Abstract
Numerous bacteria cope with the scarcity of iron in their microenvironment by synthesizing small iron-scavenging molecules known as siderophores. Mammals have evolved countermeasures to block siderophore-mediated iron acquisition as part of their innate immune response. Secreted lipocalin 2 (Lcn2) sequesters the Escherichia coli siderophore enterobactin (Ent), preventing E. coli from acquiring iron and protecting mammals from infection by E. coli. Here, we show that the iroA gene cluster, found in many pathogenic strains of Gram-negative enteric bacteria, including E. coli, Salmonella spp., and Klebsiella pneumoniae, allows bacteria to evade sequestration of Ent by Lcn2. We demonstrate that C-glucosylated derivatives of Ent produced by iroA-encoded enzymes do not bind purified Lcn2, and an iroA-harboring strain of E. coli is insensitive to the growth inhibitory effects of Lcn2 in vitro. Furthermore, we show that mice rapidly succumb to infection by an iroA-harboring strain of E. coli but not its wild-type counterpart, and that this increased virulence depends on evasion of host Lcn2. Our findings indicate that the iroA gene cluster allows bacteria to evade this component of the innate immune system, rejuvenating their Ent-mediated iron-acquisition pathway and playing an important role in their virulence.
Keywords: bacterial pathogens, host defense, innate immunity, iron, siderophores
Iron is essential for the growth of nearly all bacteria, but the insolubility of ferric iron results in extremely low bioavailability of this nutrient in most microenvironments. To satisfy their requirement for iron, many bacteria produce iron-chelating small molecules known as siderophores, which they export in their apo form and import as ferric complexes (1). Mammals, in turn, have evolved defense mechanisms that exploit the bacterial requirement for iron. During infection, a general mammalian strategy for limiting bacterial growth is to up-regulate expression of lactoferrin receptors and ferritin, which decreases the concentration of extracellular iron in serum (2). A more specific strategy involves the mammalian protein Lcn2, also known as siderocalin, neutrophil gelatinase-associated lipocalin, 24p3, or uterocalin (3–6). We have shown that Lcn2 is a bacteriostatic protein that is constitutively present in neutrophil granules and can be induced and secreted in response to activation of innate immune receptors such as Toll-like receptor 4 (TLR4) (5). Lcn2 binds several ferric siderophore complexes including the ferric complex of Ent (Fig. 1A; refs. 4 and 7), which is produced by Gram-negative enteric bacteria, including Escherichia coli, Salmonella enterica, and Klebsiella pneumoniae. Lcn2 exerts its growth-inhibitory effect by sequestering Fe(III)-bound Ent from Ent-producing bacteria, effectively starving them of iron. Indeed, Lcn2-deficient (−/−) mice are profoundly sensitive to infection by E. coli (5, 8) but not Staphylococcus aureus (5), which does not rely on Ent for iron acquisition but rather on siderophores that are predicted not to bind to Lcn2.
Fig. 1.
Schematic of the iroA gene cluster and strategy for its incorporation into E. coli H9049. (A) Chemical structures of Ent-family siderophores. (B) Schematic of iroA gene cluster. (C) Reverse-phase HPLC analysis of siderophores secreted by H9049-iroA and H9049-CAS, with monitoring at 316 nm. Peak assignments were made by MALDI-MS analysis of the corresponding fractions.
