. Observation of end points by fluorescence microscopy (Fig. 3A) indicated that much of the 10K-dextran had been lost, whereas the 70K-dextran was retained. In control reactions, the RGs retained both labels. Quantitative measurement of the ratio of the two labels was determined by flow cytometry (Fig. 3B) and indicated that the majority of RGs had undergone preferential leakage of the 10K-dextran. Similar results, indicating preferential leakage of the smaller label, were achieved by quantifying the intensity of RGs in fluorescence microscopy images of reactions with cells doubly labeled with 3K- and 70K-, 10K- and 70K-, or 10K- and 40K-dextrans (data not shown). A time course demonstrated that the ratio change was due to rapid and concurrent major leakage of the 10K label versus more minor leakage of the 40K label, rather than merely a difference in leakage rates between the two labels (Fig. 3C Upper). The concurrent loss of both labels coincides with ISVP→ISVP* conversion, as assayed by trypsin sensitivity of μ1 (Fig. 3C Lower).
The results of these experiments are consistent with formation of size-selective pores, which allow preferential leakage of smaller dextrans while retaining larger ones. As with the PEG experiments, the small amount of leakage observed with 40K- and 70K-dextrans likely reflects the polydispersity of the polymer preparations and/or a narrow distribution of pore sizes. Published estimates of the hydrodynamic radii of dextrans (37, 38) near the main size-discrimination threshold observed in these experiments (between 10K- and 40K-dextrans) indicate a median pore diameter (5–9 nm) near that suggested by the PEG experiments.
Pore-Size Distribution Is Affected by Virus Concentration.
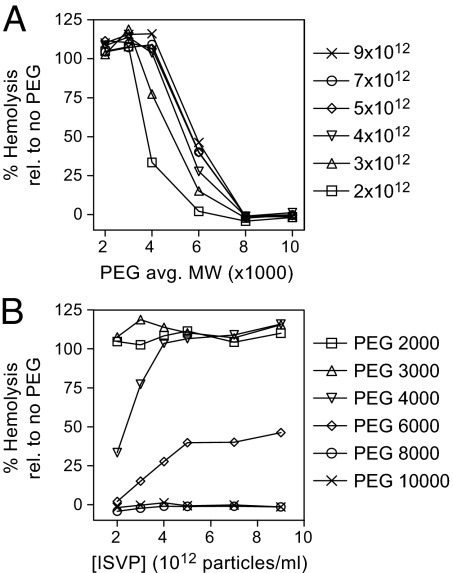
To determine whether pore-size distribution depends on virus concentration, the protective capacity of PEGs of different sizes was examined in hemolysis reactions with different concentrations of ISVPs (Fig. 4). At all virus concentrations tested, the smallest PEGs (2K and 3K) failed to inhibit hemolysis, whereas the largest ones (8K and 10K) inhibited hemolysis completely. The capacity of 4K- and 6K-PEG to inhibit hemolysis titrated with virus concentration. 4K-PEG inhibited hemolysis completely at 2 × 1012 ISVPs per milliliter, but as the virus concentration was increased, the amount of hemolysis (relative to a no-PEG control) increased until ≈50%, where it appeared to plateau, at ≈5 × 1012 ISVPs per milliliter. Similarly, 6K-PEG permitted an intermediate amount of hemolysis at 2 × 1012 ISVPs per milliliter, but as virus concentration was increased, the amount of hemolysis increased until it reached ≈100% at 4 × 1012 ISVPs per milliliter. These results indicate that, with increasing virus concentration, slightly larger PEGs are needed to provide the same level of protection from hemolysis, suggesting that pore-size distribution depends on virus concentration, but only to a limited extent.
Fig. 4.
Effect of ISVP concentration on pore size. Hemolysis reactions with the indicated ISVP concentrations (particles per milliliter) were performed in the presence of PEGs of a range of sizes. For clarity, the data are shown both as a function of PEG size (Upper) and as a function of ISVP concentration (Lower). Mean values of two experiments are shown, as relative values with respect to the no PEG control for each ISVP concentration (100%).
ISVP* Particles and Released Component μ1N Are Recruited to RBC Membranes.
