Abstract
Alphavirus vectors are being developed for possible human vaccine and gene therapy applications. We have sought to advance this field by devising DNA-based vectors and approaches for the production of recombinant vector particles. In this work, we generated a panel of alphavirus vector packaging cell lines (PCLs). These cell lines were stably transformed with expression cassettes that constitutively produced RNA transcripts encoding the Sindbis virus structural proteins under the regulation of their native subgenomic RNA promoter. As such, translation of the structural proteins was highly inducible and was detected only after synthesis of an authentic subgenomic mRNA by the vector-encoded replicase proteins. Efficient production of biologically active vector particles occurred after introduction of Sindbis virus vectors into the PCLs. In one configuration, the capsid and envelope glycoproteins were separated into distinct cassettes, resulting in vector packaging levels of 107 infectious units/ml, but reducing the generation of contaminating replication-competent virus below the limit of detection. Vector particle seed stocks could be amplified after low multiplicity of infection of PCLs, again without generating replication-competent virus, suggesting utility for production of large-scale vector preparations. Furthermore, both Sindbis virus-based and Semliki Forest virus-based vectors could be packaged with similar efficiency, indicating the possibility of developing a single PCL for use with multiple alphavirus-derived vectors.
The use of virus-derived expression vectors for gene therapy and vaccine applications increasingly is being pursued, with a number of diverse virus types and approaches. Alphaviruses are attractive candidates for such applications because of their high levels of replication and gene expression, their ability to infect a variety of diverse cell types, and the ability to manipulate cDNA clones from which infectious viral RNA may be transcribed (for review, see refs. 1 and 2). The alphavirus genome is a single-stranded, positive-sense RNA of approximately 11.7 kb and is encapsidated within an icosahedral capsid protein shell (for review, see ref. 3). Nucleocapsids, in turn, are surrounded by a host-derived lipid envelope from which the viral spike glycoproteins E1 and E2 protrude. Cytoplasmic replication of the RNA genome is mediated by four viral-encoded nonstructural proteins and proceeds through a full-length negative-sense intermediate. Subsequent positive-strand RNA synthesis results in both progeny genome RNA and an abundant, internally initiated subgenomic mRNA. The virus structural proteins are translated from the subgenomic mRNA as a polyprotein that is processed into the individual components of the virion.
The general strategy for construction of alphavirus-based expression vectors has been to substitute the viral structural protein genes with a heterologous gene, maintaining transcriptional control via the highly active subgenomic RNA promoter (1, 2, 4). As such, these vector replicons are suicide vectors, incapable of packaging progeny vector particles and causing productive infection. Vector replicon RNA can be transcribed in vitro and used directly, or the replicon RNA can be packaged into infectious vector particles by cotransfection of cultured cells with a complementing defective helper RNA, which provides the virion structural proteins in trans. Sindbis virus, Semliki Forest virus (SFV), and Venezuelan equine encephalitis virus are among the alphaviruses being exploited by using such approaches (4–7). Some potential limitations of the RNA-based vector replicon systems are utility for large-scale preparations and the generation of contaminating replication-competent virus (RCV). These issues have begun to be addressed through the conversion of alphavirus vectors into functional plasmid DNA formats that directly transcribe RNA vector replicons in vivo (8–13) and the development of split structural protein gene packaging systems (2, 7, 14). However, efficient methods for large-scale packaging of alphavirus vector particles, in the absence of RCV, remain to be developed.
In this paper, we report the development of packaging cell lines (PCLs) that can be used to produce alphavirus vector particle stocks that are free from contaminating RCV. The PCLs are stably transformed with inducible Sindbis virus structural protein expression cassettes and express these proteins only in response to input vector and subsequent synthesis of vector-encoded replicase proteins. Vector packaging was demonstrated by using a panel of different cell lines, and in one such PCL, separation of the capsid and envelope glycoprotein genes into distinct cassettes reduced the level of contaminating RCV below the limit of detection, while maintaining relatively high vector particle titers. Sindbis virus-derived PCLs were shown to package both Sindbis virus and SFV vectors efficiently and provided a method for serial propagation of vector particle seed stocks in the absence of detectable RCV. These data suggest potential utility of the PCLs for large-scale vector production and facilitating broad alphavirus applications.
MATERIALS AND METHODS
Construction of Structural Protein Expression Cassettes.