Despite the efficacy of Lcn2 at blocking Ent-mediated iron acquisition, pathogenic strains of enteric bacteria are known to harbor the gene cluster responsible for Ent production, suggesting that alternative forms of Ent might be used for iron acquisition during infection. One alternative emerged with the discovery of modified forms of Ent, termed salmochelins, in the culture broth of Sa. enterica. Production of salmochelins is associated with the iroA gene cluster (Fig. 1B), which is also found in pathogenic strains of E. coli, Salmonella spp., and K. pneumoniae (9–12). The iroA gene cluster encodes five proteins involved in modification and transport of Ent: IroB C-glucosylates Ent (13), IroE hydrolytically linearizes Ent (14, 15), IroD degrades ferric complexes of the siderophore to release iron in the bacterial cytoplasm (14, 15), and IroN and IroC transport Ent and its derivatives (16). Rather than Ent, iroA-harboring bacteria secrete a mixture of salmochelins S4 and S2 (Fig. 1A), which are macrocyclic and linearized versions of diglucosylated Ent, respectively (16, 17). We have shown that glucosylation and linearization of Ent decrease its propensity to partition into lipid bilayers, which might be advantageous for iron scavenging in a membrane-rich microenvironment (18).
Results and Discussion
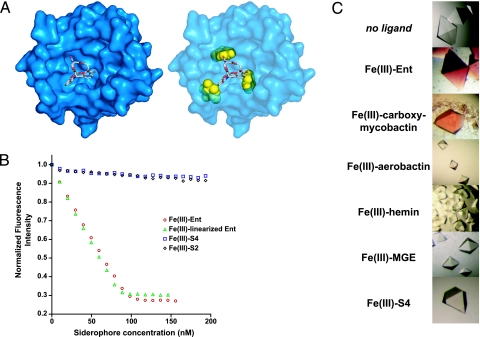
Hypothesizing that iroA-directed Ent modification would allow pathogenic bacteria to evade Lcn2-mediated host defense, we began by investigating the interaction between Ent and Lcn2. Structural analyses of siderophore-binding by Lcn2 (4, 19, 20) demonstrate significant steric clashes between even a single modeled glucose adduct on Ent in any of the catechol-binding pockets in the Lcn2 calyx (Fig. 2A). We confirmed this hypothesis by using two separate assays. The equilibrium dissociation constant (Kd) for Fe(III)-Ent binding to human or murine Lcn2 is 0.43 nM by a fluorescence quenching assay (Fig. 2B), in agreement with our previously published data (4). Under the same conditions, the absence of quenching with Fe(III)-S4 and Fe(III)-S2 demonstrates a Kd > 1 μM (Fig. 2B), which is >103-fold weaker than the affinity of the IroN homolog FepA for its siderophore ligand, Ent (21). In a separate qualitative cocrystallization binding assay, millimolar Lcn2 was mixed with excess ferric siderophore, and the degree of ligand binding was determined by the color of the crystals as judged by visual inspection. We observed red crystals when Lcn2 was mixed with Fe(III)-Ent. In contrast, colorless crystals formed when Lcn2 was mixed with either Fe(III)-S4 or Fe(III)-S2, which indicates that these ligands do not bind Lcn2 under these conditions (22) (Fig. 2C). Thus, structural and biochemical data suggest that Lcn2 does not bind Fe(III)-S4 or Fe(III)-S2 under physiological conditions.
Fig. 2.
Lcn2 does not bind Fe(III)-S4 or Fe(III)-S2. (A) View of Lcn2 crystal structure showing the predicted steric clash between Ent-bound glucose moieties (in yellow) and Lcn2. Tri-glucosyl-Ent was modeled into the binding pocket by using PyMOL (http://pymol.sourceforge.net/). (B) Fluorescence quenching curves of ferric siderophores added to 100 nM Lcn2. (C) Lcn2 cocrystallization assay. Red crystals indicate the formation of a complex between Lcn2 and the ferric siderophore, whereas colorless crystals suggest the absence of siderophore binding.
