Abstract
Huntington’s disease is a progressive neurodegenerative disorder caused by a polyglutamine [poly(Q)] repeat expansion in the first exon of the huntingtin protein. Previously, we showed that N-terminal huntingtin peptides with poly(Q) tracts in the pathological range (51–122 glutamines), but not with poly(Q) tracts in the normal range (20 and 30 glutamines), form high molecular weight protein aggregates with a fibrillar or ribbon-like morphology, reminiscent of scrapie prion rods and β-amyloid fibrils in Alzheimer’s disease. Here we report that the formation of amyloid-like huntingtin aggregates in vitro not only depends on poly(Q) repeat length but also critically depends on protein concentration and time. Furthermore, the in vitro aggregation of huntingtin can be seeded by preformed fibrils. Together, these results suggest that amyloid fibrillogenesis in Huntington’s disease, like in Alzheimer’s disease, is a nucleation-dependent polymerization.
Keywords: huntingtin, glutamine repeat, aggregation
The accumulation of polyglutamine [poly(Q)]-containing protein aggregates in neuronal intranuclear inclusions is a hallmark of several progressive neurodegenerative disorders such as Huntington’s disease (HD) (1, 2), dentatorubral pallidoluysian atrophy (1, 3), spinal bulbar muscular atrophy, (4) and the spinocerebellar ataxia types 1 (5), 3 (6), and 7 (7). Ultrastructural studies of brains of HD transgenic mice (8) revealed neuronal intranuclear inclusion that contained aggregated huntingtin protein with a granular or fibrillar morphology. Neuronal intranuclear inclusion with similar structural features also were detected in postmortem brains of HD (2) and spinocerebellar ataxia type 3 patients (6) as well as in stable and transiently transfected cell lines (9, 10). Furthermore, recombinant proteins with an expanded poly(Q) stretch (51–122 glutamines) were found to form insoluble high molecular weight protein aggregates in vitro (11). Electron micrographs of these aggregates revealed fibrillar structures with a morphology closely resembling that of scrapie prion rods and β-amyloid fibrils in Alzheimer’s disease (12, 13). These observations have led to the proposal that the poly(Q) diseases HD, dentatorubral pallidoluysian atrophy, spinal bulbar muscular atrophy, and spinocerebellar ataxia types 1, 3, and 7 could all be the result of toxic amyloid fibrillogenesis, as has been proposed for Alzheimer’s disease (14).
Although the causal relationship between neuronal intranuclear inclusion formation and HD has not been proven, the gradual deposition of amyloids in neurons that degenerate in HD would be consistent with the late onset and progressive nature of symptoms. Therefore, it would be important to understand the molecular mechanisms of amyloid formation and to explain why the huntingtin protein with an expanded poly(Q) stretch (38–100 and more residues), or an N-terminal fragment thereof, aggregates in diseased individuals, whereas the same protein with a poly(Q) tract in the normal range (6–37 residues) does not. A detailed understanding of the aggregation process could help to open new avenues for therapeutic intervention, because the selective inhibition of huntingtin aggregation may represent a feasible therapeutic strategy for HD.
Alzheimer’s disease, a late-onset, progressive neurodegenerative disorder, is characterized by the deposition of β-amyloid (Aβ) protein in the brain (15, 16), and there is strong circumstantial evidence to indicate that Aβ deposition directly contributes to the progressive neurodegeneration in this disease (17). Larrett and Lansbury (18) proposed that Aβ formation in Alzheimer’s disease patients occurs by a nucleation-dependent polymerization mechanism, based on the mechanistic resemblance of in vitro Aβ formation to protein crystallization (19), microtubule assembly (20), and sickle-cell hemoglobin fibril formation (21). The requirement that a nucleus be formed before polymerization occurs predicts certain characteristics of the aggregation process, including (i) dependence on exceeding a critical protein concentration for the initial formation of aggregates and (ii) kinetics displaying a lag phase during which nuclei are slowly forming. Furthermore, addition of an exogenous Aβ fibril, in trace amounts, leads to immediate aggregation by seeding the polymerization process.
Here we report that N-terminal huntingtin peptides with poly(Q) tracts of 37 or more residues efficiently aggregate in vitro and in transfected COS cells. Formation of huntingtin aggregates follows a kinetic mechanism, which resembles the formation of Aβ fibrils—i.e., nucleation is the rate-limiting step in the aggregation process. The consequences of this kinetic mechanism for in vivo amyloidogenesis in HD patients are discussed.
MATERIALS AND METHODS
Plasmid Constructions.