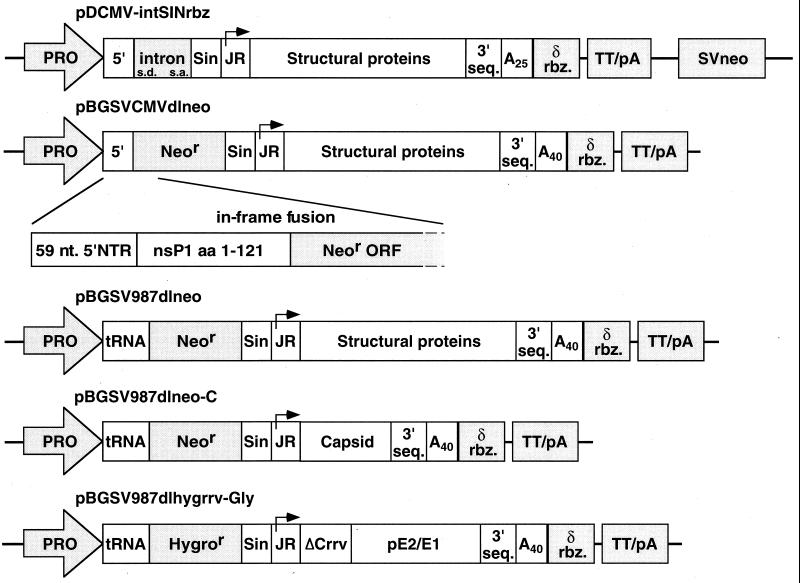
Plasmids for vector-inducible expression of the Sindbis virus structural proteins, either as a native polyprotein (C-pE2-E1) or as separate proteins (C and pE2-E1), were constructed similarly to those described by Dubensky et al. (10). Additional elements were incorporated by PCR amplification or by using synthetic oligonucleotides, including the hepatitis delta virus antigenomic ribozyme sequence (15), the simian virus 40 (SV40) small t intron sequence, or an SV40 neomycin selectable marker (see Fig. 1). Other cassettes contained the neomycin resistance gene fused with truncated nsP1 sequences at nucleotide 422. In addition, each cassette contained a cytomegalovirus (CMV) promoter linked to the Sindbis virus wild-type 5′ end, or the Rous sarcoma virus (RSV) long terminal repeat promoter linked to the Sindbis virus defective interfering RNA 5′ end tRNAAsp, as noted (6, 16). Split structural protein gene expression cassettes (Fig. 1) contained the Sindbis virus capsid protein gene, the neomycin resistance gene fusion, and the RSV promoter/5′ end tRNA sequence (pBGSV987dlneo), or the Sindbis virus glycoprotein genes, a hygromycin resistance gene fusion, the RSV promoter/tRNA 5′ end, and sequences from Ross River virus comprising the translational enhancer and the protease domain (ref. 14, pBGSV987dlhygrrv-Gly). SP6 promoter-based Sindbis virus and SFV vectors for in vitro transcription, as well as corresponding packaging defective helpers, have been described (4–6, 14). Other defective helpers were constructed to separately encode either the capsid protein (Sin-dlCap) or the envelope glycoproteins (Sin-dlGlyco) of Sindbis virus. In vitro transcription of linearized SP6 promoter-based Sindbis virus and SFV vectors encoding lacZ [SIN-βgal, ref. 10; SFV3-lacZ, ref. 5] and defective helper plasmids were performed as described (17).
Figure 1.
Schematic illustration of Sindbis virus structural protein expression cassettes used to generate PCL. Sindbis virus-derived sequences shown in white include the 5′ end (wild type or defective interfering-derived) and 3′ end cis replication elements, subgenomic RNA promoter (junction region, JR), structural protein genes (C, pE2, and E1), poly(A) tract and remaining nonstructural gene sequences deleted of nucleotides 422 (BspEI site) to 7335 (BamHI site). Shaded regions indicate other elements, including: RNA polymerase II promoter (PRO), hepatitis delta virus antigenomic ribozyme (δ rbz), bovine growth hormone transcription termination signal (TT/pA), SV40 small t antigen intron with splice donor (SD), and splice acceptor (s.a.) sites, neomycin or hygromycin phosphotransferase gene (neor or hygror, respectively), and SV40-driven neomycin phosphotransferase gene (SVneo). The fusion protein generated with neor and remaining nonstructural protein 1 (nsP1) amino acids is expanded to show more detail.
Selection, Screening, and Characterization of PCLs.
Baby hamster kidney (BHK)-21 cells transfected with expression cassette constructs were selected with G418 (GIBCO/BRL) or hygromycin (Boehringer Mannheim). Pooled foci of drug-resistant cells were cloned by limiting dilution, and individual clones were screened for packaging activity, by transfection with SIN-βgal vector RNA (10) using Lipofectin (GIBCO/BRL) or electroporation (5), as indicated in the text. Those clones with the highest levels of activity were expanded for further use. Northern and Western blot analysis of vector- specific or structural protein-specific RNA and proteins expressed in packaging cells were performed as described (10, 11). The titer of replication-incompetent SIN-βgal or SFV-βgal vector particles in clarified PCL culture supernatants was determined by infection of naive BHK-21 monolayers with serial dilutions, X-gal (5-bromo-4-chloro-3-indolyl β-d-galactoside) staining (10, 18), and counting the total number of blue cells per well at the appropriate dilution. Vector titer is designated as infectious units (IU)/ml. Contaminating RCV in culture supernatants was detected by standard plaque assay (plaque-forming units or PFU/ml) and by three serial undiluted passages in naive BHK cells.