To study how salmochelins affect the ability of Lcn2 to limit the growth of iroA-harboring bacteria, we used the new conjugative att shuttle (CAS)-CRIM system,‡‡ which allows the introduction of large genetic elements into bacterial strains that are not genetically manipulable by other methodologies. The iroA gene cluster was introduced into E. coli H9049, a nonpathogenic clinical isolate of E. coli that requires Ent production to grow in iron-deficient media (5) and does not harbor the iroA gene cluster (data not shown). We cloned the iroA locus from uropathogenic E. coli CFT073 by yeast recombination, subcloned it into a CRIM (23) vector, and integrated the iroA-CRIM plasmid into the CAS plasmid F-CAS7 (see Fig. 5, which is published as supporting information on the PNAS web site). The hybrid F-CAS7-iroA plasmid was subsequently transferred into E. coli H9049 by conjugation (Fig. 5), yielding H9049/CAS::iroA (H9049-iroA). A similar process was carried out by using the empty F-CAS7 plasmid to create the iroA− control strain, H9049-CAS. As expected, both F-CAS7 plasmids in these strains were stable in the absence of antibiotic selection (data not shown). We confirmed by HPLC and MALDI-MS analysis of cell-free culture broth that H9049-CAS excretes Ent and its hydrolytic breakdown products, whereas H9049-iroA excretes linearized, glucosylated derivatives of Ent (Fig. 1C).
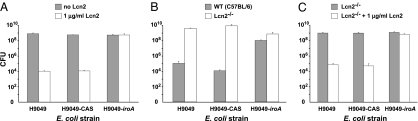
We then compared the ability of H9049, H9049-iroA, and H9049-CAS to grow in vitro in the presence of purified Lcn2 (Fig. 3A). Although all three strains grew well in the absence of Lcn2, the addition of 1 μg/ml Lcn2 to the culture media, a concentration similar to that of Lcn2 in serum after infection (5), completely inhibited the growth of parental H9049 and H9049-CAS, whereas Lcn2 had no effect on the growth of H9049-iroA. To test whether the profound insensitivity of H9049-iroA to Lcn2 would facilitate its growth in acute phase serum, we next compared the growth of H9049-iroA and H9049-CAS in media supplemented with acute phase serum from wild-type (C57BL/6) mice and their Lcn2-deficient counterparts (Fig. 3B). Indeed, whereas both strains grew well in the presence of acute phase serum from Lcn2-deficient mice, only H9049-iroA was able to grow in the presence of acute-phase serum from wild-type mice. Supplementing Lcn2−/− serum with purified Lcn2 abrogated the growth of H9049-CAS, but had no effect on the growth of H9049-iroA, demonstrating that the effect of iroA was specific for Lcn2 (Fig. 3C). These in vitro results indicate that the iroA gene cluster confers Lcn2 resistance to Ent-producing bacteria, allowing them unfettered iron acquisition that is essential for growth.
Fig. 3.
H9049-iroA is insensitive to Lcn2-mediated growth suppression in vitro. (A) Growth of H9049, H9049-CAS, and H9049-iroA in the presence and absence of Lcn2. (B) Growth of H9049, H9049-CAS, and H9049-iroA in the presence of acute phase serum from C57BL/6 vs. Lcn2−/− mice. (C) Growth of H9049, H9049-CAS, and H9049-iroA in the presence of Lcn2−/− acute phase serum with or without Lcn2.
We next investigated the effect of iroA on the virulence of E. coli H9049 during i.p. infection of mice. i.p. injection of either H9049-iroA or H9049-CAS resulted in the rapid mortality of Lcn2-deficient mice (Fig. 4A), consistent with our previously reported results (5). However, although 80% of the wild-type C57BL/6 mice survived i.p. challenge with H9049-CAS, only 10% percent of these mice survived i.p. challenge with H9049-iroA (Fig. 4B); similar results were seen with Lcn2 heterozygotes (Fig. 4C). Thus, the iroA gene cluster confers Lcn2 resistance to E. coli H9049 and profoundly augments their virulence in vivo.
Fig. 4.
The iroA gene cluster confers virulence to E. coli H9049 in a Lcn2-dependent manner. Survival curves of Lcn2−/− (A), wild-type C57BL/6 (B), and Lcn2+/− (C) mice after i.p. injection of H9049-CAS (n = 9, Lcn2−/−; n = 10, wild-type and Lcn2+/−) and H9049-iroA (n = 9, Lcn2−/−; n = 10, wild-type and Lcn2+/−).
