Abstract
RNA-specific adenosine deaminase (ADAR1) catalyzes the deamination of adenosine to inosine in viral and cellular RNAs. Two size forms of the ADAR1 editing enzyme are known, an IFN-inducible ≈150-kDa protein and a constitutively expressed N-terminally truncated ≈110-kDa protein. We have now identified alternative exon 1 structures of human ADAR1 transcripts that initiate from unique promoters, one constitutively expressed and the other IFN inducible. Cloning and sequence analyses of 5′-rapid amplification of cDNA ends (RACE) cDNAs from human placenta established a linkage between exon 2 of ADAR1 and two alternative exon 1 structures, designated herein as exon 1A and exon 1B. Analysis of RNA isolated from untreated and IFN-treated human amnion cells demonstrated that exon 1B–exon 2 transcripts were synthesized in the absence of IFN and were not significantly altered in amount by IFN treatment. By contrast, exon 1A–exon 2 transcripts were IFN inducible. Transient transfection analysis with reporter constructs led to the identification of two functional promoters, designated PC and PI. Exon 1B transcripts were initiated from the PC promoter whose activity in transient transfection reporter assays was not increased by IFN treatment. The 107-nt exon 1B mapped 14.5 kb upstream of exon 2. The 201-nt exon 1A that mapped 5.4 kb upstream of exon 2 was initiated from the interferon-inducible PI promoter. These results suggest that two promoters, one IFN inducible and the other not, initiate transcription of the ADAR1 gene, and that alternative splicing of unique exon 1 structures to a common exon 2 junction generates RNA transcripts with the deduced coding capacity for either the constitutively expressed ≈110-kDa ADAR1 protein (exon 1B) or the interferon-induced ≈150-kDa ADAR1 protein (exon 1A).
RNA editing represents an important posttranscriptional process by which transcripts are covalently modified in a manner that has the potential to alter the coding capacity of the RNA and thus the function of the encoded product (1, 2). RNA-specific adenosine deaminase (ADAR) represents one such type of RNA editing enzyme. ADAR catalyzes the C-6 deamination of adenosine to generate inosine at sites within double-stranded structures present within cellular pre-mRNAs and viral RNAs as well as synthetic double-stranded (ds)RNA substrates (1–4).
Two classes of adenosine-to-inosine (A-to-I) editing processes have been defined in naturally occurring RNA substrates of ADAR. First, highly site-specific A-to-I modifications are found at one or a few sites in cellular RNA transcripts that encode receptors for two important neurotransmitters in the vertebrate central nervous system, glutamate and serotonin. A total of eight adenosine positions so far have been identified that undergo A-to-I editing in five different glutamate receptor (GluR) subunit mRNAs (2, 3). These editing events result in changed RNA coding and subsequently altered biophysical properties and physiological functions of the GluR proteins. The serotonin-2C receptor (5-HT2CR), one of the three 5-HT2 subtype receptors linked to phospholipase C via G-protein coupling, also is regulated by RNA editing (2). A-to-I editing at four sites within the third exon of 5-HT2CR results in three amino acid substitutions that cause a 10- to 15-fold reduction in G-protein-mediated signaling (5). Second, multiple clustered modifications characteristic of adenosine deamination are found in viral RNAs. Such hypermutation deamination has most often been described in negative-stranded RNA virus genomes during lytic and persistent infections, as exemplified by measles virus RNA (2, 6), but extensive adenosine modifications are also present on the viral antisense RNA late in polyoma virus infection (7).
Protein purification and molecular cloning studies established that the ADAR enzymes constitute a multigene family of enzymes (8). So far, cDNAs for functional deaminases encoded by two human ADAR genes, ADAR1 and ADAR2, have been characterized (2, 3, 8). We isolated the ADAR1 cDNA as an interferon-inducible gene (9). The ADAR1 cDNA predicts a 1,226 amino acid protein possessing in the central region three functionally distinct copies of the highly conserved dsRNA-binding domain (dsRBD), designated as RI, RII, and RIII; these dsRBD copies are implicated in the recognition of dsRNA structures within the substrate RNAs (9–11). A repeated domain present in the N-terminal region of ADAR1 homologous to the N-terminal region of the vaccinia virus E3L protein (9) corresponds to two Z-DNA binding domains of ADAR1, designated Zα and Zβ (12).