ISVP* particles are recruited to RG membranes.
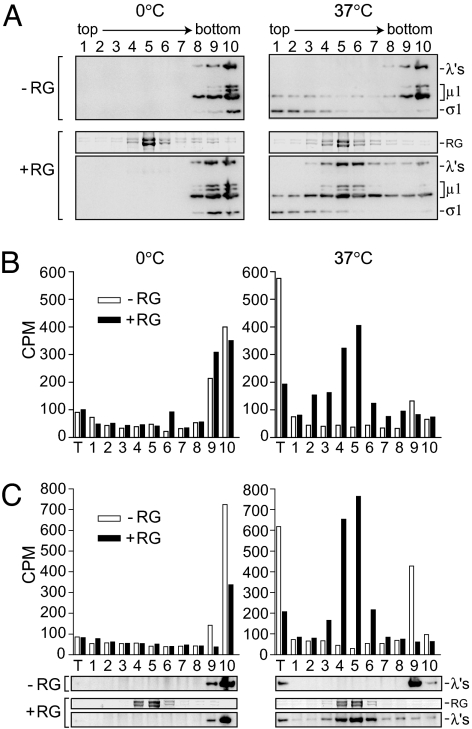
To determine which viral proteins participate in forming the pores, reactions were fractionated by density sedimentation. Reactions were performed with or without RGs and layered onto 25% isotonic Percoll. After sedimentation, the RGs formed a distinct white band near the middle of the gradient. Fractions were harvested from the top and were then analyzed by SDS/PAGE and immunoblotting (Fig. 5A). In control reactions without RGs, the particle-associated λ proteins and large μ1 species, μ1, μ1C, μ1δ, and δ (31), were found at the bottom of the gradient, indicating that both ISVPs and ISVP*s had nearly pelleted. The adhesion protein σ1, which is eluted from ISVP*s (12, 19, 24), remained near the top of the gradient, serving as an internal marker for ISVP→ISVP* conversion. In the reaction containing RGs and incubated on ice, particles again sedimented near the bottom, demonstrating that unconverted ISVPs do not associate with RG membranes. When incubated at 37°C, however, particles colocalized with RGs in the middle of the gradient, suggesting that ISVP* particles are recruited to the membranes. σ1 remained near the top of the gradient as expected. (Virions and ISVPs of some reovirus strains can bind to RBCs via σ1 attachment to cell-surface sialic acid; however, particles of the strain used in this study do not bind in this way to bovine RBCs.) Similar results with recoated virus-like particles lacking σ1 (32) confirmed that RG association is not σ1 dependent (data not shown). In addition, viral cores lacking all outer-capsid proteins sedimented to the bottom, confirming that an outer-capsid protein is required for RG association (data not shown). Quantification of bands for λ proteins and large μ1 species from experiments with [35S]Met/Cys-labeled ISVPs confirmed that essentially all of the large μ1 species remain particle associated after ISVP→ISVP* conversion, as reported previously (19, 24), and that the majority of ISVP*s colocalize with RGs in the gradient (data not shown).
Fig. 5.
Density sedimentation of RG leakage reactions. (A) Reactions performed with or without RGs were fractionated on Percoll gradients. Fractions were analyzed for virus particles by immunoblot, and gels were stained with Coomassie after transfer to indicate the location of RGs in the gradient (bands labeled “RG”). (B and C) RG leakage reactions performed with [3H]myristate-labeled ISVPs were fractionated, and each fraction was analyzed for the presence of myristoylated μ1N by scintillation counting. Two separate experiments are shown in B and C. The location of RGs and virus particles in the gradient are indicated (C Bottom) by Coomassie staining and immunoblot, respectively. T, material recovered from reaction tube.
Myristoylated fragment μ1N is also recruited to RG membranes.