Immunization, Cytotoxic T Lymphocytes (CTLs), and Antibody.
Female 6- to 8-week-old BALB/c mice (H-2d, Harlan–Sprague–Dawley) were immunized i.m. with a single dose of Sindbis virus vector particles encoding herpes simplex virus (HSV) glycoprotein B (SIN-HSVgB) via the tibialis anterior muscle. Particle preparations used for immunizations were partially purified and concentrated by sequential steps of filtration, polyethylene glycol precipitation, pelleting through 20% sucrose, and resuspension in PBS + lactose formulation buffer. At 17 days postimmunization, serum was obtained for antibody determination, and at 4 weeks, splenocytes were obtained for bulk analysis of antigen-specific CTLs as described (11, 19).
RESULTS
Derivation of Stable Alphavirus PCLs.
Previously, we described the construction of plasmid DNA-based Sindbis virus expression and packaging vectors that function in vivo (8, 10). To apply these advances toward an alphavirus vector packaging system that bypassed the need for cotransfection and provided a method for serial propagation of vector particles, we devised several strategies for generating PCLs. The role of alphavirus structural proteins in virus-induced cytopathology (20–22), coupled with down-regulation of RNA polymerase II promoters by alphavirus vectors, obviated traditional approaches for cell-based expression in PCLs. Therefore, alphavirus structural protein cassettes were designed to allow RNA amplification by the vector-encoded replicase and expression via the native subgenomic RNA promoter.
A Sindbis virus cDNA that contained 5′ and 3′ end cis sequences required for replication by vector-supplied nonstructural proteins was precisely juxtaposed within an RNA polymerase II expression cassette. The cDNA lacked most nonstructural protein gene sequences (nucleotides 422-7335; BspEI–BamHI), including the packaging sequence (23, 24), but maintained the viral subgenomic RNA junction region promoter and structural protein genes (Fig. 1). Thus, primary transcription from the packaging cassettes would produce an RNA molecule incapable of translating the Sindbis virus structural proteins before vector-induced synthesis of the subgenomic mRNA.
The basic structural protein expression cassette constructs were modified to include a selectable drug resistance marker plus other elements to enhance overall efficiency. In the first configuration, a separate SV40-driven neomycin phosphotransferase selectable marker (neor) was included in the plasmid. This construct, designated pDCMV-intSINrbz (Fig. 1), also contained the SV40 small t antigen intron (25), inserted into the region of nonstructural gene deletion, to facilitate RNA export of the primary transcript. In a second configuration, the neor marker was inserted directly within the nonstructural gene deletion site, creating an in-frame fusion between the remaining 121 aa of nsP1 and amino acid 11 of neor (pBGSVCMVdlneo, Fig. 1). Previously, we showed that a random deletion in neor, resulting in an N-terminal fusion at this site, retained protein activity (26). Linkage of the neor marker and structural protein genes to the same RNA polymerase II transcript was predicted to increase the probability of selecting functional packaging cells.
After dilution cloning of G418-resistant BHK-21 cell pools containing the packaging constructs, a higher percentage of cell lines positive for packaging activity was indeed obtained by using the pBGSVCMVdlneo cassette than with the pDCMV-intSINrbz cassette (data not shown). One clone with the highest level of packaging activity, S22 (from pDCMV-intSINrbz) and F15 (from pBGSVCMVdlneo), was chosen from each pool for further characterization. PCL clones S22 and F15 were compared for packaging activity by determining the titers of vector particles after transfection of the cells with SIN-βgal vector RNA. As shown in Table 1, packaging activities of the S22 and F15 PCLs were equivalent in several independent experiments and ranged from 1–5 × 106 IU/ml. In addition, testing for the generation of RCV also was performed. By standard plaque assay in naive BHK cells, RCV was detected consistently in supernatants from both cell lines, but the level always was found to be less than 100 PFU/ml (Table 1). Long-term stability for these PCLs was confirmed by demonstrating similar packaging activities after more than 40 serial passages (data not shown).
Table 1.
Comparison of packaging levels of SIN-βgal vector by PCL
| Cell line | Vector titer, IU/ml | RCV titer, PFU/ml |
|---|---|---|
| S22 | 1–5 × 106 | 5–100 |
| F15 | 1–5 × 106 | 5–100 |
| 987dlneo | 1–5 × 108 | ∼105 |
| 987dlsplit | 5 × 106—1 × 107 | <5* |
SIN-βgal vector RNA was transcribed in vitro and electroporated into the indicated PCLs. Culture supernatants were harvested and clarified at 48-hr posttransfection, and titers were determined for SIN-βgal vector particles (IU/ml) and contaminating RCV (PFU/ml). Detection limit of 1 PFU/2 × 106 IU.