Two immunologically related forms of the human ADAR1 deaminase were demonstrated in a variety of human cell lines by Western immunoblot and immunofluorescence microscopy analyses, by using antisera prepared against three nonoverlapping regions of the human cDNA expressed in Escherichia coli (9, 13). One, an interferon-inducible ≈150-kDa protein as estimated by SDS/PAGE, is present in both the cytoplasm and nucleus of IFN-treated human SY5Y and U cell lines. The other, a constitutively expressed ≈110-kDa protein by SDS/PAGE, is present in comparable amounts in untreated and IFN-treated cells and is found predominantly if not exclusively in the nucleus. The natural ≈150-kDa protein appears indistinguishable from the full-length recombinant protein produced from Met1 of the 1,226 ORF in transfected COS cells, and the natural ≈110-kDa protein displays similar properties on SDS/PAGE to a truncated recombinant protein initiated from the second methionine, Met-296, of the 1,226 ORF (9, 14). The full-length and truncated recombinant ADAR1 proteins display similar deaminase activity when measured with synthetic dsRNA substrates (14). The ADAR1 gene encoding these proteins maps to human chromosome 1q21.1–21.2 (15).
Little is known regarding the regulation of ADAR1 gene expression. A single ≈6.7-kb RNA is the only major ADAR1 mRNA detected in cell lines and various organs, with hybridization probes corresponding to different regions of the 15-exon ADAR1 cDNA (9–11, 16). Although the level of ADAR1 transcript is increased by treatment with both IFN-α and IFN-γ (9, 13, 17), a significant basal level of ADAR1 transcript is observed in human cell lines and organs in the absence of IFN treatment (9–11). Because of the important role that ADAR1 plays in the editing of cellular neurotransmitter receptor pre-mRNAs in the apparent absence of IFN treatment (2, 18) coupled with the unresolved origin of the constitutively expressed ≈110-kDa form of ADAR1 protein ubiquitously observed at high levels in the nucleus of animal cells in the absence of IFN treatment (9, 20), and because of the potential role of the IFN-inducible ≈150-kDa form of ADAR1 in the antiviral actions of IFN (9, 21), we have attempted to define the elements responsible for ADAR1 transcriptional control. As a step toward this goal, we isolated and characterized genomic clones of the human ADAR1 gene (14, 15). The ADAR1 gene spans about 30 kilobases and consists of 15 exons (14). Characterization of the 5′-flanking region immediately adjacent to the exon 1 that contains Met1 of the ADAR1 cDNA (herein designated as exon 1A) led to the identification of an IFN-inducible ADAR1 promoter (PI) that possesses an IFN-stimulated response element (ISRE) responsible for IFN inducibility (16). We now have identified an alternative exon 1 structure of the human ADAR transcript, designated as exon 1B, that initiates from a second promoter not regulated by IFN, designated PC. These results reveal a surprisingly complex organization of the ADAR1 gene and provide insights regarding the regulation of expression of ADAR1 in human cells.
EXPERIMENTAL PROCEDURES
ADAR1 Promoter Cloning.
Two kinds of libraries, a λ-phage library and a P1-phage library, were screened to obtain genomic clones of human ADAR1 (14). Clone λ176 was isolated from a genomic library in the λ phage vector EMBL-3 SP6/T7 prepared from human placenta DNA (CLONTECH); clones P1–249, P1–652, and P1–959 were isolated from a genomic library in the P1 phage vector pAD10SacBII vector prepared from human foreskin fibroblast DNA (22). Genomic inserts were characterized by restriction mapping and Southern blot analysis (14, 23, 24). Restriction fragments of genomic clones were subcloned into the pBluescript SK plasmid (Stratagene) for detailed restriction mapping and DNA sequencing.
Southern Gel-Blot Analysis.
Southern gel-blot analysis (25) of λ- and P1-genomic clone DNA were performed as previously described (9, 14).
Sequence Analysis.
Plasmid subclones were sequenced by the Sanger dideoxynucleotide procedure (26) by using t7 sequenase version 2.0 and protocols from United States Biochemical. Sequences were analyzed by using the University of Wisconsin Genetics Computer Group programs on a Silicon Graphics (Mountain View, CA) IRIS 4D/340VGX computer.
Determination of the 5′-cDNA Region.