During ISVP→ISVP* conversion, μ1 undergoes an autocatalytic cleavage, generating a 4-kDa N-terminally myristoylated peptide, μ1N, that is released from particles (23, 24). This cleavage appears to be required for hemolysis activity as well as for cell entry during infection (24). To determine whether μ1N is recruited to RG membranes, [3H]myristate-labeled ISVPs were incubated in hemolysis-like reactions, which were fractionated as described above. Fractions were assayed for the presence of μ1N by scintillation counting (two experiments are shown in Fig. 5 B and C). Since μ1N is hydrophobic (12, 25, 39), the tubes in which the hemolysis-like reactions were performed were additionally washed and then boiled with sample buffer to recover any material that had adhered to the tube. In control reactions incubated on ice, most of the 3H label was found at the bottom of the gradient, consistent with the μ1N region remaining particle associated. When incubated at 37°C in the absence of membranes, a large portion of the 3H label was recovered in the reaction tube, whereas most of the ISVP* particles were found near the bottom of the gradient, demonstrating that a large portion of μ1N had eluted from ISVP*s as expected. In the presence of membranes, however, both ISVP* particles and μ1N colocalized with RGs in the middle of the gradient. These results suggest that released μ1N is recruited to the membranes and may participate in forming the pores.
Discussion
The mechanism by which reovirus penetrates the membrane during cell entry is poorly understood. In this study, we present evidence for pore-forming activity in a model membrane. The functional pore diameter was estimated to be near 4–9 nm, by measuring translocation of molecules of different sizes both into RBCs (Fig. 1) and out of resealed ghosts (Figs. 2 and 3). The intermediate behavior of some of the markers may reflect polydispersity of the polymers and/or formation of pores with a narrow range of sizes. Consistent with the latter, the apparent pore size changes to a limited extent as a function of virus concentration (Fig. 4). This may suggest some flexibility in the pore architecture, as seen with several pore-forming toxins (35, 40).
Previously, reovirus ISVPs were reported to form multisized, ion-conducting channels in planar lipid bilayers, which were estimated to have a maximum diameter of ≈3 nm (41). The relationship between that finding and the other documented membrane-perforation activities of ISVPs, such as hemolysis, has been unclear. Our new results indicate that pore formation is a requisite step in the hemolysis pathway, leading to water influx and subsequent lysis. Thus, hemolysis does not reflect gross disruption of the RBC membrane by reovirus components directly.
The data in Fig. 5 implicate ISVP* particles and/or released fragment μ1N in pore formation. The μ1N findings are consistent with other evidence that both its autocleavage (24) and its myristoylation (unpublished data) are required for hemolysis, as well as for infectivity. Efficient association of ISVP* particles with RG membranes may suggest an additional role for other parts of μ1 in pore formation. ISVP*s are known to aggregate (19), so perhaps only a fraction are directly associated with the RG membranes, or are recruited via membrane-associated μ1N, whereas others are recruited in aggregates. The reovirus outer capsid contains 600 copies of μ1, arranged in trimers. Recent work indicates that μ1 trimers in solution are metastable and can dissociate to form monomers with hydrophobic sequences exposed (42). Less is known about the postconversion form of μ1 on particles, or of the released components. Higher-order oligomers of released components, such as might be expected to form the pores, have yet to be identified.
The significance of pore formation during infection remains an important question. The observed pores are only ≈1/10th as wide as the ISVP* or core particle that enters the cytoplasm (6). Potentially, pore formation in a cellular membrane could provide access to a host factor required for enlargement of the pore or other membrane-disrupting activity. Alternatively, pore formation in a vacuole (e.g., endosome or lysosome) might lead to osmotic swelling and lysis of that compartment. The trypanolytic factor apolipoprotein L-I forms pores in the trypanosome lysosomal membrane, causing chloride-ion influx and subsequent swelling of the lysosome (43). In addition, DNA delivery by polycationic transfection reagents has been reported to occur by increased proton buffering in the endosome, coupled influx of chloride ions, and subsequent osmotic swelling and lysis (44). At a minimum, formation of such pores in an endo- or lysosomal membrane might be expected to neutralize the low pH of that compartment, reducing the activity of acid-dependent hydrolases and perhaps making the environment more hospitable for ensuing steps.
In conclusion, we propose that formation of small pores is a discrete, intermediate step in the reovirus membrane-penetration pathway. This model brings the cell-entry mechanism of reovirus into striking congruence with that of poliovirus, which also includes deployment of a myristoylated autocleavage fragment and formation of a size-selective pore (45–47). Although poliovirus has been proposed to extrude genetic material through the pore, leaving the virus particle behind (48), the parallels in the membrane-interaction machinery suggest that the entry mechanisms of these viruses may be more similar than previously thought and, furthermore, may reflect a general paradigm of cell entry by nonenveloped viruses.