RCV also was undetected by blind serial passage in naive BHK cells.
Previously, cotransfection experiments by Bredenbeek et al. (6) demonstrated that Sindbis virus defective helper RNAs modified to contain a naturally occurring defective interfering RNA 5′ end tRNAAsp sequence, in place of the wild-type viral 5′ end, produced higher levels of packaged vector. Therefore, the 5′ region of pBGSVCMVdlneo was substituted with a corresponding fragment containing the tRNAAsp sequence, to generate the packaging construct pBGSV987dlneo (Fig. 1, see Materials and Methods). G418-resistant 987dlneo packaging cell clones obtained after selection were found to produce significantly higher levels of packaged vector particles than either the F15 or S22 PCL. Repeated transfections of SIN-βgal vector RNA into 987dlneo PCL clone 11 produced vector particle titers in the range of 1–5 × 108 IU/ml (Table 1). However, similar to transient cotransfection results (6), the level of RCV in these vector preparations was found to be significantly higher. Regardless, the modification in 987dlneo suggested a strategy for PCL improvement, provided that the RCV could be eliminated.
Sindbis Virus Structural Protein Expression Is Tightly Regulated in Vector PCLs.
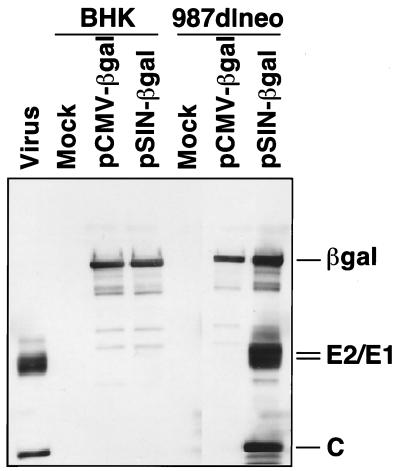
As structural protein expression in the alphavirus PCLs requires prior synthesis of subgenomic mRNA, induction should occur only after introduction of vector RNA and subsequent translation of the replicase proteins. To confirm this vector-induced structural protein gene expression, we examined Sindbis virus-specific protein and RNA products in packaging cells. Parental BHK-21 cells and 987dlneo PCL were transfected with the Sindbis virus-derived DNA vector pSIN-βgal (induced) or with a control CMV-βgal plasmid (uninduced), and total RNA and protein lysates were prepared. As shown in Fig. 2, Western blot detection of Sindbis virus capsid and envelope glycoprotein expression was observed only after transfection of packaging cells with pSIN-βgal vector, but not with the control CMV-βgal vector. Northern blot analysis also confirmed the presence of a subgenomic structural gene mRNA only after the transfection of PCL with pSIN-βgal vector (data not shown). Interestingly, the level of primary transcripts in the uninduced PCL was sufficiently low that poly(A) selection of total RNA was required to obtain a strong signal by Northern blot. The RNA and protein data indicated that the PCLs appear to function in the predicted manner.
Figure 2.
Western blot analysis of Sindbis virus structural proteins expressed in PCLs. Cell lysates were obtained from parental BHK-21 cells and 987dlneo PCLs 48 hr after transfection with a SIN-βgal DNA vector (induced), a CMV-βgal vector (uninduced), or mock transfection. Equivalent cell lysates were separated by SDS/PAGE and probed with a mixture of Sindbis virion and β-gal-specific rabbit polyclonal antisera. Positive control lysates were obtained from wild-type Sindbis virus-infected BHK-21 cells.
Sindbis Virus-Derived PCLs Function with Both Sindbis Virus and SFV Vectors.
We wanted to determine whether other alphavirus vector RNAs could be packaged by using the Sindbis virus-derived PCLs, to expand their utility. Therefore, in vitro-transcribed SFV vector RNA expressing β-galactosidase, SFV3-lacZ (5), was transfected into F15 or 987dlneo PCL, in parallel with the corresponding SIN-βgal vector RNA (Table 2). Culture supernatants were harvested 48 hr posttransfection, and the levels of packaged β-gal vector particles were determined. Interestingly, the titers of SFV3-LacZ and SIN-βgal vector particles were similar. The level of RCV detected in these preparations also was found to be similar. To further substantiate the observed packaging of SFV vector RNA by Sindbis virus structural proteins, we cotransfected either SIN-βgal or SFV3-LacZ vector RNAs into BHK-21 cells together with a Sindbis virus defective helper RNA encoding the structural proteins. The vector particle titers produced after cotransfection again were similar for SIN-βgal and SFV3-LacZ (Table 2). These data clearly indicated that Sindbis virus structural proteins were able to efficiently package SFV vector RNA, suggesting an enhanced utility of the Sindbis virus-derived PCL.
