Abstract
Methylobacterium sp. strain CM4, an aerobic methylotrophic α-proteobacterium, is able to grow with chloromethane as a carbon and energy source. Mutants of this strain that still grew with methanol, methylamine, or formate, but were unable to grow with chloromethane, were previously obtained by miniTn5 mutagenesis. The transposon insertion sites in six of these mutants mapped to two distinct DNA fragments. The sequences of these fragments, which extended over more than 17 kb, were determined. Sequence analysis, mutant properties, and measurements of enzyme activity in cell-free extracts allowed the definition of a multistep pathway for the conversion of chloromethane to formate. The methyl group of chloromethane is first transferred by the protein CmuA (cmu: chloromethane utilization) to a corrinoid protein, from where it is transferred to H4folate by CmuB. Both CmuA and CmuB display sequence similarity to methyltransferases of methanogenic archaea. In its C-terminal part, CmuA is also very similar to corrinoid-binding proteins, indicating that it is a bifunctional protein consisting of two domains that are expressed as separate polypeptides in methyl transfer systems of methanogens. The methyl group derived from chloromethane is then processed by means of pterine-linked intermediates to formate by a pathway that appears to be distinct from those already described in Methylobacterium. Remarkable features of this pathway for the catabolism of chloromethane thus include the involvement of a corrinoid-dependent methyltransferase system for dehalogenation in an aerobe and a set of enzymes specifically involved in funneling the C1 moiety derived from chloromethane into central metabolism.
Attention has been focused on chloromethane and bromomethane because of their role as sources of stratospheric chlorine and bromine, the primary agents of ozone destruction. Chloromethane (CH3Cl) is the most abundant halocarbon in the atmosphere and is responsible for 15–20% of chlorine-catalyzed ozone destruction in the stratosphere (1). It is released at an estimated global rate of 3.5–5 × 106 tons per year, primarily from natural sources, and less than 1% of the global chloromethane flux is caused by industrial production of the compound (reviewed in ref. 1). For bromomethane, current estimates of ocean emission and natural formation during combustion of vegetation fall in the range of 8 × 104 tons per year, slightly more than the amount annually emitted by the use of this compound in soil fumigation (2). On a molar basis, bromine is 40–100 times more effective than chlorine in depleting ozone (3). Thus, chloromethane and bromomethane contribute about equally to an estimated 40% of the total global loss of stratospheric ozone.
Green plants (4) and soil bacteria (5–9) represent terrestrial sinks for chloromethane and bromomethane. Evidence for bacterial degradation of halogenated methanes in seawater was also reported (10). Microbial metabolism of monohalomethanes includes oxidative (11) and hydrolytic (12) cometabolic processes, as well as mineralization by methylotrophic bacteria that use chloromethane as a growth substrate. The homoacetogenic bacterium Acetobacterium dehalogenans (13) is the only known strictly anaerobic representative of the latter group (14). Anoxic dehalogenation of chloromethane by this organism was shown to be catalyzed by enzymes that transfer the methyl group of chloromethane by means of a corrinoid protein to H4folate to yield chloride and CH3-H4folate, an intermediate of the acetyl-CoA pathway (15). In contrast, the reactions by which some recently isolated strictly aerobic methylotrophic bacteria (8) use chloromethane as a growth substrate have yet to be elucidated. The physiological properties of the wild-type and of chloromethane utilization-negative mutants of a representative strain, Methylobacterium sp. CM4, led us to propose that this organism metabolizes chloromethane by initial dehalogenation by means of a methyl transfer reaction (16).
Here we report on the sequence of two large DNA fragments containing at least four genes essential for chloromethane metabolism in strain CM4. We present experimental evidence that this aerobic bacterium is able to catalyze transfer of the methyl moiety of chloromethane to H4folate by means of a corrinoid intermediate to yield CH3-H4folate, a key intermediate of methylotrophic metabolism.
MATERIALS AND METHODS
Materials.
Reagents for molecular biology were obtained from Fermentas (Vilnius, Lithuania) and Boehringer Mannheim. (6S)-5,6,7,8-tetrahydrofolic acid trihydrochloride (H4folate) and (6S)-5-methyltetrahydrofolate (CH3-H4folate) were purchased from Schircks Laboratorium (Jona, Switzerland). ATP, S-adenosyl methionine, and methylcobalamin were from Sigma, and NADPH:FMN oxidoreductase was from Boehringer Mannheim. All other chemicals were reagent grade or better and were purchased from Fluka.
Bacterial Strains.
Bacterial strains in this study included Methylobacterium sp. strain CM4, which grows with chloromethane (8), and miniTn5 (17) insertion mutants, whose phenotypes have been described (16). Escherichia coli K12 strain DH5α (GIBCO/BRL Life Technologies) was used as a host in DNA work.
DNA Manipulations.