Escherichia coli Sure (Stratagene) was used as host strain in plasmid constructions. Recombinant λ phage from stock 91974 (22) was used as source of HD exon 1 DNA with different numbers of CAG repeats. PCR-amplified fragments encoding the entire exon 1 portion of huntingtin (amino acids 1–90; GenBank accession no. L13292) with poly(Q) tracts of various lengths were prepared as described (11), taking advantage of the instability of the CAG repeat during phage propagation in E. coli. After digestion with BamHI and SalI, PCR products were cloned into the BamHI-SalI site of the pGEX-5X-1 vector (Amersham Pharmacia) to generate the pCAG plasmid series. The pCAG plasmids encoding GST-HDex1 fusion proteins with 20, 27, 32, 37, 39, 40, 42, 45, 51, and 83 glutamines were subsequently digested with BamHI and XhoI, and the HD exon1 fragments were subcloned into the BamHI-XhoI site of the pTL1 vector (23) to generate the pTL1-CAG plasmid series. Sequence analysis of the resulting clones indicated that one of them underwent expansion of the CAG repeat from the 83 units in the original pCAG construct to 93 units in the final construct.
Antibodies.
The huntingtin-specific polyclonal antibody HD1 has been described (11). The polyclonal antibody AG51 was generated by injecting HDex1-Q51 fibrils, isolated as described below, into a rabbit. The resulting immune serum was passed two times over a Sepharose 4B-glutathione S-transferase (GST) column to remove traces of contaminating anti-GST antibodies. A polyclonal anti-GST antibody was purchased from Amersham Pharmacia.
Protein Purification and Tryptic Digestions.
The GST-HDex1 fusion proteins were overexpressed in E. coli Sure cells and affinity-purified on glutathione-agarose beads (24). Purified proteins were dialyzed overnight at 4°C against TNEG buffer (40 mM Tris⋅HCl, pH 8.0/0.1 M NaCl/0.1 mM EDTA/5% glycerol), frozen in liquid N2, and stored at −80°C. Protein concentration was determined by the Bio-Rad assay using BSA as standard.
For in vitro aggregation studies, the GST-HDex1 proteins were digested with trypsin (modified version, Boehringer Mannheim), resulting in release of a poly(Q)-containing HD exon 1 peptide (HDex1p) of the structure shown in Fig. 1A. Tryptic digestions of the fusion proteins were carried out at 37°C in TNEG buffer containing 1 mM CaCl2 at an enzyme/substrate rate of 1:30 (wt/wt). Digestion was terminated by adjusting the protein mixtures to 2% SDS and 50 mM DTT followed by heating at 98°C for 3 min. The degree of proteolysis was determined by using SDS/PAGE and immunoblotting with anti-AG51 or anti-GST antibodies.
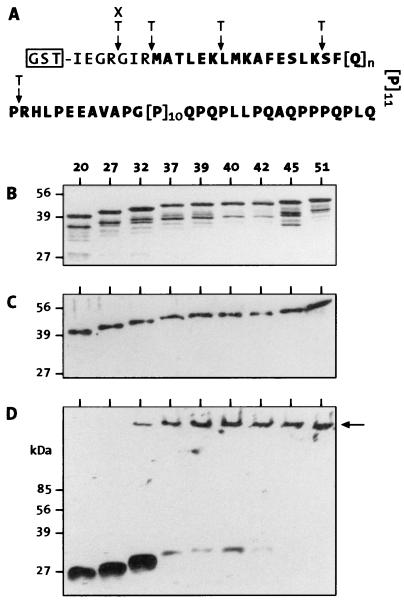
Figure 1.
SDS/PAGE analysis of purified GST-HDex1 proteins and their tryptic digestion products. (A) Structure of the GST fusion proteins showing potential factor Xa (X) and trypsin (T) cleavage sites. The amino acid sequence corresponding to exon 1 of huntingtin is shown in bold. (B) SDS/12.5% PAGE gel stained with Coomassie blue. Five micrograms of each fusion protein, containing poly(Q) tracts of 20, 27, 32, 39, 40, 42, 45, or 51 residues, was loaded in each lane. (C and D) Immunoblot analysis of GST-HDex1 proteins before (C) and after (D) trypsin digestion. Incubation with trypsin was carried out for 24 h at a protein concentration of 0.8 mg/ml. Eighty nanograms of each fusion protein was loaded in each lane. ← marks the origin of electrophoresis.
Fibril Isolation.
One milliliter of a 1 mg/ml solution of GST-HDex1-Q45 or -Q51 proteins was incubated for 24 h at 37°C with trypsin (50 μg). The resulting HDex1p fibrils were collected by centrifugation at 50,000 × g for 30 min, washed two times with H2O, and resuspended in a minimal volume of 5 mM Tris⋅HCl (pH 8.0). For use in seeding experiments, the fibrils were sonicated for 2 min to create fresh surfaces for monomer addition.