Table 2.
Packaging of Sindbis virus and SFV vectors with Sindbis virus structural proteins
| Packaging method | SIN-βgal vector | SFV-lacZ vector |
|---|---|---|
| F15 PCL | 5 × 106 IU/ml | 4 × 106 IU/ml |
| 5–100 PFU/ml | 5–100 PFU/ml | |
| 987dlneo PCL | 3 × 108 IU/ml | 3 × 108 IU/ml |
| 2 × 105 PFU/ml | 9 × 104 PFU/ml | |
| SIN DH co-txf | 4 × 108 IU/ml | 8 × 107 IU/ml |
| 4 × 104 PFU/ml | 1 × 104 PFU/ml |
SIN-βgal vector RNA was transcribed in vitro and electroporated into the indicated PCLs or cotransfected (co-txf) into BHK cells with defective helper (DH) RNA. Culture supernatants were harvested and clarified at 24 (DH co-txf) or 48 hr (PCL) posttransfection, and titers were determined for SIN-βgal vector particles (IU/ml) and contaminating RCV (PFU/ml). Detection limit of 1 PFU/2 × 106 IU.
Development of Alphavirus PCLs with Reduced Potential for RCV.
One possible method for large-scale production of alphavirus vector particles is sequential low multiplicity of infection of increasingly larger PCL cultures. Such serial propagation of alphavirus vector particles requires that no RCV be produced during the particle amplification process. With this goal in mind, we made additional modifications to the prototype PCL to reduce the potential for such an occurrence. Previous RNA cotransfection studies by us and others (2, 7, 14) demonstrated that separation of the alphavirus capsid and envelope glycoprotein genes into distinct defective helper RNAs reduced the generation of RCV below the limits of detection, while providing efficient vector packaging (>108 IU/ml). Therefore, a similar split structural protein gene strategy was adopted for the PCL expression cassettes.
As shown in Fig. 1, a Sindbis virus capsid gene cassette, pBGSV987dlneo-C, was assembled in the same basic neor-fusion configuration as pBGSV987dlneo. A similar nsP1 fusion approach also was used for the glycoprotein gene cassette, by using the hygromycin phosphotransferase selectable marker (hygr). In pBGSV987dlhygrrv-Gly, hygr was fused in-frame to the remaining nsP1 amino acids 1–121. To derive stable PCLs containing both cassettes, BHK-21 cells first were transfected with the capsid gene construct and then subjected to G418 selection. Positive cell lines were identified by Western blot analysis after dilution cloning and induction with SIN-βgal vector RNA. One capsid cell line subsequently was transfected with the glycoprotein gene construct and selected by using hygromycin. After dilution cloning, several positive split structural protein gene PCLs then were identified by a functional packaging assay and two with the highest levels of activity (referred to as 987dlsplit PCL clones 9 and 24) were characterized further.
Transfection of the two split structural protein gene PCLs with SIN-βgal vector RNA in several experiments produced vector particle titers in the range of 5 × 106 to 1 × 107 IU/ml (Table 1). Importantly, no RCV was detected in several experiments by either plaque assay or blind serial passage of culture supernatants in naive BHK cells. Similar to the genomic PCL, Northern and Western blot analysis revealed that capsid-specific and glycoprotein-specific subgenomic mRNAs and proteins were detected only after induction with a Sindbis virus-derived vector, but not with a control vector (data not shown).
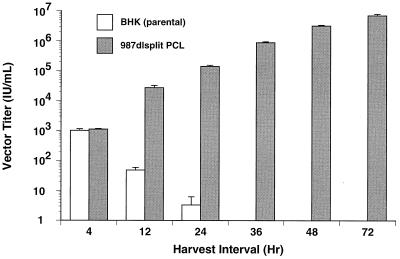
We next wanted to test the utility of the split structural protein gene PCL for amplification of vector particles. To accomplish this, a seed stock of SIN-βgal vector particles first was generated by vector cotransfection with split structural protein gene defective helper RNAs and shown to be free of detectable contaminating RCV. These particles then were used to infect in triplicate 987dlsplit PCL clone 24 and parental BHK-21 cells at a low multiplicity of infection (0.2). Production of new vector particles was determined by collecting each supernatant for titration at the indicated times and refeeding the same monolayers with an equal volume of fresh media. As shown in Fig. 3, vector particle production occurred only in the PCL and was continuous throughout the 72-hr time course of the experiment. Equally important was that no detectable RCV occurred in any of the 72-hr samples. Similar results also were observed with 987dlsplit PCL clone 9 (data not shown). These data clearly validate the utility of such PCL strategies for alphavirus vector systems.
Figure 3.