The 5′-region of the ADAR1 cDNA (GenBank accession no. U18121) was obtained by the 5′-rapid amplification of cDNA ends (RACE) procedure (27) by using the Marathon–Ready cDNA system (CLONTECH) according to the manufacturer’s recommendations. An uncloned library of adaptor-ligated cDNA prepared from human placenta was used to amplify the 5′-end of the ADAR1 cDNA. First-round PCR was performed with an ADAR1 exon 2 minus-sense primer (E2 minus 273) corresponding to nucleotides 290–273 (5′-TGACTTCCGAGATGCACG-3′) of exon 2, numbered from the 5′-end of exon 2 as nucleotide 1 (14), and the plus-sense anchor primer AP1 supplied by CLONTECH. Nested PCR then was performed with the CLONTECH plus primer AP2 and either the original minus primer or the new cDNA-specific minus (E2 minus 48) primer (5′-GGTATCTGAGCTGTCTGTGC-3′) corresponding to ADAR1 antisense nucleotides 67 to 48 of exon 2 (14, 15). The 5′-RACE cDNA products were cloned into the pBluescript SK plasmid, sequenced, and the results compared with the sequences obtained for the genomic clones.
Reverse Transcription–PCR (RT-PCR) Analysis.
RT-PCR was performed essentially as described (28, 29). RNA isolated from U cells either untreated or IFN-treated by using the Tri-Reagent (Molecular Research Center, Cincinnati) according to the manufacturer’s recommendation was reverse transcribed by using random hexamer oligonucleotides as the primer and Moloney murine leukemia virus reverse transcriptase (New England Biolabs) at 37°C. PCR reactions (29) were performed by using native Taq DNA polymerase (Fisher). The random-primed U cell cDNA was amplified with the following two primer pairs: exon 1A plus 170 (nucleotides 170–193), 5′-AATGCCTCGCGGGCGCAATGAATC-3′ and exon 2 minus 273 (nucleotides 290–273); exon 1B plus 12 (nucleotides 12–29), 5′-GAGAAGGCTACGTGGTGG-3′, and exon 2 minus 273 (nucleotides 290–273). Exon 1A and 1B numbering is as in Fig. 1.
Figure 1.
Nucleotide sequence of the ADAR1 5′-RACE-derived cDNAs and comparison to corresponding genomic DNA sequences. Sequences of the two unique 5′-RACE-derived cDNAs are designated 5′-RACE-1A and 5′-RACE-1B. The previously described exon 1 (14) corresponds to the 5′-RACE-1A sequence, with the ATG translation initiation codon in bold font. The newly identified exon 1 corresponds to the 5′-RACE-1B sequence. The bold sequence shown by the lower lines corresponds to the 5′ portion of exon 2; the position of the exon 2 nested minus primer at nucleotides 48 to 67 is indicated by the underlined sequence. Nucleotide numbering begins with 1 at the 5′ end of each exon. Genomic DNA sequence was obtained from a subclone of the λ176 genomic clone for exon 1A and exon 2, and from a subclone of the P1–249 genomic clone for exon 1B (see Fig. 2B).
Northern Gel-Blot Analysis.
Northern gel-blot analysis of total RNA isolated from human amnion U cells was performed as described previously (9, 13, 16).
Construction of Reporter Gene Plasmids.
The pCAT-Basic promoter-less plasmid (Promega) containing the chloramphenicol acetyltransferase (CAT) gene was used for construction of the reporter gene plasmids for analysis of the ADAR1 gene promoter function following standard cloning procedures (23) as previously described (16, 24, 30). Deletions (Fig. 5) were subsequently made from the parent 5.8-kb BamHI- BamHI plasmid by using appropriate restriction enzymes; the structures of ADAR1 promoter deletion constructions were confirmed by restriction analysis and by sequencing. The pCAT-0.6-kb PI S/X plasmid was the previously described 591-bp Sac/Xho construct (16).
Figure 5.
Identification of a constitutively active ADAR1 gene promoter ≈9-kb upstream of the interferon-inducible promoter. Genomic DNA restriction fragments derived from the 5.8-kb BamHI fragment (Fig. 2) that includes exon 1B were inserted into the promoter-less pCAT-Basic plasmid. Promoter activities observed in human U cells transfected with the indicated CAT reporter plasmids are shown as percent of the conversion of [14C]chloramphenicol to the acetylated derivatives. Open bars refer to cells left untreated, and hatched bars refer to cells treated with IFN-α. pCAT-Control, the CAT reporter gene linked to the simian virus 40 promoter and enhancer; pCAT-Basic, the promoter-less plasmid vector without inserted human genomic DNA; pCAT-0.6Pi (S/X), the IFN-inducible PI promoter present within the 591-bp Sac-Xho genomic DNA fragment inserted into the CAT-Basic plasmid (16).