Preparation of genomic DNA, restriction enzyme digestions, ligations, and transformations were performed by using standard procedures (18). Plasmid pBluescript-KSII(+) (Stratagene) was used for cloning. DNA fragments from mutants of strain CM4 containing a miniTn5 insertion were cloned by selection of transformants for kanamycin resistance (25 μg/ml).
Sequence Analysis.
The cloned genomic DNA was sequenced on both strands by using PCR methods with fluorescent dideoxynucleotide terminators and an ABI-Prism automatic sequencer (Perkin–Elmer). The precise site of insertion of the minitransposon was determined for all mutants. The sequences of cluster I (9,658 nt, accession no. AJ011316) and cluster II (8,457 nt, accession no. AJ011317) were assembled from sequence fragments obtained from DNA cloned from the different mutants with the GCG sequence analysis package (Version 8.1, Genetics Computer Group, Madison, WI). Similarity searches were performed by using gapped blast and psi–blast programs (19) against public protein and gene databases.
N-Terminal Sequencing.
The 67-kDa protein induced during growth with chloromethane (16) was partially purified, and the N-terminal sequence of the corresponding protein band on SDS/PAGE was determined by Edman degradation using an Applied Biosystems 476A automatic sequencer.
Preparation of Cell-Free Extract.
Methylobacterium sp. strain CM4 and mutants were grown as described (16). Bacteria were harvested at an OD600 of 0.5 to 0.7 (10,000 × g for 15 min) and resuspended (1 g wet cells per ml) in 50 mM Tris-SO4 buffer (pH 7.2) containing 5 mM DTT. Cells were disrupted by two passages through a French pressure cell (120 MPa, 4°C), and DNaseI (50 μg/ml final) was added to the suspension, which was centrifuged (35000 × g for 45 min) to remove cell debris. The resulting supernatant was cleared from membrane components by ultracentrifugation (160,000 × g for 45 min), and the cell-free extract obtained (≈15 mg protein/ml) was flash-frozen in liquid nitrogen and stored at −20°C.
Activity Measurements with Crude Extracts.
Solutions were made anoxic by degassing with N2 plus H2 [95:5 (vol/vol)]. Enzyme reactions, manipulations, and measurements were performed under the same atmosphere. Assays of enzymatic activity were done at 30°C in a 3-ml volume in 12.4-ml serum flasks with gas-tight rubber stoppers. The H4folate-dependent dehalogenation of chloromethane was measured by following the consumption of chloromethane by gas chromatography (20) by using a Henry constant of 0.43, calculated by interpolation to 30°C of the published value for chloromethane (21). Dehalogenation was also determined by monitoring the chloromethane-dependent formation of CH3-H4folate from H4folate. CH3-H4folate was separated from the incubation mixture by HPLC (14) and detected spectrophotometrically at 320 nm. The incubation mixture contained cell-free extract (0.8–4 mg/ml protein) in 100 mM Tris-SO4 (pH 7.8) buffer, 5 mM DTT, 2.4 mM H4folate [of which 1 mM was biologically available (22)], and 1 mM titanium(III)citrate. Cell-free extracts dialyzed against 100 mM Tris-SO4 (pH 7.8) were also used to check whether any endogenous cofactors in the extracts were involved in dehalogenase activity. The dehalogenation reaction was initiated by the addition of 0.5 mM chloromethane gas (based on the liquid phase volume) through the rubber stopper with a gas-tight syringe. For determination of methylcobalamin:H4folate methyltransferase activity, the assay mixture contained 0.5 mM methylcobalamin instead of chloromethane. The numbers reported are from representative individual experiments that were performed at least twice and all yielded very similar results.
Determination of Protein Concentration.
Protein was determined by the method of Bradford (23) by using a commercial dye reagent (Bio-Rad) with BSA as a standard.
RESULTS
Identification of Genes Involved in Chloromethane Utilization.
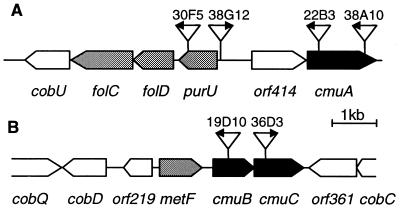
We previously isolated miniTn5 transposon insertion mutants of Methylobacterium sp. strain CM4 that were unable to grow with chloromethane (16). Thus, nine Cmu− (chloromethane utilization negative) mutants were obtained that were still able to grow with methanol, methylamine, or formate. Conversely, 73 transposon mutants defective in the utilization of methanol, methylamine, methanol plus methylamine, or formate could still grow with chloromethane (16). This suggested that chloromethane was metabolized in Methylobacterium sp. CM4 by reactions different from those involved in the metabolism of methanol and methylamine. The genes whose insertional inactivation caused loss of the ability to grow with chloromethane were isolated by selection of the kanamycin resistance gene present on the minitransposon (17). The DNA fragments carrying a transposon insertion were sequenced from all Cmu− mutants. Transposon insertion mutants, arbitrarily labeled in order of their detection (16), were found in four apparently unlinked DNA regions, which were termed cluster I (mutants 30F5, 38G12, 22B3, and 38A10), cluster II (mutants 19D10 and 36D3), cluster III (mutant 11G7), and cluster IV (mutant 27B11). The Cmu− mutant 27C10 was not analyzed in detail because it appeared to carry a partial duplication of the transposon. A schematic representation of the 14 ORFs identified in the DNA sequences of clusters I and II is shown in Fig. 1.