Vector particle amplification by a PCL. Parental BHK-21 cells and split structural protein gene PCL clone 24 were infected in triplicate with an RCV-free stock of SIN-βgal vector particles at a multiplicity of infection of 0.2. After a 1-hr infection period, the inoculum was removed, and cell monolayers were washed and replenished with fresh media. At the indicated intervals, culture supernatants were harvested for vector titer determination and replaced with fresh media. The titer of SIN-βgal vector particles produced during each period was quantitated by infecting naive BHK-21 cells and staining with X-gal (5-bromo-4-chloro-3-indolyl β-d-galactoside), as described in Materials and Methods.
Vector Particle Immunization Results in Potent Antigen-Specific Immune Responses.
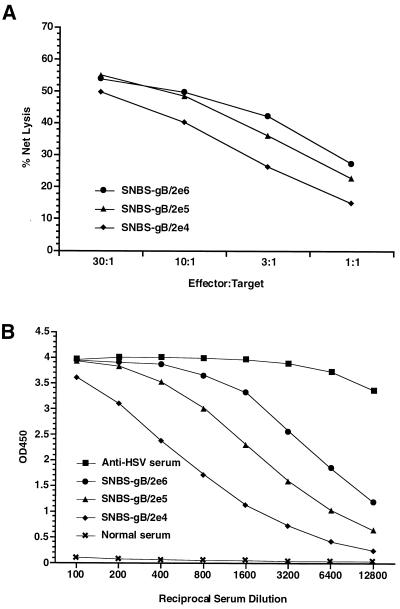
To demonstrate ability of alphavirus vector particle preparations free of contaminating RCV to induce potent antigen-specific immune responses, we immunized BALB/c mice i.m. with single doses of SIN vector particles expressing HSV glycoprotein B (SIN-HSVgB) and tested for the induction of HSVgB-specific CTL and antibody responses. As shown in Fig. 4, a single immunization with SIN-HSVgB vector particles at doses ranging from 2 × 104 to 2 × 106 IU resulted in the induction of robust HSVgB-specific antibody and CTL. Furthermore, the immune response induced by each of these doses was at a sufficient level for protection from lethal HSV challenge (ref. 11, data not shown). Thus, similar to previous HSVgB immunization data with plasmid DNA-launched alphavirus replicons (11), vector particles are extremely efficient, and by inference, more efficacious than conventional plasmid DNA vaccines in the HSV model.
Figure 4.
Induction of HSVgB-specific CTL (A) and antibody (B) responses in mice immunized with SIN-HSVgB vector particles. BALB/c mice were immunized once i.m. with the indicated doses of a SIN-HSVgB vector preparation free of RCV. Serum was obtained at 17 days postimmunization and tested for antibody by ELISA. Splenocytes from individual mice were obtained at week 4, stimulated in vitro with retroviral vector-transduced BC10ME (H-2d) cells expressing HSVgB, and tested separately for specific CTL lysis by using BC10ME target cells expressing either HSVgB or β-gal. Percent specific target cell lysis was calculated as [(experimental release − spontaneous release)/(maximum release − spontaneous release)] × 100, and graphs show specific lysis after subtraction of β-gal target cell background. Values shown within each dosage group represent the mean of four mice.
DISCUSSION
Application of viral-based vectors for human vaccines or gene therapy will require efficient means for their large-scale manufacture. In this work, we have generated stable PCLs that efficiently produce alphavirus vector particles without contaminating RCV. These PCLs should lead to broad advances in alphavirus vector technology by providing a system for large-scale amplification of vector particles. Two characteristics of alphavirus replication precluded the generation of alphavirus PCL analogous to retrovirus (27, 28) models: (i) alphaviruses maintain an RNA-based replication cycle that occurs entirely in the host cytoplasm, without an intermediate DNA stage; and (ii) alphavirus infection is lytic, thus suggesting a requirement for an inducible system. Previous demonstrations of RNA polymerase II-based expression of Sindbis virus vectors and their complementing packaging constructs (8–10), together with the Sindbis virus replicase-induced reporter system of Olivo et al. (29), provided initial clues of how to overcome these obstacles.
As described in this paper, vector-inducible alphavirus PCLs with a variety of different configurations have been developed. The first PCL contained the Sindbis virus structural protein genes in a single expression cassette. Although functional, the prototype PCLs were plagued by unacceptable levels of RCV. A significant increase in packaging activity of the PCL was observed with the 5′ end tRNAAsp modification. Unfortunately, this increase also was accompanied by an even greater increase in RCV. The increased level of contaminating RCV produced by the original 987dlneo cell line may have been caused by a higher abundance of cytoplasmic RNA, copackaging of the vector and helper RNA molecules (30), or other undefined mechanisms. In addition, it is unclear to what extent the RCV contributed to the increased level of vector particles. However, RCV does not appear to be an issue with the split structural protein gene PCL. Modification of the Sindbis virus PCL into a split structural protein gene configuration somewhat paralleled the evolution of retrovirus vector PCLs, whereby the required gene products were expressed from individual cassettes (31–33). In the case of alphavirus vector PCL, expression of the capsid protein and envelope glycoproteins from distinct cassettes rather than as the native polyprotein reduced the level of contaminating RCV to below the limit of detection. Furthermore, only in the context of the split structural protein gene cassettes was it possible to demonstrate PCL-based propagation and amplification of the replication-incompetent vector particles. Our results suggest that low multiplicity of infection of PCL cultures with vector particles is amenable to large-scale manufacture.