Materials and Methods
Virions, ISVPs, and Recoated Cores.
Virions of reovirus T1L were made as described (19) and stored at 4°C in virion buffer (VB) (150 mM NaCl/20 mM MgCl2/10 mM Tris, pH 7.5). [3H]Myristate-labeled virions (25) and [35S]Met/Cys-labeled virions (20) were made as described. Chymotrypsin (CHT) ISVPs were made by digesting virions with 200 μg/ml TLCK-treated α-CHT (Sigma, St. Louis, MO) for 10–15 min at 32 or 37°C, or for 15 min at 23°C. Digestion was stopped by addition of 1 mM PMSF and incubation on ice for at least 10 min. Trypsin ISVPs were made by digesting virions with trypsin (Sigma) (100 μg/ml) for 30 min at 32°C. Digestion was stopped by addition of 300 μg/ml soybean trypsin inhibitor (Sigma) and incubation on ice for at least 15 min. T1L cores and cores recoated with WT T1L μ1 and σ3 were made as described (32), except that virions were digested for 3–4 h, cores were purified by banding on CsCl step gradients (1.30–1.53 g/cm3), and cores were recoated by incubating with insect cell lysates for ≈4 h, followed by banding on CsCl step gradients (1.30–1.45 g/cm3).
RBCs and RGs.
Immediately before use, citrated bovine calf RBCs from Colorado Serum (Denver, CO) were washed repeatedly in cold PBS supplemented with 2 mM MgCl2 (PBS-Mg). Ghosts were made by lysing cells hypotonically in 10–20 vol of cold lysis buffer (5 mM sodium phosphate, pH 7), and washing three to four times in lysis buffer until the pelleted ghosts were white. For each wash, ghosts were pelleted at 16,000 × g for 5–10 min. Ghosts resuspended in 2 vol of lysis buffer were resealed by addition of 1/9 vol of 10× resealing buffer (1.2 M KCl/300 mM NaCl/10 mM MgCl2/100 mM sodium phosphate, pH 7), and incubation on ice for 15 min, then at 37°C for 45–60 min. Resealed ghosts (RGs) were washed once or twice in cold PBS-Mg. RGs loaded with fluorescent labels were made as described above, except label was added and the mixture was incubated on ice for 15 min before addition of resealing buffer. Labels from Molecular Probes (Invitrogen, Carlsbad, CA) were diluted in lysis buffer, and included at the following final concentrations in the resealing reaction: CF, 0.03 mM; fluorescein-3K-dextran, 0.1 mg/ml; and Texas red-3K-dextran, fluorescein-10K-dextran, Alexa Fluor 647–10K-dextran, fluorescein-40K-dextran, and fluorescein-70K-dextran, 0.3 mg/ml. Loaded RGs were prepared in the dark, resuspended in an approximately equal volume of PBS-Mg, and used the same day. For gradient experiments, empty RGs were resuspended in an ≈1/2 vol of PBS-Mg, and stored at 4°C for up to 2 weeks.
Hemolysis.
Twenty-microliter reactions were incubated at 37°C in hemolysis buffer [25% VB, 70% Tris buffer (10 mM Tris, pH 7.4), 5% PBS-Mg, 200 mM CsCl], with 5% RBCs, with or without 30 mM PEG (Sigma), and 3 × 1012 CHT ISVPs per milliliter, unless otherwise noted. For time courses, each time point represents a separate tube, which was removed to an ice bath. For end points, tubes were removed at 40 min. Reactions were centrifuged at 380 × g for 3–5 min at 4°C to pellet unlysed cells. 10 μl of the supernatant was added to 90 μl VB, and absorbance at 405 nm (A405) was measured in a microplate reader (Molecular Devices, Sunnyvale, CA). Percentage of hemolysis was calculated as [(A405 (sample) − A405 (blank))/(A405 (detergent) − A405 (blank))] × 100%, where the blank reaction contained all components except ISVPs and PEG, and the detergent reaction was the same as the blank, except containing 1% Triton X-100 (Sigma). For PEG washout, reactions were centrifuged for 1 min (yielding first supernatant), washed by gently resuspending in 19 μl of hemolysis buffer containing 30 mM PEG, centrifuged for 1 min (yielding second supernatant), resuspended in 19 μl hemolysis buffer with no PEG, and centrifuged for 1 min (yielding third supernatant). Ten microliters of each supernatant was assayed by A405 as above.