Figure 1.
Schematic view of gene clusters I (A) and II (B) of Methylobacterium sp. CM4. Genes encoding methyltransferases are black, genes encoding putative pterine-dependent enzymes of C1 metabolism are shaded, and genes encoding enzymes of cobalamin biosynthesis or proteins with unknown function are white. The position and orientation of the transposon insertions in the genome of Cmu− mutants is also shown.
Most of the encoded polypeptides displayed significant sequence identity to proteins of known functions (Table 1). With respect to their possible role in metabolism, the proteins encoded in DNA clusters I and II fell into four groups: methyltransferases, pterine-dependent enzymes, proteins associated with cobalamin biosynthesis, and proteins of unknown function. Similarity searches of the protein sequences encoded in clusters III and IV (data not shown) did not provide insights as to their association with chloromethane transformation and are not discussed further here.
Table 1.
Genes and ORFs in DNA regions associated with chloromethane utilization
| Gene orf | Length, aa | Calculated Mr, kDa | Gene
|
Inferred function | Sequence comparison of representative hit: % protein sequence identity* | Identity %† | |
|---|---|---|---|---|---|---|---|
| Begins | Ends | ||||||
| Cluster I | |||||||
| cobU | 342 | 34.9 | 1,502 | 474 | Cobalamin biosynthesis | CobU (P. denitrificans) (P29935) | 57 |
| folC | 467 | 49.8 | 2,942 | 1,539 | Folylglutamate synthetase | FolC (E. coli) (P08192) | 32 |
| folD | 306 | 32.4 | 3,865 | 2,945 | 5,10-methylene-H4folate dehydrogenase/5,10-methenylH4folate cyclohydrolase | FolD (E. coli) (P24186) | 49 |
| purU | 287 | 32.7 | 4,849 | 3,986 | 10-formyl-H4folate hydrolase | PurU (Corynebacterium sp.) (Q46339) | 47 |
| orf414 | 414 | 43.7 | 5,628 | 6,872 | Unknown | Orf (Mycobacterium tuberculosis) (P72042) | 31 (143 aa) |
| cmuA | 617 | 67.0 | 6,897 | 8,750 | Methyltransferase/ | MtbA (M. barkeri) (O30640) | 24/32‡ |
| corrinoid protein | MtmC (M. barkeri) (O30641) | ||||||
| Cluster II | |||||||
| cobQ | >424 | ND | <1 | 1,275 | Cobalamin biosynthesis | CobQ (P. denitrificans) (P29932) | 59 |
| cobD | 330 | 34.2 | 2,275 | 1,283 | Cobalamin biosynthesis | CobD (P. denitrificans) (P21634) | 55 |
| orf219 | 219 | 24.9 | 3,345 | 2,686 | Unknown | (Synechocystis sp.) (Q55963) | 32 (117 aa) |
| metF | 320 | 34.3 | 3,507 | 4,469 | 5,10-methylene-H4folate reductase | Orf (Saccharomyces cerevisiae) (P53128) | 24 (156 aa) |
| cmuB | 311 | 33.3 | 4,703 | 5,638 | Methyl transfer | MtrH (M. thermoautotrophicum) (P80187) | 30 |
| cmuC | 378 | 41.2 | 5,635 | 6,771 | Methyl transfer | MtaA (M. barkeri) (Q48949) | 28 (104 aa) |
| orf361 | 361 | 37.5 | 7,971 | 6,886 | Cobalamin biosynthesis | MTH808 (M. thermoautotrophicum) (O26899) | 35 |
| cobC | >162 | ND | >8,456 | 7,968 | Cobalamin biosynthesis | CobC (P. denitrificans) (P21633) | 38 |
ND, not determined.
*Accession numbers from SwissProt or Trembl databases shown in parentheses.
Sequence identity is over the entire length of the shorter of the two compared sequences, except where noted.
MtbA sequence (339 aa) can be aligned to residues 7–353 of CmuA, and the MtmC sequence (217 aa) can be aligned to residues 401–607 of CmuA, respectively.