Analysis of in Vitro Aggregate Formation.
The GST-HDex1 proteins (before or after trypsin digestion) were fractionated by using SDS/12.5% PAGE, electrotransferred to nitrocellulose (Schleicher & Schuell; BA 83), and probed with anti-AG51 serum (1:1,000 dilution). Immunodetection was carried out with the enhanced chemiluminescence (ECL) Western blot system (Amersham Pharmacia). Filtration of GST-HDex1 proteins and their tryptic cleavage products through a 0.2-μm cellulose acetate membrane (Schleicher & Schuell; OE66) was performed as detailed (25) by using a BRL dot-blot filtration unit. Fibril formation by trypsinated GST-HDex1 proteins was also examined by electron microscopy using a Philips CM100 electron microscope as described (11).
Analysis of Aggregate Formation in COS Cells.
COS-1 cells were plated to 40% confluence in 9-cm dishes and transfected with 5 μg of the various pTL1-CAG constructs by the calcium phosphate coprecipitation technique. Cells were harvested 42 h after transfection and solubilized in lysis buffer containing 0.5% Nonidet P-40 (26). Insoluble material was pelleted by centrifugation in an Eppendorf microcentrifuge at 14,000 rpm for 5 min. The pellet was taken up in 100 μl of 20 mM Tris⋅HCl (pH 8.0)/15 mM MgCl2 and incubated for 1 h at 37°C with DNaseI (50 μg) to achieve maximal resuspension. Aliquots of these pellet fractions and the corresponding supernatant fractions were then fractionated by SDS/12.5% PAGE, transferred to nitrocellulose, and probed with the anti-HD1 antibody (1:1,000 dilution).
Isolation of HDex1p Aggregates from COS Cells and Immunogold Labeling.
COS-1 cells transfected with the pTL1-CAG51 construct were harvested 40 h posttransfection, and an insoluble fraction (0.5 ml) was prepared as described above, except that the suspension was sonicated with 30 1-sec bursts before treatment with DNaseI. After DNase digestion, the volume of the sample was increased to 4 ml with 10 mM Tris⋅HCl (pH 7.5) and mixed with 1.48 g of CsCl. Four milliliters of this solution was layered onto 4.2 ml of dense CsCl solution (0.52 g of CsCl per ml), and 4.2 ml of less dense CsCl solution (0.25 g of CsCl per ml) was then layered on the top. Centrifugation was in a Beckman SW40 rotor at 30,000 rpm for 24 h at 4°C. Fractions of 0.5 ml were collected from the bottom of the tube, and an aliquot of each fraction (25 μl) was analyzed with the filter retardation assay. The HDex1p aggregates banded at a density of ≈1.307 g/cm3. The peak fraction was dialyzed for 3 h against TBS (150 mM NaCl/20 mM Tris⋅HCl, pH 7.5) to remove the CsCl. For immunogold labeling of the aggregates, the sample was incubated for 1 h at room temperature with anti-AG51 IgG at 20 μg/ml. The resulting immunocomplex was centrifuged for 90 min at 67,000 × g, and after two TBS washes, the pelleted material was taken up in 50 μl of TBS, sonicated for 30 sec, and incubated for 2 h with 1 μl of goat anti-rabbit IgG gold conjugate (British BioCell). After centrifugation in a Microfuge for 10 min at 12,000 rpm, the resuspended pellet was negatively stained with 1% uranyl acetate for electron microscopic analysis.
RESULTS
Purification of GST-HDex1 Fusion Proteins.
We used exon 1 of the HD gene (HDex1) with CAG repeats in the normal and pathological range for the production of GST-HDex1 fusion proteins in E. coli. The structure of the GST-HDex1 proteins is shown in Fig. 1A. Fusion proteins with 20, 27, 32, 37, 39, 40, 42, 45, and 51 glutamines were purified under native conditions by affinity chromatography (24). SDS/PAGE analysis of the purified proteins revealed monomeric GST-HDex1 proteins with the expected shifts in migration relative to poly(Q) repeat length (Fig. 1B). These bands were also detected by immunoblot analysis using the polyclonal anti-huntingtin antibodies AG51 (Fig. 1C) and HD1 (data not shown). AG51, raised against purified HDex1-Q51 fibers, specifically recognizes the proline-rich region at the C terminus of the HDex1 protein (E.S., unpublished data). Therefore, this antibody failed to detect the C-terminal GST-HDex1 degradation products seen in the Coomassie blue-stained gel (compare Fig. 1 B and C).
HDex1p Aggregation Is Poly(Q) Repeat Length-Dependent.