Another possible utility for the PCL technology is the packaging of diverse alphavirus vectors. This prospect was supported by the demonstration of efficient SFV vector packaging by Sindbis virus-derived PCLs. Molecular characterization of the Sindbis virus packaging sequence has identified a region within nsP1, between nucleotides 945 and 1076 (3, 23, 24), and this sequence is shared by all packaged Sindbis virus defective interfering RNAs that have been studied (34). Other genome sequences additionally may contribute to packaging, albeit at a much lower efficiency (6). In contrast, the region likely responsible for SFV packaging appears to reside within nsP2 (nucleotides 2737 to 2993), as suggested by common sequences among SFV-defective interfering RNAs and by identifying sequences that inhibit capsid assembly (35, 36). Because the primary sequence of Sindbis virus involved in packaging differs from that of SFV, the ability of Sindbis virus capsid protein to interact efficiently with both Sindbis virus and SFV vector RNAs may suggest involvement of a common structural motif.
Continued improvements to the alphavirus PCL should result in vector titers that approach those of wild-type virus, which is greater than 109 PFU/ml. One area of our focus is the optimization of nucleocytoplasmic transport and stability of primary alphavirus RNA transcripts. Data obtained by using Sindbis virus DNA vectors, as well as the packaging cassettes from this paper, indicated that functional alphavirus transcripts are not detected in the cytoplasm of all cells (ref. 10, B.A.B., unpublished work). In addition, a long-range goal of our work is the stable introduction of a vector cassette into the PCL. The lytic nature of alphavirus replication will require the use of a tightly controlled inducible promoter, a noncytopathic virus variant (refs. 37 and 38, S.P., unpublished data), or the exploitation of a parental cell line, such as mosquito cells, in which alphaviruses establish a persistent rather than lytic infection. At any rate, the current PCLs described in this paper represent a promising innovation for the eventual clinical application of alphavirus vectors.
Acknowledgments
A portion of this work was supported by National Institutes of Health Grant AI11377 to S. Schlesinger.
ABBREVIATIONS
- SFV
Semliki Forest virus
- SIN
Sindbis virus
- RCV
replication-competent virus
- PCL
packaging cell line
- SV40
simian virus 40
- CMV
cytomegalovirus
- IU
infectious units
- PFU
plaque-forming units
- CTL
cytotoxic T lymphocyte
- HSV
herpes simplex virus
- HSVgB
HSV glycoprotein B
- BHK
baby hamster kidney
References
- 1.Huang H V. Curr Opin Biotechnol. 1996;7:531–535. doi: 10.1016/s0958-1669(96)80057-7. [DOI] [PubMed] [Google Scholar]
- 2.Frolov I, Hoffman T A, Pragai B M, Dryga S A, Huang H V, Schlesinger S, Rice C M. Proc Natl Acad Sci USA. 1996;93:11371–11377. doi: 10.1073/pnas.93.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strauss J H, Strauss E G. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong C, Levis R, Shen P, Schlesinger S, Rice C M, Huang H V. Science. 1989;243:1188–1191. doi: 10.1126/science.2922607. [DOI] [PubMed] [Google Scholar]
- 5.Liljeström P, Garoff H. Biotechnology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 6.Bredenbeek P J, Frolov I, Rice C M, Schlesinger S. J Virol. 1993;67:6439–6446. doi: 10.1128/jvi.67.11.6439-6446.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pusko P, Parker M, Ludwig G V, Davis N L, Johnston R E, Smith J F. Virology. 1997;239:389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- 8.Driver D A, Latham E M, Polo J M, Belli B A, Banks T A, Chada S, Brumm D, Chang S M W, Mento S J, Jolly D J, Dubensky T W., Jr Ann NY Acad Sci. 1995;772:261–264. doi: 10.1111/j.1749-6632.1995.tb44754.x. [DOI] [PubMed] [Google Scholar]
- 9.Herweijer H, Latendresse J S, Williams P, Zhang G, Danko I, Schlesinger S, Wolff J A. Hum Gene Ther. 1995;6:1161–1167. doi: 10.1089/hum.1995.6.9-1161. [DOI] [PubMed] [Google Scholar]