The products of cmuA, cmuB, and cmuC in clusters I and II showed sequence similarity to methyltransferases or corrinoid-binding proteins from archaea (Table 1). The C-terminal part of CmuA was found to be most similar to MtmC, the 29-kDa corrinoid protein which, when methylated by methylamine, acts as the substrate of the methyltransferase catalyzing the methylation of coenzyme M in Methanosarcina barkeri (24). The C-terminal part of CmuA also showed similarity to many other corrinoid-binding proteins, including methionine synthases (25). The N-terminal part of CmuA showed considerable sequence identity to MtbA, the 36-kDa methyltransferase that transfers the methyl group from MtmC to coenzyme M (24, 26). The similarity in sequence between the two proteins extended over the entire length of MtbA (Table 1). It thus appears that CmuA, whose calculated molecular mass is 67 kDa, represents an unprecedented fusion of two proteins that are expressed as separate but closely associated polypeptides in methyl transfer systems of methanogenic archaea. CmuB, the second methyltransferase-like protein suggested from sequence analysis, showed most sequence identity (30%, Table 1) with subunit MtrH of the membrane-associated N5-methyl-tetrahydromethanopterin:coenzyme M methyltransferase complex from Methanobacterium thermoautotrophicum, which catalyzes transfer of the methyl group of N5-methyltetrahydromethanopterin to the corrinoid protein MtrA (27). CmuB, unlike CmuA, also showed low but significant pairwise identity (23%) to the CH3-H4folate-binding domain of MetH from E. coli (residues 337–648 in the protein sequence).
CmuC, the third methyltransferase-like putative protein, was most similar to MtaA, another corrinoid:coenzyme M methyltransferase characterized in M. barkeri (26) (Table 1). CmuC was 19% identical to the N-terminal domain of CmuA and displayed low identity (14%) over its entire length to MtbA and MtaA from M. barkeri and to DcuP from E. coli in multiple alignments.
The second group of proteins detected in clusters I and II were similar to enzymes involved in interconversion pathways of one-carbon compounds. A Cmu− phenotype was observed in mutant 30F5, in which the ORF encoding a protein with strong similarity to bacterial 10-formyl-H4folate hydrolases (Table 1) was disrupted. Accordingly, the gene was named purU. Proteins similar to bacterial FolD and FolC (Table 1), enzymes involved in the metabolism of one-carbon compounds, are encoded by genes downstream of purU. It is noteworthy that cell-free extracts of mutant 38G12, in which the transposon insertion is located upstream of the purU gene (Fig. 1), lack a protein of about 35 kDa that is induced by chloromethane (16). This protein could therefore be PurU (32.7 kDa calculated molecular mass) and perhaps also FolD (32.4 kDa; see Table 1). Finally, the gene tentatively named metF in cluster II codes for a protein similar to enzymes of the 5,10-methylene-H4folate reductase family in part of its sequence. The role in chloromethane degradation of this and other ORFs detected in the DNA sequence of clusters I and II, however, remains uncertain because no mutants are yet available in which these genes have been knocked out.
In Vitro Dehalogenation of Chloromethane.
Transposon insertions into genes cmuA and cmuB (Fig. 1) led to a dechlorination-negative phenotype in the corresponding mutants that could neither dehalogenate chloromethane nor grow with this compound. The other Cmu− mutants released chloride from chloromethane in a resting cell assay (16), but were unable to grow with chloromethane. The dechlorination-negative phenotype of cmuA and cmuB mutants strongly indicated that the proteins encoded by these genes were directly involved in chloromethane dehalogenation.
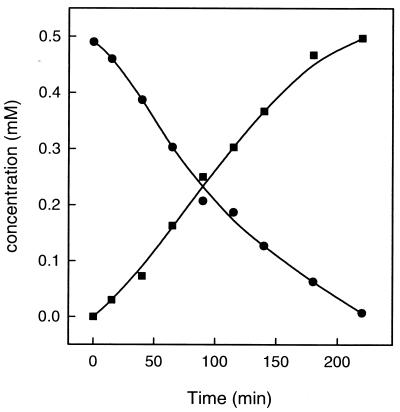
In previous work with strain CM4, chloromethane dehalogenation activity could only be detected in cell suspensions (16). The inferred function of several ORFs (Table 1) suggested that assay mixtures containing H4folate and chloromethane (Table 2) might allow activity measurements in cell-free extracts of Methylobacterium sp. CM4, as previously observed in chloromethane dehalogenation in A. dehalogenans (14). Indeed, chloromethane was consumed with the concomitant formation of CH3-H4folate from H4folate by cell-free extracts of the chloromethane-grown wild-type strain CM4 (Fig. 2) at 0.5% of the in vivo chloromethane degradation rate. The data presented in Table 2 demonstrated that the dehalogenation activity was not present in extracts of cells grown with methanol, confirming the previously observed inducibility of chloromethane utilization in strain CM4 (16). CH3-H4folate formation was strictly dependent on chloromethane and H4folate. The chloromethane dehalogenase activity converting chloromethane and H4folate to CH3-H4folate was stimulated by the nonphysiological reductant titanium(III)citrate (Table 2). Most notably, low molecular weight components of known corrinoid protein reactivation systems, such as ATP, as well as GTP, S-adenosyl-methionine, FMNH2, and FADH2 were without effect on the dehalogenase activity of Methylobacterium sp. CM4 (data not shown). In contrast, chloromethane dehalogenase activity in cell-free extracts of the strict anaerobe A. dehalogenans requires the addition of ATP, presumably to maintain the cobalt ion of the corrinoid cofactor in the reduced Co(I) state (14, 15, 28).