To determine the length of the poly(Q) chain required for HDex1p aggregate formation in vitro, the GST-HDex1 proteins at a concentration of 800 μg/ml (≈20 μM) were incubated with trypsin under conditions designed to totally remove the GST tag from the fusion proteins within the first 30 min of incubation. Relevant trypsin cleavage sites within the GST-HDex1 fusion proteins are shown in Fig. 1A. After incubation for 24 h at 37°C, proteins were denatured by boiling in 2% SDS and analyzed by SDS/PAGE and immunoblotting using the AG51 antibody (Fig. 1D). Cleavage of GST-HDex1-Q20, -Q27, and -Q32 resulted in the production of soluble, apparently monomeric HDex1 peptides migrating in a 12.5% gel at approximately 27, 30, and 32 kDa, respectively. By contrast, cleavage of GST-HDex1 proteins with 37 or more glutamine repeats yielded almost exclusively SDS-insoluble high molecular weight aggregates that did not enter the stacking gel. These aggregates also were detected by the HD1 antibody but not by an antibody directed against GST (data not shown). A small but significant amount of aggregated protein also was evident for HDex1-Q32, but we have never observed HDex1p aggregates by SDS/PAGE and immunoblotting with the 20- and 27-repeat constructs. Similar results were obtained when the GST-HDex1 proteins were digested with factor Xa protease (data not shown).
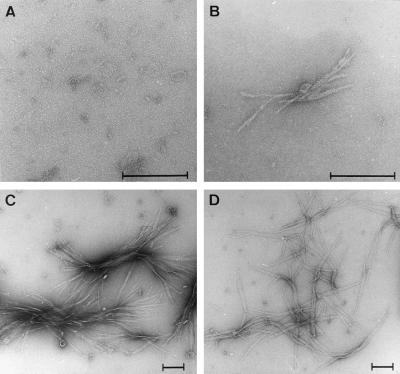
Electron microscopy of the trypsin-digested GST-HDex1-Q37 and -Q45 preparations showed numerous clusters of large polymers with the typical ribbon-like morphology observed previously for the HDex1-Q51 protein (11) (Fig. 2 C and D). The length of these fibrils varied considerably, from 100 nm up to several micrometers. In the trypsin-treated sample of GST-HDex1-Q32, only a few small (<400 nm) fibrillar structures could be detected (Fig. 1B), whereas no or only occasional very short (<40 nm) fibers were detected in the 27-repeat sample (Fig. 2A), consistent with the results seen by Western blot analysis.
Figure 2.
Electron microscope pictures of HDex1 fibrils. Samples of the trypsin-digested GST-HDex1 proteins analyzed in Fig. 1D were negatively stained with 1% uranyl acetate and viewed by using electron microscopy. (A) HDex1-Q27. (B) HDex1-Q32. (C) HDex1-Q37. (D) HDex1-Q45. (Bar = 200 nm.)
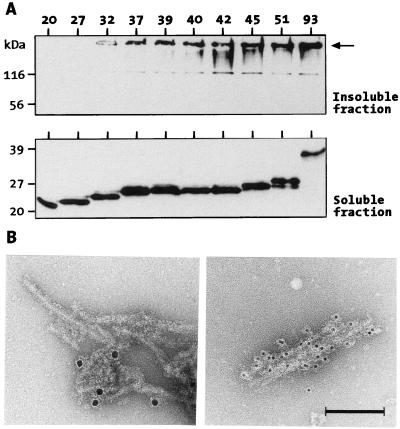
Formation of poly(Q)-containing HDex1p aggregates also was investigated in a cell culture model. HDex1 proteins with 20, 27, 32, 37, 39, 40, 42, 45, 51, and 93 glutamines (without a GST tag) were transiently expressed in COS-1 cells, and after cell lysis, the soluble proteins were separated from insoluble material by centrifugation. Both cell fractions were analyzed by using SDS/PAGE and immunoblotting using the HD1 antibody (Fig. 3A). HDex1p aggregates that remained near the top of the stacking gel were detected only in the insoluble fraction, and as with the trypsin-treated GST-HDex1 proteins described above, aggregation in COS cells occurred in a poly(Q) repeat length-dependent manner. No SDS-resistant aggregates were detected in the insoluble fraction from cells transfected with the HDex1-Q20 and -Q27 constructs. Western blot analysis of the corresponding soluble fractions, on the other hand, revealed that all HDex1 proteins were expressed at similar levels, giving rise to prominent protein bands migrating on SDS/PAGE at about 23–35 kDa. Although not shown in Fig. 3A, no higher molecular weight HDex1p aggregates were detected in the soluble fractions, indicating that they are not an artifact of SDS/PAGE.
Figure 3.