Leakage from RGs.
Hemolysis-like reactions with RGs were performed in buffer containing 62–65% VB and 400 mM CsCl, with 25% RG preparation, and 3 × 1012 CHT ISVPs per milliliter. For end points, reactions were incubated at 37°C for 30 min, chilled on ice, and diluted in cold VB for flow cytometry. For time courses, aliquots were taken from the reaction and immediately diluted in cold VB and analyzed by flow cytometry.
Flow Cytometry.
RG leakage was assayed by flow cytometer (FACSCalibur; BD Biosciences, San Jose, CA). Forward scatter voltage was set to E00 (unmultiplied), and side scatter voltage was set to 300. All parameters were detected in log mode. Events with both forward scatter and side scatter values >10 were collected. For end point and time-course experiments, 50,000 and 10,000 events were collected from each sample, respectively. Analysis of RGs made with either Alexa Fluor 647–10K-dextran alone, fluorescein-40K-dextran alone, or both labels together, indicated that there was no significant spectral overlap between these fluors in the acquisition channels. Analysis of RGs made with different concentrations of each label indicated that at the concentration used in leakage experiments, the signal from each label was within the linear range of detection, and that the labels were not self-quenching. Data were analyzed with CellQuest software (BD Biosciences) and MFI software (E. Martz, University of Massachusetts, Amherst, MA). All calculations were performed with data transformed to linear fluorescence values. Percentage of leakage was calculated as [1 − (TFI+isvp, 37°C − Background)/(TFI−isvp, 37°C − Background)] × 100%, where TFI is the total fluorescence intensity in the indicated sample, and Background is defined as the sum signal from the events in the (−isvp, 0°C) sample with signal ≤100. Label ratios in double-labeled RGs were calculated individually for each event.
Fluorescence Microscopy.
To visualize RG leakage by fluorescence microscopy, 1- to 2-μl reaction end points were spotted on a slide, covered with a coverslip, and imaged immediately. Images were colleted digitally by using a Nikon Optishot 2 epifluorescence upright microscope equipped with a Photometrics CoolSnap-cf camera (Roper Scientific, Tucson, AZ) and MetaVue v5.0 software (Molecular Devices). Fluorescence intensity of RGs was quantified from raw images by using ImageQuant (GE Healthcare, Piscataway, NJ), with histogram peak background correction. Images were scaled and overlaid for presentation by using ImageQuant and Photoshop (Adobe Systems, San Jose, CA).
Protease Sensitivity.
Trypsin sensitivity of μ1 was assayed by digestion with 100 μg/ml trypsin for 45 min on ice, followed by incubation with soybean trypsin inhibitor (300 μg/ml) for at least 15 min. Samples were centrifuged at 16,000 × g for 30 min, and pelleted particles were resuspended in 1× Laemmli sample buffer for SDS/PAGE. For PEG experiments, hemolysis supernatants were diluted 10-fold with cold VB before addition of trypsin, as the PEG was found to interfere with trypsin activity.
Density Sedimentation.
RG leakage reactions performed with empty RGs and 5 × 1012 trypsin ISVPs per milliliter were layered atop 400 μl of gradient material [25% Percoll (GE Healthcare) in 1× VB with 400 mM CsCl]. In some experiments, 26.5% Percoll in 1.2× VB with 470 mM CsCl was used; no difference in results was observed. Gradients were centrifuged at 30,000 × g for 15 min at 4°C in a TLA100.2 rotor (Beckman, Fullerton, CA). Fractions were collected from the top of the gradient by using a pipette with wide-mouth tips, and pipeted directly into either 4× Laemmli sample buffer or Ready-Safe scintillation fluid (Beckman). To recover material that had adhered to the tube during the RG-leakage reaction, tubes were washed once with cold VB and then boiled with 1× Laemmli sample buffer.