Table 2.
Components required for dehalogenation of chloromethane by cell-free extracts of Methylobacterium sp. CM4
| Growth substrate | Assay mixture* | Maximum rate (nmol/min⋅mg protein)
|
|
|---|---|---|---|
| CH3Cl consumption | CH3-H4folate formation | ||
| MeOH | Complete | <0.1 | <0.1 |
| CH3Cl | Complete | 3.9 | 3.9 |
| CH3Cl | Without CH3Cl | — | <0.1 |
| CH3Cl | Without H4folate | 0.3 | <0.1 |
| CH3Cl | Without Ti(III)Citrate | 2.6 | 1.7 |
*See Materials and Methods.
Figure 2.
Disappearance of chloromethane (circles) and formation of CH3-H4folate from chloromethane and H4folate (squares) by cell-free extracts of chloromethane-grown Methylobacterium sp. CM4. Chloromethane was determined by gas chromatography and CH3-H4folate by HPLC (see Materials and Methods). The assay mixture contained 2.4 mg protein, 0.5 mM chloromethane, 1 mM H4folate, and 1 mM titanium(III)citrate.
Enzyme Activities in Cell-Free Extracts of Cmu− Mutants.
Methylcobalamin could replace chloromethane as a methyl donor in the formation of CH3-H4folate from H4folate catalyzed by cell-free extracts of strain CM4 grown with chloromethane (Table 3). This suggested that the transformation of chloromethane and H4folate to CH3-H4folate and chloride in strain CM4 resulted from two sequential methyl transfer reactions involving a methylated corrinoid intermediate (Fig. 3). Such sequential methyl transfer reactions were previously documented in enzyme systems of methanogens catalyzing the formation of methyl-CoM from coenzyme M and methanol or methylamine (29), and most likely also operate in the chloromethane dehalogenase of A. dehalogenans (15). In these systems, methylcobalamin presumably acts as a surrogate for the physiological, protein-bound methyl-corrinoid. This may explain the approximately 3-fold lower specific activity of the methylcobalamin:H4folate methyltransferase (methyltransferase II) activity, as compared with the chloromethane dehalogenase activity representing the overall rate of the transformation of chloromethane to CH3-H4folate by methyltransferase I and methyltransferase II reactions (Fig. 3).
Table 3.
Methyltransferase activities in cell-free extracts of Methylobacterium sp. CM4 wild-type and Cmu− mutants
| Strain | Gene affected by miniTn5 insertion | Initial rate of CH3-H4folate formation (nmol/min⋅mg protein)
|
|
|---|---|---|---|
| From CH3Cl | From CH3B12 | ||
| Wild type | — | 2.6* | 0.8* |
| 30F5 | purU | 1.7 | 0.5 |
| 38G12 | purU (upstream) | 2.1 | 0.8 |
| 22B3 | cmuA | <0.1 | 1.0 |
| 38A10 | cmuA | <0.1 | 0.7 |
| 19D10 | cmuB | <0.1 | <0.1 |
| 36D3 | cmuC | 2.2 | 0.7 |
Grown with 20 mM methanol and 2% vol/vol CH3Cl.
Initial rate of CH3-H4folate formation in extracts from wild-type bacteria grown with methanol was <0.1 nmol/min⋅mg.
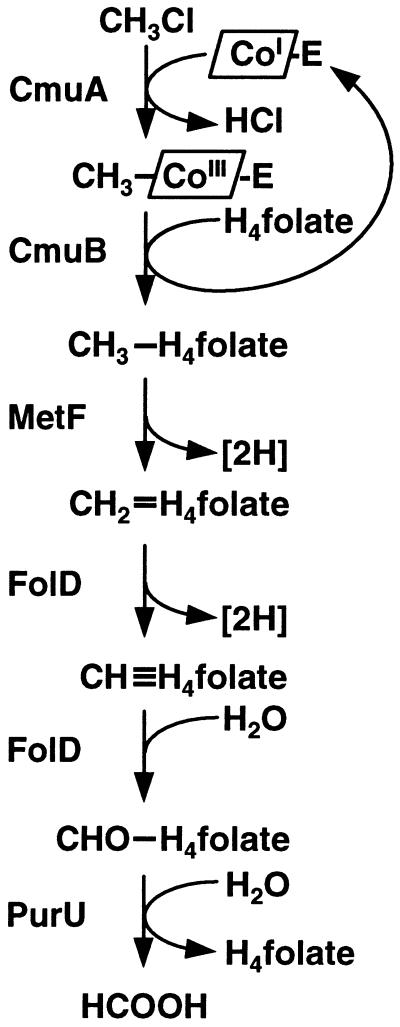
Figure 3.