(A) HDex1p expression and aggregation in COS cells. COS-1 cells transfected with HDex1-Q20, -Q27, -Q32, -Q37, -Q39, -Q40, -Q45, -Q51, and -Q93 constructs were harvested 42 h after transfection and lysed, and the crude cell lysates were separated by centrifugation into an insoluble (pellet) and a soluble (supernatant) fraction (see Materials and Methods). Ten micrograms of total protein from each soluble fraction and 10 μl of each insoluble fraction were run on separate SDS gels and immunoblotted with anti-HD1 serum. ← in the Upper gel marks the origin of electrophresis. (B) Electron micrographs of negatively stained immunogold-labeled HDex1p aggregates. (Left) Protein aggregates formed from purified GST-HDex1-Q51 protein after cleavage with factor Xa. (Right) Aggregated HDex1 protein isolated from HDex1-Q51-expressing COS-1 cells. The sizes of the gold particles were 10 and 5 nm, respectively. (Bar = 100 nm.)
Purification of insoluble HDex1p aggregates by centrifugation in a CsCl density gradient yielded fractions containing aggregates of fibrillar structure. These structures, which were not detected in a control gradient containing insoluble material from untransfected cells, were morphologically similar to those obtained from GST-HDex1-Q51 protein after proteolytic cleavage with faxtor Xa. The anti-huntingtin antibody AG51 was found to decorate the fibrils using a goat anti-rabbit gold colloid secondary antibody (Fig. 3B).
Formation of HDex1p Aggregates Is Nucleation-Dependent.
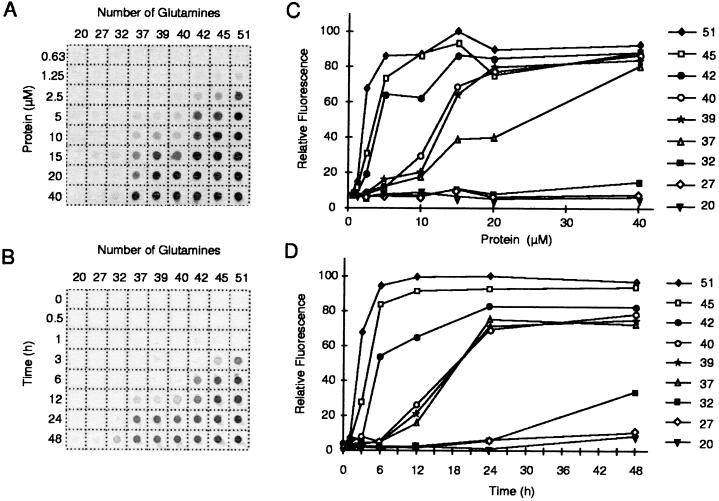
In a typical nucleation-dependent polymerization, polymer is not observed until the monomer concentration exceeds a certain critical concentration. Above that level, polymerization does not proceed immediately but after a defined time period, or lag time (18). Consistent with these predictions, we found that the in vitro aggregation of poly(Q)-containing HDex1 peptides depends on both protein concentration and time. Fig. 4 A and C shows the effect of protein concentration on HDex1p aggregate formation using GST-HDex1 proteins with poly(Q) tracts of 20–51 residues. After trypsin treatment to remove the GST moiety from the fusion proteins, the digestion products were incubated for 24 h to permit aggregation and then filtered through a cellulose acetate membrane. The HDex1p aggregates retained on the filter were detected by immunoblotting with the AG51 antibody. By using this assay, none of the GST-HDex1 proteins analyzed yielded detectable amounts of aggregates below 1.25 μM. At higher concentrations, substantial aggregation was evident for HDex1-Q51, -Q45, -Q42, -Q40, -Q39, -Q37, and, to a much lesser but significant extent, also for HDex1-Q32, whereas HDex1-Q27 and -Q20 showed no detectable aggregation over the entire concentration range examined (0.63–40 μM).
Figure 4.
(A) Concentration dependence of HDex1p aggregation. GST-HDex1 proteins with poly(Q) tracts of different lengths were incubated at the indicated concentrations with trypsin for 24 h at 37°C. Aliquots (200 ng) of each protein were then diluted into 0.2 ml of 2% SDS/50 mM DTT, boiled for 3 min, and filtered through a cellulose acetate membrane. Captured aggregates were detected by incubation with anti-AG51 serum (1:1,000), followed by incubation with alkaline phosphatase-conjugated anti-rabbit secondary antibody and the fluorescent substrate AttoPhos. (B) Time course of HDex1p aggregation. The various GST-HDex1 proteins were incubated at a concentration of 20 μM with trypsin. At the indicated times, aliquots (200 ng) of each protein were removed and analyzed by the filter retardation assay as in A. (C and D) Quantitative analysis of the dot-blot results shown in A and B, respectively. The relative amount of aggregate for each sample was quantitated on a Fuji-Imager (LAS 2000). For each experiment, the dot with the highest signal intensity was arbitrarily set as 100. The data reported are representative for three independent experiments using, in part, different GST-HDex1p preparations.