The iroA gene cluster renders bacteria insensitive to Lcn2-mediated Ent sequestration, and is a bacterial strategy to evade this specific effector mechanism of the innate immune system. Other examples of bacterial evasion of immune effectors include the chemical modifications of LPS that prevent antimicrobial cationic peptides from binding to the bacterial cell wall (24), efflux pumps that eliminate antimicrobial peptides, and secreted proteases that cleave immunoglobulins, complement and other serum proteins (25). In addition, bacteria have evolved mechanisms to avoid detection by innate immune receptors: TLR5 evasion by the flagellin molecule for strains of Campylobacter, Helicobacter, and Bartonella (26); and LPS modifications that alter TLR4 recognition, as seen in Salmonella, Pseudomonas, Yersinia, and other bacteria (27–29). Furthermore, several bacteria use complex secretion systems (molecular syringes) to inject effector molecules into the host cell cytoplasm as a means to subvert the host cell machinery or block signaling pathways (30).
Among these innate immune evasion strategies, iroA-mediated Lcn2 evasion is unique, because iroA-harboring bacteria have targeted a secreted small molecule for chemical alteration and acquired a complete metabolic system for export, uptake and degradation to preserve iron acquisition. To prevent Ent from binding Lcn2 without compromising its ability to bind iron, an unusual C-glycosyltransferase (IroB) appends glucose to carbon atoms on two of the three catechol rings of Ent. Because glucosylated Ent derivatives are bulkier than Ent, proteins from the Ent gene cluster are unable to transport and degrade these salmochelins. Three additional proteins are required for salmochelin-mediated iron acquisition: a predicted exporter for apo-salmochelin efflux (IroC), a porin for ferric-salmochelin import (IroN), and an esterase to degrade ferric-salmochelins (IroD), releasing iron in the bacterial cytoplasm. The fifth protein encoded by the iroA cluster, IroE, is probably not required for Lcn2 evasion, but improves the iron-scavenging ability of salmochelin S4 by linearizing this siderophore to the more hydrophilic salmochelin S2. Such an elaborate “second layer” of secondary metabolism attests to the importance of Ent-mediated iron acquisition for enteric bacteria, and to the efficacy of Lcn2 in thwarting Ent-mediated iron acquisition. This result may be paralleled for other Lcn2-binding siderophores, such as the carboxymycobactins generated by mycobacteria, which are predicted to include both Lcn2-binding and nonbinding iso forms (20).
Our detailed understanding of this innate immune evasion mechanism reveals a level of complexity that we predict will be a commonality. Although iroB is the only gene required to abrogate Lcn2 binding, four additional genes are needed to accommodate the IroB modification and preserve Ent-mediated iron acquisition. Thus, the innate immune system has targeted functions that require multigenic compensation in order for the pathogen to preserve its vital function.
Our data demonstrate that the iroA gene cluster, a potent virulence factor in some pathogenic strains of E. coli, Salmonella spp, and K. pneumoniae, mediates bacterial virulence through Lcn2 evasion. Indeed, molecular epidemiological studies have associated the presence of iroN in isolates of E. coli and Salmonella with virulence (9, 31, 32), although it is not known how many of these harbor the entire iroA gene cluster. Our results warrant further investigation of the prevalence of the iroA cluster in pathogenic enteric bacteria and the function of Lcn2 as an innate barrier to microbial infection.
A recent report demonstrated the efficacy of targeting bacterial virulence factors with small molecules for the human pathogen, Vibrio cholerae (33). Unlike traditional antibiotics such as those that inhibit cell wall or protein synthesis, novel “virulence-specific” antibiotics would target pathogenic bacteria while sparing commensal bacteria. Our study has identified another bacterial virulence factor that may be a relevant drug target. Inhibition of iroA function could selectively attenuate the virulence of iroA-harboring bacteria, rendering them susceptible to Lcn2 without affecting iroA-deficient commensals. Further efforts to determine the contribution of each iroA gene to virulence likely will identify new members of the emerging class of virulence-specific antibiotic targets.