Proposed pathway for the oxidation of chloromethane to formate based on DNA sequence data and biochemical analysis. CmuA, methyltransferase I; CmuB, methyltransferase II; MetF, putative 5,10-methylene-H4folate reductase; FolD, putative 5,10-methylene-H4folate dehydrogenase/5,10-methenyl-H4folate cyclohydrolase; PurU, putative 10-formyl-H4folate hydrolase. The corrinoid protein acting as the primary methyl acceptor and thought to be part of CmuA is indicated by CoI-E.
The two mutants, 22B3 and 38A10, that carried insertions in cmuA were defective in the dehalogenation reaction (proposed to be initiated by methyltransferase I, Fig. 3), but still capable of catalyzing the methyltransferase II reaction (Table 3). This suggested that cmuA encoded methyltransferase I, but not methyltransferase II. Moreover, the 67-kDa protein previously noted as being induced during growth with chloromethane (16) was shown to be CmuA by determination of its N-terminal sequence (XGKMTSRERMFAXTM), suggesting further an important role of CmuA in chloromethane degradation.
The inactivation of the cmuB gene in mutant 19D10 resulted in the loss of both dehalogenase and methyltransferase II activity (Table 3). Thus, CmuB appeared to be required for both methyltransferase reactions that lead from chloromethane by means of a putative methylated corrinoid protein to CH3-H4folate (Table 3; Fig. 3). Alternatively, the cmuB mutant may still be able to perform the initial dehalogenation reaction (catalyzed by methyltransferase I), but not the subsequent transfer of the methyl group from the corrinoid-binding protein to H4folate. In this case, methylated corrinoid protein would be produced in amounts stoichiometric to those of methyltransferase I in cell-free extracts, but the dehalogenation reaction would remain undetected because of the low amounts of this protein in the assay.
Mutants of Methylobacterium sp. CM4 disrupted in purU and in cmuC were unable to grow with chloromethane, but exhibited wild-type levels of both dehalogenase and methyltransferase II activity (Table 3). These mutants are thus unaffected in the dehalogenation reaction but are deficient in some later step of chloromethane metabolism.
DISCUSSION
The results presented here lead us to propose the corrinoid-dependent pathway for chloromethane catabolism shown in Fig. 3. This pathway implies that the dehalogenation reaction proceeds with the Co(I) of a corrinoid protein acting as primary acceptor for the methyl group of chloromethane. It requires methyltransferase I to form methylated corrinoid protein from chloromethane, and methyltransferase II for the transfer of the methyl group to the pterine cofactor.
The amino acid sequence of CmuA (Table 1) supports the view that this protein not only encodes methyltransferase I activity detected in cell-free extracts, but also acts as the corrinoid-binding protein indicated in the model (Table 3). We thus hypothesize that the CmuA protein carrying a methylated corrinoid serves as the methyl-donating substrate for CH3-H4folate formation from H4folate by the methyltransferase II encoded by cmuB.
Sequence alignments of the C-terminal domain of the CmuA protein with that of the corrinoid-binding domain of methionine synthase of E. coli, whose structure has been solved (30), allows one to speculate further on the properties of the CmuA protein (Fig. 4). The most striking feature of the CmuA sequence, when compared with the motifs that were defined for cobalamin-dependent methionine synthases (25, 30), is residue Gln-504, equivalent to His-759 in E. coli (Fig. 4). The three residues, His-759, Asp-757, and Ser-810 in E. coli methionine synthase, form a ligand catalytic triad with the histidine residue as the lower axial ligand of the “base-off/His-on” corrinoid (25). His-759 in E. coli was shown to be essential for enzyme turnover (31). It is unclear in what way the corresponding glutamine in CmuA can be isofunctional to this residue. No other sequence in the database so far matches the corrinoid-binding motif so closely as CmuA, but lacks the histidine residue. A glutamine residue, however, was recently described to be the axial ligand of the nickel porphinoid F430 of methyl-coenzyme M reductase of M. thermoautotrophicum (32), and the manually aligned sequence of the AcsD corrinoid iron–sulfur protein of Clostridium thermoaceticum also features a Gln residue in register with His-759 of methionine synthases (S. Ragsdale, personal communication). A glutamine at position 504 in CmuA is expected to contribute much weaker ligation to a corrinoid-bound cobalt than a histidine residue. As a consequence, it is also expected to render the reduction potential of the Co(II)/Co(I) couple less negative and thus to stabilize the corrinoid bound by CmuA in its Co(I) state. A reactive Co(I) species in CmuA would readily react with chloromethane, which is known to be a good corrinoid alkylating agent (33). This could contribute toward maintaining the methyltransferase I in an active form by preventing oxidation of the cobalt to Co(II). In support of this idea, the ATP- and/or reductant-dependent reactivation system essential for activity of methyltransferases from anaerobes (15, 34, 35) and of methionine synthase (36) was not required for chloromethane dehalogenase activity in cell-free extracts of strain CM4 (Table 3). The sequence similarity of CmuA to E. coli methionine synthase does not include the C-terminal “AdoMet” domain of methionine synthase involved in reactivation of the cobalt center (36). In addition, none of the other protein sequences deduced from the genes of cluster I or II in strain CM4 (Table 1) showed any detectable similarity to those involved in the reductive activation of methyltransferases.