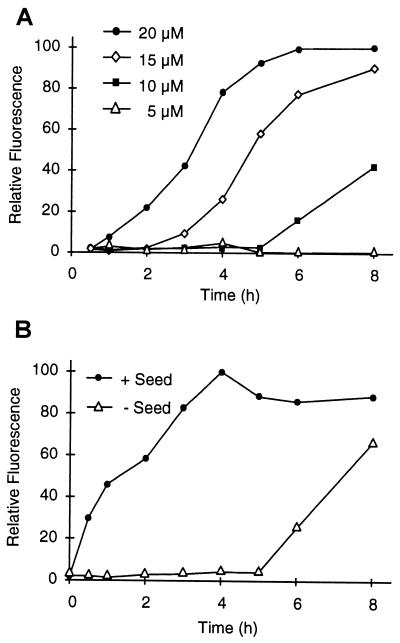
The time course of in vitro aggregation by using the nine poly(Q)-containing fusion proteins is shown in Fig. 4 B and D. Aggregation was initiated by addition of trypsin to a 20 μM solution of each protein, and after incubation for various times (0–48 h), aggregated protein was captured by filtration. Under the experimental conditions used, the GST moiety was completely (>95%) removed from each fusion protein within the first 30 min of incubation (data not shown). As expected, HDex1-Q20 and -Q27 showed no aggregation at any time point. For the longer HDex1-Q32, -Q37, -Q39, -Q40, -Q42, -Q45, and -Q51 peptides aggregate formation was evident after a lag phase, the length of which increased with decreasing glutamine chain length. For example, HDex1-Q51 and -Q45 aggregated after 1–3 h, resulting in a rapid accumulation of nonfilterable protein, whereas HDex1-Q32 under identical conditions required 1–2 days to develop aggregates that could be detected by our filter retardation assay. Reducing the concentration of the fusion proteins dramatically increased the observed lag time, as shown in Fig. 5A for HDex1-Q45. For this protein, assays conducted at concentrations of 20, 15, and 10 μM yielded apparent lag times of approximately 1, 3, and 5 h, respectively, suggestive of an exponential dependence of the length of the lag time on protein concentration. Analogous results were obtained for the HDex1-Q51 and -Q40 proteins (data not shown).
Figure 5.
HDex1p aggregation requires a nucleation event. The aggregation assay was performed as in Fig. 4 by using dot-blot filtration on a cellulose acetate membrane and subsequent immunodetection of the captured aggregates. For each experiment shown, 100% represents maximum fluorescence intensity. (A) Effect of protein concentration on the kinetics of HDex1p aggregation. GST-HDex1-Q45 at 5, 10, 15, and 20 μM was incubated with trypsin at 37°C. At the indicated times, aliquots (200 ng) of each protein mixture were removed, denatured and reduced, and filtered. (B) HDex1p aggregate formation can be seeded by preformed fibrils. HDex1p aggregation was performed with GST-HDex1-Q45 at 10 μM in the presence or absence of added HDex1-Q45 fibrils (0.15 equivalent, 1.5 μM). A sample containing the seed alone was incubated in parallel, and the amount of aggregate captured by filtration at each time point was subtracted from the reported values. The results shown are from a single experiment performed in duplicate.
The time lag of 5 h exhibited by HDex1-Q45 at 10 μM could be eliminated by adding a relatively small amount (15% per mol) of preformed HDex1-Q45 fibrils to the digestion mixture (Fig. 5B), consistent with nucleation being the rate-limiting step of HDex1p aggregate formation. An identical result was obtained with purified HDex1-Q51 fibrils used as a heterologous seed (data not shown).
DISCUSSION
In this paper we show that the in vitro aggregation of N-terminal, poly(Q)-containing huntingtin peptides is self-initiated and, like a crystallization (19) or Aβ formation (14), follows a nucleation-dependent pathway. This means that both protein concentration and incubation time are important for promoting the self-assembly of huntingtin aggregates. Furthermore, formation of ordered huntingtin aggregates is highly poly(Q) repeat length-dependent, with a threshold and progressivity for poly(Q) repeat length remarkably similar to that found in HD. The pathological threshold in HD is 38–41 glutamines, with no case of HD having been reported with fewer than 38 glutamines, nor any individual with more than 41 glutamines having been found to remain free from HD (27). It is possible that 38 glutamines represents the pathogenic threshold, but because of the influence of genetic and possibly other modifiers, some individuals with 38–41 repeats do not develop HD within their lifetime. Our data show that poly(Q) tracts with 37 or more glutamines readily self-assemble into insoluble HDex1p aggregates in vitro and in transfected COS-1 cells. In contrast, poly(Q) tracts with 27 or fewer glutamines did not show any evidence of ordered fibril formation under any of the conditions tested. Constructs with 32 glutamines did form some higher ordered structures, but the kinetics of this aggregation was considerably slower than that for 37 glutamines (Fig. 4). Perutz (28) proposed that expansion of poly(Q) repeats beyond a certain length may lead to a phase change from random coils to hydrogen-bonded hairpins that self-associate to form a β-sheet structure. Our in vitro and in vivo experiments with HDex1 proteins containing poly(Q) tracts in the normal and pathological range support this hypothesis and suggest that the structural transition caused by expansion occurs between 32 and 37 glutamines.