SDS/PAGE and Immunoblotting.
Equivalent amounts of each fraction were subjected to SDS/PAGE with 10% acrylamide gels as described (31). Proteins were transferred to nitrocellulose and detected with a mixture of rabbit T1L-virion-specific serum (49) (1:1,000 dilution) and rabbit T1L-σ1-specific serum (19) (1:1,000), followed by rabbit-specific donkey IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch, West Grove, PA) (1:1,000 to 1:5,000). Antibody binding was detected with Western Lightning chemiluminescence reagents (PerkinElmer, Wellesley, MA) and exposure to film, or scanning on a Typhoon (GE Healthcare) in chemiluminescence mode.
Acknowledgments
We thank E. C. Freimont for technical support; A. L. Odegard (Harvard Medical School) for [3H]myristate-labeled virions; D. E. Higgins (Harvard Medical School) for Texas red-3K-dextran; and K. Chandran, S. C. Harrison, and J. J. Perez for thoughtful discussions and review of the manuscript. This work was supported in part by National Institutes of Health Grants R01 AI46440 (to M.L.N.) and F31 AI064142 (to M.A.A.).
Abbreviations
- ISVP
infectious subvirion particle
- RG
resealed RBC ghost
- CF
carboxyfluorescein
- VB
virion buffer
- CHT
chymotrypsin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Harrison SC. Adv Virus Res. 2006;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kielian M, Rey FA. Nat Rev Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weissenhorn W, Dessen A, Calder LJ, Harrison SC, Skehel JJ, Wiley DC. Mol Membr Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Nat Struct Biol. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tao Y, Farsetta DL, Nibert ML, Harrison SC. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 6.Nibert ML, Schiff LA. In: Fields Virology. 4th Ed. Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 1679–1728. [Google Scholar]
- 7.Nibert ML, Furlong DB, Fields BN. J Clin Invest. 1991;88:727–734. doi: 10.1172/JCI115369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogle JM. Annu Rev Microbiol. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. J Virol. 2005;79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baer GS, Dermody TS. J Virol. 1997;71:4921–4928. doi: 10.1128/jvi.71.7.4921-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodkin D, Nibert ML, Fields BN. J Virol. 1989;63:4676–4681. doi: 10.1128/jvi.63.11.4676-4681.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nibert ML. Cambridge, MA: Harvard Univ; 1989. Ph.D. thesis. [Google Scholar]
- 13.Sturzenbecker LJ, Nibert ML, Furlong DB, Fields BN. J Virol. 1987;61:2351–2361. doi: 10.1128/jvi.61.8.2351-2361.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang C-T, Zweerink HJ. Virology. 1971;46:544–555. doi: 10.1016/0042-6822(71)90058-4. [DOI] [PubMed] [Google Scholar]
- 15.Silverstein SC, Schonberg M, Levin DH, Acs G. Proc Natl Acad Sci USA. 1970;67:275–281. doi: 10.1073/pnas.67.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS. J Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borsa J, Copps TP, Sargent MD, Long DG, Chapman JD. J Virol. 1973;11:552–564. doi: 10.1128/jvi.11.4.552-564.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joklik WK. Virology. 1972;49:700–715. doi: 10.1016/0042-6822(72)90527-2. [DOI] [PubMed] [Google Scholar]
- 19.Chandran K, Farsetta DL, Nibert ML. J Virol. 2002;76:9920–9933. doi: 10.1128/JVI.76.19.9920-9933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandran K, Parker JS, Ehrlich M, Kirchhausen T, Nibert ML. J Virol. 2003;77:13361–13375. doi: 10.1128/JVI.77.24.13361-13375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drayna D, Fields BN. J Virol. 1982;41:110–118. doi: 10.1128/jvi.41.1.110-118.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borsa J, Long DG, Sargent MD, Copps TP, Chapman JD. Intervirology. 1974;4:171–188. doi: 10.1159/000149856. [DOI] [PubMed] [Google Scholar]