- 10.Dubensky T W, Jr, Driver D A, Polo J M, Belli B A, Latham E M, Ibanez C E, Chada S, Brumm D, Banks T A, Mento S J, et al. J Virol. 1996;70:508–519. doi: 10.1128/jvi.70.1.508-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hariharan M J, Driver D A, Townsend K, Brumm D, Polo J M, Belli B A, Catton D J, Hsu D, Mittelstaedt D, McCormack J E, et al. J Virol. 1998;72:950–958. doi: 10.1128/jvi.72.2.950-958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berglund P, Smerdou C, Fleeton M N, Tubulekas I, Liljestrom P. Nat Biotech. 1998;16:562–565. doi: 10.1038/nbt0698-562. [DOI] [PubMed] [Google Scholar]
- 13.Polo J M, Dubensky T W., Jr Nat Biotech. 1998;16:517–518. doi: 10.1038/nbt0698-517. [DOI] [PubMed] [Google Scholar]
- 14.Frolov I, Frolova E, Schlesinger S. J Virol. 1997;71:2819–2829. doi: 10.1128/jvi.71.4.2819-2829.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrotta A T, Been M D. Nature (London) 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 16.Monroe S S, Schlesinger S. Proc Natl Acad Sci USA. 1983;80:3279–3283. doi: 10.1073/pnas.80.11.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice C M, Levis R, Strauss J H, Huang H V. J Virol. 1987;61:3809–3819. doi: 10.1128/jvi.61.12.3809-3819.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacGregor G R, Mogg A E, Burke J F, Caskey C T. Somat Cell Mol Genet. 1987;13:253–265. doi: 10.1007/BF01535207. [DOI] [PubMed] [Google Scholar]
- 19.Chada S, DeJesus C E, Townsend K, Lee W T L, Laube L, Jolly D J, Chang S M W, Warner J F. J Virol. 1993;67:3409–3417. doi: 10.1128/jvi.67.6.3409-3417.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elgizoli M, Dai Y, Kempf C, Koblet H, Michel M R. J Virol. 1989;63:2921–2928. doi: 10.1128/jvi.63.7.2921-2928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frolov I, Schlesinger S. J Virol. 1994;68:1721–1727. doi: 10.1128/jvi.68.3.1721-1727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Steeg H, Kasperaitis M, Voorma H O, Benne R. Eur J Biochem. 1984;138:473–478. doi: 10.1111/j.1432-1033.1984.tb07940.x. [DOI] [PubMed] [Google Scholar]
- 23.Weiss B, Nitschko H, Ghattas I, Wright R, Schlesinger S. J Virol. 1989;63:5310–5318. doi: 10.1128/jvi.63.12.5310-5318.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frolova E, Frolov I, Schlesinger S. J Virol. 1997;71:248–258. doi: 10.1128/jvi.71.1.248-258.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy V B, Ghosh P K, Lebowitz P, Piatak M, Weissman S M. J Virol. 1979;30:279–296. doi: 10.1128/jvi.30.1.279-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Townsend K, Sallberg M, O’Dea J, Banks T, Driver D, Sauter S, Chang S M, Jolly D J, Mento S J, Milich D R, Lee W T L. J Virol. 1997;71:3365–3374. doi: 10.1128/jvi.71.5.3365-3374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mann R, Mulligan R C, Baltimore D. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 28.Wantanabe S, Temin H M. Mol Cell Biol. 1983;3:2241–2249. doi: 10.1128/mcb.3.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olivo P D, Frolov I, Schlesinger S. Virology. 1994;198:381–384. doi: 10.1006/viro.1994.1046. [DOI] [PubMed] [Google Scholar]
- 30.Geigenmuller-Gnirke U, Weiss B, Wright R, Schlesinger S. Proc Natl Acad Sci USA. 1991;88:3253–3257. doi: 10.1073/pnas.88.8.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosselman R A, Hsu R-Y, Bruszewski J, Hu S, Martin F, Nicolson M. Mol Cell Biol. 1987;7:1797–1806. doi: 10.1128/mcb.7.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danos O, Mulligan R C. Proc Natl Acad Sci USA. 1988;85:6460–6464. doi: 10.1073/pnas.85.17.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markowitz D, Goff S, Bank A. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monroe S S, Schlesinger S. J Virol. 1984;49:865–872. doi: 10.1128/jvi.49.3.865-872.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jalanko A, Söderlund H. Virology. 1985;141:257–266. doi: 10.1016/0042-6822(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 36.Kääriäinen L, Pettersson R F, Keränen S, Lehtovaara P, Söderlund H, Ukkonen P. Virology. 1981;113:686–697. doi: 10.1016/0042-6822(81)90197-5. [DOI] [PubMed] [Google Scholar]
- 37.Weiss B, Rosenthal R, Schlesinger S. J Virol. 1980;33:463–474. doi: 10.1128/jvi.33.1.463-474.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dryga S A, Dryga O A, Schlesinger S. Virology. 1997;228:74–83. doi: 10.1006/viro.1996.8364. [DOI] [PubMed] [Google Scholar]