A molecular model of HD pathogenesis must account for the late onset of the disease, the pathogenic poly(Q) threshold, and the inverse relationship between poly(Q) length and disease age of onset. As discussed above, the pathogenic threshold in HD shows a striking correlation with the poly(Q) repeat length at which the HDex1p constructs readily formed higher ordered fibrillar aggregates. Our demonstration that the kinetics of this process requires the formation of a nucleus and that in situ nucleus formation is time- and protein concentration-dependent may in part provide a mechanism that accounts for the late onset of the disease. Consistent with nucleation being the rate-determining step in HDex1p aggregation, addition of exogenous seed fibrils to a solution of HDex1-Q45 protein eliminated the observed lag time (Fig. 5B). This suggests that both homologous and/or heterologous seeds may be important in accelerating huntingtin aggregation in vivo. Our data further indicate that the length of the lag time is extremely sensitive to protein concentration. For example, a 50% reduction of the HDex1-Q45 protein concentration (10 vs. 20 μM) increased the lag time from about 1 to 5 h (Fig. 5A). Therefore, the length of time required to form poly(Q) aggregates in HD will depend on the time it takes to reach the critical concentration of the huntingtin molecule that is capable of aggregation. There is mounting evidence from immunocytochemistry of poly(Q) inclusions in postmortem brains (1, 2) and from transient and stable transfected cell lines (9) that this is an N-terminal fragment of huntingtin. Therefore, the proteolytic cleavage event that yields this “toxic” fragment would also provide a rate-limiting step in HD onset.
The age of onset and the severity of HD is inversely correlated to the length of the poly(Q) expansion (29). The majority of adult-onset cases have expansions ranging from 41 to 55 units, whereas expansions of 70 and above invariably cause the juvenile form of the disease. Therefore, the kinetics of the initial pathogenic step in HD must progress more rapidly as the glutamine repeats increase in length. Consistent with this, we have found that the rate of aggregate formation in vitro directly correlates with repeat length: the longer the poly(Q) tract, the faster the aggregation rate. Similarly, the protein concentration required for aggregate formation decreased with an increase of the poly(Q) repeat length. For example, HDex1-Q51 readily aggregated at a protein concentration as low as 2.5 μM, whereas with the 32-repeat construct aggregation was barely detected even at a 16-fold higher concentration (40 μM).
The tight correlation between the threshold for aggregation as determined in these in vitro experiments and the expansion into the mutant range strongly argues for poly(Q) aggregation being the critical event in the initiation and progression of the symptoms of HD. Although the formation of ordered huntingtin aggregates has not formally been proven to be the cause of HD pathogenesis (for discussion, see refs. 30 and 31), we propose that inhibition of nucleus formation is a feasible therapeutic strategy. Potential targets include a reduction in the intracellular concentration of the N-terminal huntingtin fragment that is capable of aggregation. This reduction could be effected either by specifically down-regulating the expression of the HD gene or by inhibiting the proteolytic cleavage of the huntingtin protein. Alternatively, drugs that specifically interfere with nucleus formation could be screened. This strategy has been shown to be effective in the inhibition of sickle-cell hemoglobin fibrillization and also in the treatment of anemia (21). Furthermore, it has been demonstrated recently that β-sheet breaker peptides inhibit the Aβ fibrillogenesis in a rat brain model (32). Therefore, the future challenge is to find small molecules, capable of crossing the blood-brain barrier, that specifically prevent the nucleus formation of poly(Q) tracts.