Materials and Methods
Cloning the iroA Gene Cluster.
A genomic fragment containing the iroA gene cluster from E. coli CFT073 was isolated by yeast recombinational cloning with a previously described two-step procedure (34). The fragment is flanked by EcoRI and HindIII recognition sites in the genomic sequence, where the EcoRI site is ≈0.4 kb from the start codon of iroN and the HindIII site is ≈0.9 kb from the start codon of iroB. The capture vector pMF145 was constructed by recombination of four DNA fragments in Sa. cerevisiae: a fragment from pLLX8 containing markers selectable in E. coli (bla) and counterselectable in yeast (CYH2), the pLLX13 vector linearized by NheI, and two targeting sequences with homology to genomic regions flanking the EcoRI and HindIII sites and to the pLLX8-derived fragment. The pLLX8 fragment was PCR-amplified by using the primers T5 (5′-TTTTCTAGAACGCGTTTAATTAAAATCTAAAGTATATATGAGTAAAC-3′) and T6 (5′-CCCTCTAGAGTTAACGTTTAAACAAAAAACGGTGAAAATGGGTGATAG-3′). The targeting sequences were PCR amplified from E. coli CFT073 genomic DNA by using the following primer pairs: TS1-F (5′-TCCCTATATTACCCTGTTATCCCTAGCGTAACTATCGATCTCGAGcgcatcttgatgatgtcagctc-3′) and TS1-R (5′-CATATATACTTTAGATTTTAATTAAACGCGTTCTAGAAAAgcatcgtaatggtgtcacctac-3′); TS2-F (5′-CATTTTCACCGTTTTTTGTTTAAACGTTAACTCTAGAGGGcttgcggtctggcaactgcaag-3′) and TS2-R (5′-AGGGATAACAGGGTAATATAGAGATCTGGTACCCTGCAGGAGCTCtgccccgtcaacctgaccaggg-3′). All four fragments were transformed into lithium acetate-treated Sa. cerevisiae CRY1–2 and cells were plated on uracil-deficient media. DNA was isolated from yeast transformants and used to transform into E. coli Top10 (Invitrogen, Carlsbad, CA), and cells were plated on LB-agar containing 100 μg/ml ampicillin. The identity of recovered pMF145 clones was confirmed by restriction digest. To create pMF146 by capturing the iroA-containing genomic fragment with pMF145, lithium acetate-treated Sa. cerevisiae CRY1–2 was transformed with MluI-linearized pMF145 and lightly sheared E. coli CFT073 genomic DNA, and cells were plated on uracil-deficient media containing 2.5 μg/ml cycloheximide. Plasmid DNA was isolated from transformants by resuspending colonies in 15 μl of zymolyase buffer (10 mM sodium phosphate, pH 7.8/1 M sorbitol/2.5 mg/ml zymolyase) and incubating for 50 min at 37°C, and used to transform E. coli Top10 (Invitrogen). The identity of pMF146 clones was confirmed by diagnostic PCR and restriction digest.
CAS-CRIM-Based Strain Construction.
E. coli H9049 has been described (5). Luria–Bertani (LB) broth was used for growing E. coli K12 derivatives and E. coli H9049. Tetracycline (Tet) was used to select exconjugants containing F-CAS plasmids and their derivatives at 12.5 μg/ml for both E. coli K12 derivatives and H9049 derivatives. Kanamycin (Kan) was used for selecting the integration of the CRIM plasmids onto the CAS plasmids at 10 μg/ml. Erythromycin was used at 400 μg/ml for selection of exconjugants. Diaminopimelic acid (DAP) was supplied at a final concentration of 0.4 μM/ml for DAP− derivatives, which require DAP for growth.