Cell Maintenance and Interferon Treatment.
Human amnion U cells were maintained as previously described (13, 30). IFN-α treatment was with 1,000 units per ml by using Sendai virus-induced leukocyte IFN generously provided by K. Cantell (Helsinki, Finland) or recombinant IFN-αA/D (PBL Biomedical Laboratories, New Brunswick). Parallel cultures were left untreated as controls.
Transfection and Reporter Assays.
U cells were transfected by the DEAE-dextran-chloroquine phosphate transfection method by using 10 μg of the CAT reporter gene construct and 5 μg of the internal reference plasmid pRSV2-βgal as previously described (16, 30, 31). Treatment with IFN-α, cell harvest, extract preparation, and CAT and β-galactosidase assays were performed as previously described (16, 30). CAT activity was quantified after thin-layer chromatography by use of a Bio-Rad GS-525 molecular imager system. CAT activity values, normalized by β-galactosidase activity to control for variation in transfection efficiency, were calculated as percent conversion of [14C]chloramphenicol to the 14C-acetylated derivatives. Transfections were repeated three to five times in independent experiments to permit calculation of a mean value and standard deviation.
Materials.
Unless otherwise specified, materials and reagents were as described previously (9, 14–16).
RESULTS
Isolation and Characterization of 5′-RACE cDNA Clones of Human ADAR1.
To obtain additional 5′-UTR sequence for human ADAR1, 5′-RACE was carried out with nested primers and an adaptor-ligated cDNA library prepared from human placenta. The ADAR1 minus primer corresponding to antisense nucleotides 290 to 273 of exon 2 (E2 minus 273) and the plus AP1 primer were first used, followed by the nested primer pair ADAR1 antisense nucleotides 67 to 48 of exon 2 (E2 minus 48) or E2 minus 273 and the plus AP2 primer. The 5′-RACE cDNAs were subcloned and sequenced. Unexpectedly, two kinds of sequences were obtained for the 5′-RACE cDNA clones that diverged from each other exactly at the exon 1–exon 2 junction (Fig. 1). In addition to the 5′-RACE-1A sequence that matches exactly the previously identified exon 1 (herein designated as exon 1A) sequence obtained for cDNA prepared from human kidney and human U cells (14, 16), a second 5′-RACE cDNA sequence was obtained. This 5′-RACE cDNA sequence was designated as exon 1B; the longest 5′-RACE-1B cDNA clone extended 107 nucleotides upstream of the exon 2 junction (Fig. 1).
Physical Map Position of Exon 1B.
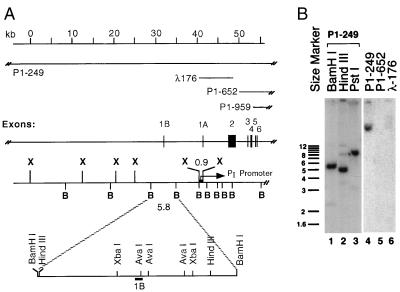
Overlapping phage λ and P1 clones containing the human ADAR1 gene were characterized by restriction mapping and Southern blot analysis to map the position of exon 1B (14). A composite map of the ADAR1 gene was determined (Fig. 2A). The newly identified exon 1B fragment hybridized to a 5.8-kb BamHI fragment present within the P1–249 genomic clone but not in the P1–652 or λ176 genomic clones (Fig. 2B). The precise exon 1B-intron junction was determined by sequencing plasmid subclones. The 210-nt exon 1A was previously shown to be separated from exon 2 by a 5.4-kb intron (14, 32). Exon 1B was positioned ≈9-kb upstream of exon 1A. The 5′-end cDNA sequences of ADAR1 obtained from the 5′-RACE-1A and -1B clones isolated from the human placenta library corresponded exactly to the genomic sequence obtained from human placenta and human foreskin fibroblast DNA subclones (Fig. 1).
Figure 2.