ABBREVIATIONS
- HD
Huntington’s disease
- Aβ
β-amyloid
- GST
glutathione S-transferase
References
- 1.Becher M W, Kotzuk J A, Sharp A H, Davies S W, Bates G P, Price D L, Ross C A. Neurobiol Dis. 1997;4:1–11. doi: 10.1006/nbdi.1998.0168. [DOI] [PubMed] [Google Scholar]
- 2.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 3.Igarashi S, Koide R, Shimohata T, Yamada M, Hayashi Y, Takano H, Date H, Oyake M, Sato T, Sato A, et al. Nat Genet. 1998;18:111–117. doi: 10.1038/ng0298-111. [DOI] [PubMed] [Google Scholar]
- 4.Li M M, Kobayashi Y, Merry D E, Yamamoto M, Tanaka F, Doyu M, Hashizume Y, Fischbeck K H, Sobue G. Ann Neurol. 1998;44:249–254. doi: 10.1002/ana.410440216. [DOI] [PubMed] [Google Scholar]
- 5.Skinner P J, Koshy B T, Cummings C L, Klement I A, Helin K, Servadio A, Zoghbi H Y, Orr H T. Nature (London) 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 6.Paulson H L, Perez M K, Trottier Y, Trojanowski J Q, Subramony S H, Das S S, Vig P, Mandel J-L, Fischbeck K H, Pittman R N. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- 7.Holmberg M, Duyckaerts C, Durr A, Cancel G, Gourfinkel-An I, Damier P, Faucheux B, Trottier Y, Hirsch E C, Agid Y, Brice A. Hum Mol Genet. 1998;7:913–918. doi: 10.1093/hmg/7.5.913. [DOI] [PubMed] [Google Scholar]
- 8.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 9.Lunkes A, Mandel J-L. Hum Mol Genet. 1998;7:1355–1361. doi: 10.1093/hmg/7.9.1355. [DOI] [PubMed] [Google Scholar]
- 10.Perez M K, Paulson H L, Pendse S J, Saionz S J, Bonini N M, Pittman R N. J Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 12.Caughey B, Chesebro B. Trends Cell Biol. 1997;7:56–62. doi: 10.1016/S0962-8924(96)10054-4. [DOI] [PubMed] [Google Scholar]
- 13.Caputo C B, Fraser P E, Sobel I E, Krischner D A. Arch Biochem Biophys. 1992;292:199–205. doi: 10.1016/0003-9861(92)90068-8. [DOI] [PubMed] [Google Scholar]
- 14.Harper J H, Lansbury P T. Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 15.Kosik K S. J Cell Biol. 1994;127:1501–1504. doi: 10.1083/jcb.127.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schellenberg G D. Proc Natl Acad Sci USA. 1995;92:8552–8559. doi: 10.1073/pnas.92.19.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pike C J, Burdick D, Walencewicz A J, Glabe C G, Cotman C W. J Neurosci. 1993;13:1676–1687. doi: 10.1523/JNEUROSCI.13-04-01676.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarrett J T, Lansburry P T. Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 19.Blow D M, Chayen N E, Lloyd L F, Saridakis E. Protein Sci. 1994;3:1638–1643. doi: 10.1002/pro.5560031003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andreu J M, Timasheff S N. Methods Enzymol. 1986;130:47–59. doi: 10.1016/0076-6879(86)30007-7. [DOI] [PubMed] [Google Scholar]
- 21.Eaton W A, Hofrichter J. Science. 1995;268:1142–1143. doi: 10.1126/science.7539154. [DOI] [PubMed] [Google Scholar]
- 22.Sathasivam K, Amaechi I, Mangiarini L, Bates G P. Hum Genet. 1997;99:692–695. doi: 10.1007/s004390050432. [DOI] [PubMed] [Google Scholar]
- 23.Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub M-P, Durand B, Lanotte M, Berger R, Chambon P. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 25.Wanker, E. E., Scherzinger, E., Heiser, V., Sittler, A., Eickhoff, H. & Lehrach, H. (1999) Methods Enzymol., in press. [DOI] [PubMed]
- 26.Sittler A, Wälter S, Wedemeyer N, Hasenbank R, Scherzinger E, Eickhoff H, Bates G P, Lehrach H, Wanker E E. Mol Cell. 1998;2:427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- 27.Rubinsztein D C, Leggo J, Coles R, Almqvist E, Biancalana V, Cassiman J-J, Chotai K, Connarty M, Crauford D, Curtis A, et al. Am J Hum Genet. 1996;59:16–22. [PMC free article] [PubMed] [Google Scholar]
- 28.Perutz M. Curr Opin Struct Biol. 1996;6:848–858. doi: 10.1016/s0959-440x(96)80016-9. [DOI] [PubMed] [Google Scholar]
- 29.The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 30.Saudou F, Finkbeiner S, Devys D, Greenberg M E. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 31.Klement I A, Skinner P J, Kaytor M D, Yi H, Hersch S M, Clark H B, Zoghbi H Y, Orr H T. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 32.Soto C, Sigurdsson E M, Morelli L, Kumar R A, Castano E M, Frangione B. Nat Med. 1998;4:822–826. doi: 10.1038/nm0798-822. [DOI] [PubMed] [Google Scholar]