The HindIII site outside of the multiple-cloning region of the previously described CRIM vector pAH70 (23) was eliminated by a primer-directed mutagenesis strategy with a previously described whole-plasmid amplification procedure (35) to yield pMF152. The EcoRI-HindIII fragment containing the iroA gene cluster was then subcloned from pMF146 into pMF152, yielding pMF154. pMF154 was integrated into the F-CAS7 (pLZ119) plasmid at the attHK site as described (23). One single copy integrant was saved, which harbors the hybrid F-CAS7-iroA plasmid.
The hybrid F-CAS7-iroA plasmid was mated first into an E. coli DAP− auxotroph, BW29829. The intermediate exconjugants were then mated with H9049. Mating was done by reviving the donor and recipient on the appropriated media (LB supplemented with Tet or DAP or both) and incubating them overnight. The donor and the recipient strains were streaked onto the same LB plate supplemented with DAP, incubated for 4 h at 37°C, and then mixed by streaking and incubated at 37°C for another 4–6 h. The mating mixtures were then streaked onto an appropriate selection media. If BW29829 (erythromycin-resistant) was the recipient, DAP, erythromycin, and Tet were added for the selection of the exconjugants. If BW29829 containing a CAS plasmid was a donor, only Tet was used for the selection.
Analysis of Secreted Siderophores.
Overnight cultures of H9049-iroA and H9049-CAS grown in LB broth were diluted 1:100 into M63-glycerol medium (16) and incubated for 18 h at 37°C in a rotary shaker at 225 rpm. After cells were removed from the culture broth by centrifugation, the 1 ml of the cell-free culture fluid was lyophilized to dryness and resuspended in 200 μl of 20% acetonitrile in 0.1% aqueous trifluroacetic acid. One hundred-microliter aliquots were analyzed by reverse-phase HPLC, monitoring at 316 nm, with a gradient of 0–40% acetonitrile in 0.1% aqueous trifluoroacetic acid. After MALDI-MS analysis of the peaks corresponding to siderophore species, we assigned their identities as Ent ([M+H]+ m/z: calculated 670.1, observed 670.2), linearized Ent ([M+H]+ m/z: calculated 688.2, observed 688.3), (DHB-Ser)2 ([M+H]+ m/z: calculated 465.1, observed 465.2), linearized MGE ([M+H]+ m/z: calculated 850.2, observed 850.3), and S2 ([M+H]+ m/z: calculated 1,012.3, observed 1,012.5).
Lcn2-Siderophore Cocrystallization Assay.
Recombinant Lcn2, prepared as described (22) was cocrystallized with test siderophores as described (20). Fe(III)-bound siderophores were added at a 2-fold molar excess and equilibrated with protein at neutral pH before crystallization. In this crystal form, the concentration of the protein (and, therefore, specifically bound ligand) is ≈25 mM. Therefore, a positive result in this assay corresponds to a dramatically more colored crystal in a less colored background, where the color is due to the UV-vis absorption of the ferric catecholate complexes. Negative results correspond to an even color level across crystal and field, which can include unbound siderophore.
Lcn2-Siderophore Fluorescence Assay.
Ent, linearized Ent, S4, and S2 were obtained as described (18). Tryptophan fluorescence quenching of recombinant Lcn2 was measured on a Jobin Yvon fluoroLOG-3 fluorometer with 3-nm slit widths in Tris-buffered saline (pH 7.4) plus 32 μg/ml ubiquitin (Sigma, St. Louis, MO). Measurements were made at a protein concentration of 100 nM. Fluorescence values were corrected for dilution upon addition of ferric-siderophore. Fluorescence data were analyzed by nonlinear regression analysis of fluorescence response versus ferric-siderophore concentration by using a one-site binding model as implemented in DYNAFIT (36).
In Vitro Growth Assay.
For in vitro growth measurements, ≈104 cfu log-phase E. coli H9049, H9049-CAS, and H9049-iroA were added in 100 μl media per well in 96-well plates. The bacteria were grown in RPMI medium 1640 with 10% heat-inactivated FBS or acute phase serum from Lcn2−/− or C57BL/6 mice. Recombinant mouse Lcn2 was added at the indicated concentration, and growth was measured as cfu from serial dilutions plated onto on LB agar.