- 23.Nibert ML, Odegard AL, Agosto MA, Chandran K, Schiff LA. J Mol Biol. 2005;345:461–474. doi: 10.1016/j.jmb.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 24.Odegard AL, Chandran K, Zhang X, Parker JS, Baker TS, Nibert ML. J Virol. 2004;78:8732–8745. doi: 10.1128/JVI.78.16.8732-8745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nibert ML, Schiff LA, Fields BN. J Virol. 1991;65:1960–1970. doi: 10.1128/jvi.65.4.1960-1967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith RE, Zweerink HJ, Joklik WK. Virology. 1969;39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 27.Liemann S, Chandran K, Nibert ML, Harrison SC. Cell. 2002;108:283–295. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooper JW, Fields BN. J Virol. 1996;70:672–677. doi: 10.1128/jvi.70.1.672-677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooper JW, Fields BN. J Virol. 1996;70:459–467. doi: 10.1128/jvi.70.1.459-467.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lucia-Jandris P, Hooper JW, Fields BN. J Virol. 1993;67:5339–5345. doi: 10.1128/jvi.67.9.5339-5345.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nibert ML, Fields BN. J Virol. 1992;66:6408–6418. doi: 10.1128/jvi.66.11.6408-6418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandran K, Walker SB, Chen Y, Contreras CM, Schiff LA, Baker TS, Nibert ML. J Virol. 1999;73:3941–3950. doi: 10.1128/jvi.73.5.3941-3950.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandran K, Nibert ML. J Virol. 1998;72:467–475. doi: 10.1128/jvi.72.1.467-475.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomita T, Noguchi K, Mimuro H, Ukaji F, Ito K, Sugawara-Tomita N, Hashimoto Y. J Biol Chem. 2004;279:26975–26982. doi: 10.1074/jbc.M402676200. [DOI] [PubMed] [Google Scholar]
- 35.Lang S, Palmer M. J Biol Chem. 2003;278:38167–38173. doi: 10.1074/jbc.M303544200. [DOI] [PubMed] [Google Scholar]
- 36.Sugawara N, Tomita T, Kamio Y. FEBS Lett. 1997;410:333–337. doi: 10.1016/s0014-5793(97)00618-2. [DOI] [PubMed] [Google Scholar]
- 37.Kuga S. J Chromatogr. 1981;206:449–461. [Google Scholar]
- 38.Scherrer R, Gerhardt P. J Bacteriol. 1971;107:718–735. doi: 10.1128/jb.107.3.718-735.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jayasuriya AK, Nibert ML, Fields BN. Virology. 1988;163:591–602. doi: 10.1016/0042-6822(88)90300-5. [DOI] [PubMed] [Google Scholar]
- 40.Sharpe JC, London E. J Membr Biol. 1999;171:209–221. doi: 10.1007/s002329900572. [DOI] [PubMed] [Google Scholar]
- 41.Tosteson MT, Nibert ML, Fields BN. Proc Natl Acad Sci USA. 1993;90:10549–10552. doi: 10.1073/pnas.90.22.10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Chandran K, Nibert ML, Harrison SC. J Virol. 2006 doi: 10.1128/JVI.01343-06. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, Nolan DP, Lins L, Homble F, Vanhamme L, Tebabi P, Pays A, Poelvoorde P, et al. Science. 2005;309:469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 44.Sonawane ND, Szoka FC, Jr, Verkman AS. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 45.Danthi P, Tosteson M, Li Q, Chow M. J Virol. 2003;77:5266–5274. doi: 10.1128/JVI.77.9.5266-5274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tosteson MT, Chow M. J Virol. 1997;71:507–511. doi: 10.1128/jvi.71.1.507-511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gromeier M, Wetz K. J Virol. 1990;64:3590–3597. doi: 10.1128/jvi.64.8.3590-3597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belnap DM, Filman DJ, Trus BL, Cheng N, Booy FP, Conway JF, Curry S, Hiremath CN, Tsang SK, Steven AC, et al. J Virol. 2000;74:1342–1354. doi: 10.1128/jvi.74.3.1342-1354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Virgin HW IV, Bassel-Duby R, Tyler KL. J Virol. 1988;62:4594–4604. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]