Figure 4.
Alignment of the sequences from CmuA, MtmC of M. barkeri (SwissProt accession no. O30641), MetH of E. coli (P13009), and MutA the human methylmalonyl-CoA mutase (P22033), in the conserved C-terminal sequence region of CmuA around the corrinoid-binding site. This site was defined from a structure-based sequence fingerprint for cobalamin-dependent methionine synthases and mutases (25). Most conserved residues in these motifs (indicated by shaded boxes) are labeled by dots below the alignment. Positions in which amino acids were identical or similar to the CmuA sequence in at least two other sequences were boxed in black or shaded, respectively, using the program boxshade.
The reactions in the second part of the proposed chloromethane utilization pathway (Fig. 3) lead from CH3-H4folate to formate. Indication for a H4folate-dependent pathway specific for the conversion of CH3-H4folate derived from chloromethane to formate is of interest in light of recent findings on C1 metabolism of Methylobacterium extorquens AM1 (37). This organism possesses a dephospho-tetrahydromethanopterin-mediated C1 transfer pathway that is essential for growth with C1 compounds and brings about the conversion of formaldehyde to carbon dioxide. The sequences of many proteins involved in this pathway most closely resemble those of enzymes that participate in reduction of carbon dioxide to methane in methanogenic archaea. Dephospho-tetrahydromethanopterin was previously thought to be unique to methanogenic and sulfate-reducing archaea (38). In parallel to the tetrahydromethanopterin-mediated pathway, M. extorquens AM1 appears to operate an H4folate-dependent pathway for the oxidation of formaldehyde to carbon dioxide. With the exception of the gene for NADP-dependent methylene-H4folate dehydrogenase (mtdA), however, the genes encoding the enzymes of this pathway are still unknown (37).
The proposed pterine-dependent pathway for the conversion of chloromethane to formate in Methylobacterium sp. CM4 (Fig. 3) represents yet a third variant of the reactions for interconverting C1 compounds in Methylobacterium. Because the Cmu− mutant with a disrupted copy of the purU gene still grew with methanol or methylamine, this pathway would be specific for processing a methylated pterine-based cofactor derived from chloromethane. Although the nature of the pterin cofactor in this pathway needs to be determined, we have found that purified CmuB protein catalyses methyl transfer from methylcobalamin by using H4folate but not tetrahydromethanopterin as the methyl group acceptor (unpublished data).
In conclusion, our biochemical and genetic data suggest that growth of the strict aerobe Methylobacterium sp. CM4 with chloromethane is based on a specific catabolic pathway involving corrinoid-dependent enzymes that was hitherto unknown in organisms with an aerobic lifestyle. The similarity in sequence of CmuA and CmuB to other proteins of related function in methanogenic archaea (Table 1) is interesting from an evolutionary standpoint, because it extends the emerging notion that genes involved in methylotrophy and methanogenesis share a common origin (37), and that strictly anaerobic archaea and aerobic bacteria may use similar reactions to exploit C1 substrates for metabolism.
Acknowledgments
This work was supported by Grant 0-20-436-97 from the Swiss Federal Institute of Technology (Zurich).