Mouse i.p. Injections and Infection Model.
The Lcn2-deficient mice have been described (5). These mice were back-crossed for eight generations onto to the C57BL/6 background to generate homozygote and heterozygote controls. Genotype was confirmed by PCR analysis of genomic DNA. The C57BL/6 mice that were used for breeding and controls were purchased from The Jackson Laboratory (Bar Harbor, ME). Lcn2-deficient mice were age- and sex-matched with C57BL/6 or Lcn2+/− controls. The mice were housed in a specific pathogen-free facility, and all procedures were approved by the Institutional Animal Care and Use Committee at the Institute for Systems Biology. Acute phase serum was produced by i.p. injection of 109 cfu heat-killed E. coli into Lcn2−/− and wild-type mice, and after 24 h mice were euthanized and bled. For infections with live bacteria, log phase E. coli H9049, H9049-CAS, and H9049-iroA were washed, resuspended in PBS to the desired concentration and injected i.p. in 0.5 ml per mouse by using a 30 gauge needle. All injected doses were verified by counting cfu. For measuring the bacterial burden, heparinized blood samples and homogenized liver and spleen were diluted in PBS and plated on LB agar to determine cfu.
Supplementary Material
Acknowledgments
We thank Esteban Martinez-Garcia for help with yeast recombinational cloning, April Clark and David Rodriguez for animal care, and Shizou Akira (Osaka University, Osaka, Japan) for providing the original Lcn2-deficient mice. This work was supported in part by National Institutes of Health Grants AI062859 (to K.D.S.), AI42738 and GM20011 (to C.T.W.), AI052286 (to A.A.), AI059432 (to R.K.S.), GM62662 (to B.L.W.), AI11744-28 (to K.N.R.), and GM065400 (to D.R.L); a grant from the Howard Hughes Medical Institute (to D.R.L.); a fellowship from the Hertz Foundation (to M.A.F.); and a fellowship from the Jane Coffin Childs Memorial Fund (to H.L.).
Abbreviations
- Ent
enterobactin
- Lcn2
lipocalin 2
- S2
salmochelin S2
- S4
salmochelin S4.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
The CAS-CRIM system is a new tool for high-throughput functional genomic studies of bacteria, which permits easy gene manipulation in E. coli K-12 and efficient transfer from E. coli to other bacteria including pathogens (L.Z., Y.Y., and B.L.W., unpublished data). In brief, this system is based on conditional-replication, integration, and modular (CRIM) (23) plasmids and a series of highly engineered, self-mobilizable CAS plasmids that contain multiple attachment (attB) sites for integration of various CRIM plasmids. The resulting CAS-CRIM hybrid plasmid can be readily transferred to recipient bacteria by conjugation, which is facilitated by use of Dap- (diaminopimelic acid-requiring) E. coli.
References
- 1.Crosa JH, Walsh CT. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jurado RL. Clin Infect Dis. 1997;25:888–895. doi: 10.1086/515549. [DOI] [PubMed] [Google Scholar]
- 3.Devireddy LR, Teodoro JG, Richard FA, Green MR. Science. 2001;293:829–834. doi: 10.1126/science.1061075. [DOI] [PubMed] [Google Scholar]
- 4.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 5.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Ryon J, Nilsen-Hamilton M. Mol Reprod Dev. 1997;46:507–514. doi: 10.1002/(SICI)1098-2795(199704)46:4<507::AID-MRD9>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 7.Raymond KN, Dertz EA, Kim SS. Proc Natl Acad Sci USA. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger T, Togawa A, Duncan GS, Elia AJ, You-Ten A, Wakeham A, Fong HE, Cheung CC, Mak TW. Proc Natl Acad Sci USA. 2006;103:1834–1839. doi: 10.1073/pnas.0510847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumler AJ, Norris TL, Lasco T, Voight W, Reissbrodt R, Rabsch W, Heffron F. J Bacteriol. 1998;180:1446–1453. doi: 10.1128/jb.180.6.1446-1453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumler AJ, Tsolis RM, van der Velden AW, Stojiljkovic I, Anic S, Heffron F. Gene. 1996;183:207–213. doi: 10.1016/s0378-1119(96)00560-4. [DOI] [PubMed] [Google Scholar]
- 11.Fischbach MA, Lin H, Liu DR, Walsh CT. Nat Chem Biol. 2006;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 12.Foster JW, Hall HK. J Bacteriol. 1992;174:4317–4323. doi: 10.1128/jb.174.13.4317-4323.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischbach MA, Lin H, Liu DR, Walsh CT. Proc Natl Acad Sci USA. 2005;102:571–576. doi: 10.1073/pnas.0408463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin H, Fischbach MA, Liu DR, Walsh CT. J Am Chem Soc. 2005;127:11075–11084. doi: 10.1021/ja0522027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu M, Valdebenito M, Winkelmann G, Hantke K. Microbiology. 2005;151:2363–2372. doi: 10.1099/mic.0.27888-0. [DOI] [PubMed] [Google Scholar]
- 16.Hantke K, Nicholson G, Rabsch W, Winkelmann G. Proc Natl Acad Sci USA. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bister B, Bischoff D, Nicholson GJ, Valdebenito M, Schneider K, Winkelmann G, Hantke K, Sussmuth RD. Biometals. 2004;17:471–481. doi: 10.1023/b:biom.0000029432.69418.6a. [DOI] [PubMed] [Google Scholar]
- 18.Luo M, Lin H, Fischbach MA, Liu DR, Walsh CT, Groves JT. ACS Chem Biol. 2006;1:29–32. doi: 10.1021/cb0500034. [DOI] [PubMed] [Google Scholar]
- 19.Goetz DH, Willie ST, Armen RS, Bratt T, Borregaard N, Strong RK. Biochemistry. 2000;39:1935–1941. doi: 10.1021/bi992215v. [DOI] [PubMed] [Google Scholar]
- 20.Holmes MA, Paulsene W, Jide X, Ratledge C, Strong RK. Structure (London) 2005;13:29–41. doi: 10.1016/j.str.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Annamalai R, Jin B, Cao Z, Newton SM, Klebba PE. J Bacteriol. 2004;186:3578–3589. doi: 10.1128/JB.186.11.3578-3589.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 23.Haldimann A, Wanner BL. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 25.Finlay BB, McFadden G. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 26.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. Proc Natl Acad Sci USA. 2005;102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst RK, Adams KN, Moskowitz SM, Kraig GM, Kawasaki K, Stead CM, Trent MS, Miller SI. J Bacteriol. 2006;188:191–201. doi: 10.1128/JB.188.1.191-201.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller SI, Ernst RK, Bader MW. Nat Rev Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 29.Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Mol Microbiol. 2004;52:1363–1373. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- 30.Stebbins CE, Galan JE. Nature. 2001;412:701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- 31.Johnson JR, Scheutz F, Ulleryd P, Kuskowski MA, O'Bryan TT, Sandberg T. J Clin Microbiol. 2005;43:3895–3900. doi: 10.1128/JCM.43.8.3895-3900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dozois CM, Daigle F, Curtiss R., III Proc Natl Acad Sci USA. 2003;100:247–252. doi: 10.1073/pnas.232686799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Science. 2005;310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 34.Wolfgang MC, Kulasekara BR, Liang X, Boyd D, Wu K, Yang Q, Miyada CG, Lory S. Proc Natl Acad Sci USA. 2003;100:8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai JR, Fischbach MA, Liu DR, Walsh CT. Proc Natl Acad Sci USA. 2006;103:5314–5319. doi: 10.1073/pnas.0601038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuzmic P. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.