Footnotes
References
- 1.Harper D B. Mycol Res. 1998;102:769–787. [Google Scholar]
- 2.Yvon-Lewis S A, Butler J H. Geophys Res Lett. 1997;24:1227–1230. [Google Scholar]
- 3.Montzka S A, Butler J H, Myers R C, Thomson T M, Swanson T H, Clarke A D, Lock L T, Elkins J W. Science. 1996;272:1318–1322. doi: 10.1126/science.272.5266.1318. [DOI] [PubMed] [Google Scholar]
- 4.Jeffers P M, Wolfe N L, Nzengung V. Geophys Res Lett. 1998;25:43–46. [Google Scholar]
- 5.Connell Hancock T L, Costello A M, Lidstrom M E, Oremland R S. Appl Environ Microbiol. 1998;64:2899–2905. doi: 10.1128/aem.64.8.2899-2905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shorter J H, Kolb C E, Crill P M, Kerwin R A, Talbot R W, Hines M E, Harriss R C. Nature (London) 1995;377:717–719. [Google Scholar]
- 7.Miller L G, Connell T L, Guidetti J R, Oremland R S. Appl Environ Microbiol. 1997;63:4346–4354. doi: 10.1128/aem.63.11.4346-4354.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doronina N V, Sokolov A P, Trotsenko Y A. FEMS Microbiol Lett. 1996;142:179–183. [Google Scholar]
- 9.Hines M E, Crill P M, Varner R K, Talbot R W, Shorter J H, Kolb C E, Harriss R C. Appl Environ Microbiol. 1998;64:1864–1870. doi: 10.1128/aem.64.5.1864-1870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodwin K D, Lidstrom M E, Oremland R S. Environ Sci Technol. 1997;31:3188–3192. [Google Scholar]
- 11.Oremland R S, Miller L G, Culbertson C W, Connell T L, Jahnke L. Appl Environ Microbiol. 1994;60:3640–3646. doi: 10.1128/aem.60.10.3640-3646.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keuning S, Janssen D B, Witholt B. J Bacteriol. 1985;163:635–639. doi: 10.1128/jb.163.2.635-639.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufmann F, Wohlfarth G, Diekert G. Eur J Biochem. 1998;253:706–711. doi: 10.1046/j.1432-1327.1998.2530706.x. [DOI] [PubMed] [Google Scholar]
- 14.Messmer M, Wohlfarth G, Diekert G. Arch Microbiol. 1993;160:383–387. [Google Scholar]
- 15.Wohlfarth G, Diekert G. Curr Opin Biotechnol. 1997;8:290–295. doi: 10.1016/s0958-1669(97)80006-7. [DOI] [PubMed] [Google Scholar]
- 16.Vannelli T, Studer A, Kertesz M, Leisinger T. Appl Environ Microbiol. 1998;64:1933–1936. doi: 10.1128/aem.64.5.1933-1936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Lorenzo V, Timmis K N. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 18.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Greene/Wiley; 1987–1998. [Google Scholar]
- 19.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mägli A, Messmer M, Leisinger T. Appl Environ Microbiol. 1998;64:646–650. doi: 10.1128/aem.64.2.646-650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gossett J M. Environ Sci Technol. 1987;21:202–208. [Google Scholar]
- 22.Wohlfarth G, Geerligs G, Diekert G. Eur J Biochem. 1990;192:411–417. doi: 10.1111/j.1432-1033.1990.tb19242.x. [DOI] [PubMed] [Google Scholar]
- 23.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 24.Burke S A, Lo S L, Krzycki J A. J Bacteriol. 1998;180:3432–3440. doi: 10.1128/jb.180.13.3432-3440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drennan C L, Dixon M M, Hoover D M, Jarrett J T, Goulding C W, Matthews R G, Ludwig M L. In: Vitamin B12 and B12 Proteins. Kräutler B, Arigoni D, Golding B T, editors. Weinheim, Germany: Wiley–VCH; 1998. pp. 133–155. [Google Scholar]
- 26.Harms U, Thauer R K. Eur J Biochem. 1996;235:653–659. doi: 10.1111/j.1432-1033.1996.00653.x. [DOI] [PubMed] [Google Scholar]
- 27.Harms U, Thauer R K. Eur J Biochem. 1997;250:783–788. doi: 10.1111/j.1432-1033.1997.00783.x. [DOI] [PubMed] [Google Scholar]
- 28.Messmer M, Reinhardt S, Wohlfarth G, Diekert G. Arch Microbiol. 1996;165:18–25. [Google Scholar]
- 29.Thauer R K. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 30.Drennan L C, Huang S, Drummond J T, Matthews R G, Ludwig M L. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 31.Jarrett J T, Amaratunga M, Drennan C L, Scholten J D, Sands R H, Ludwig M L, Matthews R G. Biochemistry. 1996;35:2464–2475. doi: 10.1021/bi952389m. [DOI] [PubMed] [Google Scholar]
- 32.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer R K. Science. 1997;278:1457–1462. doi: 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]
- 33.Schrauzer G N, Deutsch E. J Am Chem Soc. 1969;91:3341–3350. doi: 10.1021/ja01040a041. [DOI] [PubMed] [Google Scholar]
- 34.Menon S, Ragsdale S W. Biochemistry. 1998;37:5689–5698. doi: 10.1021/bi9727996. [DOI] [PubMed] [Google Scholar]
- 35.Daas P J H, Wassenaar R W, Willemsen P, Theunissen R J, Keltjens J T, van der Drift C, Vogels G D. J Biol Chem. 1996;271:22339–22345. doi: 10.1074/jbc.271.37.22339. [DOI] [PubMed] [Google Scholar]
- 36.Jarrett J T, Hoover D M, Ludwig M L, Matthews R G. Biochemistry. 1998;37:12649–12658. doi: 10.1021/bi9808565. [DOI] [PubMed] [Google Scholar]
- 37.Chistoserdova L, Vorholt J A, Thauer R K, Lidstrom M E. Science. 1998;281:99–102. doi: 10.1126/science.281.5373.99. [DOI] [PubMed] [Google Scholar]
- 38.White R H. Biochim Biophys Acta. 1998;1380:257–267. doi: 10.1016/s0304-4165(97)00148-7. [DOI] [PubMed] [Google Scholar]