Genomic organization of the human ADAR1 gene. (A) Organization of the exons and introns within the 5′ region of the ADAR1 gene. The 5′-most seven exons are indicated to scale by filled boxes numbered 1B to 6; introns and the 5′ flanking regions are indicated by the solid lines. PI corresponds to the IFN-inducible promoter upstream of exon 1A. The entire gene spans approximately 39 kb in length and contains 16 exons including exon 1B. Shown to scale are the P1-phage genomic clone P1–249, the overlapping λ-phage genomic clone λ176, and the 5′-portion of the overlapping P1 clones P1–652 and P1–959. The restriction map shows cleavage sites for BamHI (B) and XhoI (X) spanning the ≈60-kb region upstream of exon 6. For the 5.8-kb BamHI fragment that includes exon 1B, cleavage sites for AvaI, HindIII, and XbaI are included; no PstI sites are present in the fragment. (B) Southern gel-blot of P1- and λ-phage genomic clones, either uncut or digested with BamHI, HindIII, or PstI as indicated. The probe was 32P-labeled exon 1B.
Effect of Interferon Treatment on the Expression of Exon 1A- and Exon 1B-Containing ADAR1 Transcripts.
The effect of IFN-α treatment on the expression of ADAR1 mRNA species was examined by quantitative RT-PCR to distinguish between exon 1A- and exon 1B-containing transcripts. cDNAs prepared from U cell RNA preparations were used as templates for amplification by PCR, either with the E1B plus 12 and E2 minus 273 primer pair for detection of transcripts possessing the exon 1B–exon 2 junction or with the E1A plus 170 and E2 minus 273 primer pair for detection of the transcripts with the exon 1A–exon 2 junction (Fig. 3A). Amplification of templates was determined after increasing cycle number, ranging from 10 to 35, by directly measuring incorporation of 32P-labeled primer into reaction products that were resolved by electrophoresis (Fig. 3B). Comparable amounts of the exon 1B–exon 2 product were detectable by PCR with samples prepared from untreated and IFN-α-treated cells as the amplification cycle number was increased (Fig. 3B Top). By contrast, the amount of exon 1A–exon 2 specific product obtained was very low with template cDNA prepared with RNA from untreated relative to IFN-treated human U cells (Fig. 3B Middle). As a control, when the same cDNA preparations were examined by using β-actin specific primers, the amount of actin product obtained was similar for samples prepared from untreated as compared with IFN-α treated cells (Fig. 3B Bottom). The observed sizes of the ADAR1 and β-actin products were consistent with sizes predicted from their cDNA sequences.
Figure 3.
Quantitative RT-PCR analysis of ADAR1 mRNA expression in human amnion U cells. RNA isolated from monolayer cultures of U cells either left untreated or treated with IFN-α for 24 h was analyzed by RT-PCR as described in Experimental Procedures. (A) Schematic showing the locations of the alternative exons 1A and 1B and exon 2, with exons denoted by boxes and introns by solid lines; the exon-specific oligomer primers are denoted by arrows. (B) PCR products fractionated by gel electrophoresis after the indicated number of amplification cycles. cDNA templates were prepared by using RNA prepared from either IFN-α treated or untreated U cells. PCR reactions contained 0.25 μM of primer, by using the primer pairs E1B plus 12 and 32P-labeled E2 minus 273, E1A plus 170 and 32P-labeled E2 minus 273, or β-actin plus and minus primers as indicated.
Northern gel-blot analyses with hybridization probes specific to either exon 1A or exon 1B were performed to estimate the size of the corresponding transcripts. Both the exon 1A and exon 1B probes detected a single major transcript of about ≈6.7-kb with RNA isolated from human U cells (Fig. 4). The transcript detected with the exon-1A probe was IFN-inducible as predicted (Fig. 3B; 14, 16). By contrast, ≈6.7-kb transcript detected with the exon 1-B probe was present in comparable amounts in untreated and interferon-treated cells. The integrity and amounts of β-actin RNA and 18S and 28S ribosomal RNAs were comparable between the RNA samples prepared from untreated and IFN-α-treated cells (data not shown).
Figure 4.
Northern gel-blot analysis of ADAR1 mRNA expression in human amnion U cells. Northern gel-blot analysis of RNA from human U cells. Details are as described in Materials and Methods. The probe was 32P-labeled exon 1A-exon 2 junction (16) or 32P-labeled exon 1B. The positions of the two ribosomal RNAs are indicated on the sides of the blots. Lanes 1 and 3–5, RNA from untreated cells; lanes 2 and 6–8, RNA from cells treated with IFN-α for 24 h. Samples of 6 μg (lanes 1–3, 6), 12 μg (lanes 4, 7), and 24 μg (lanes 5, 8) of total RNA were analyzed.
Identification of a Constitutively Active Promoter Flanking Exon 1B Unique from the Interferon-Inducible Promoter Flanking Exon 1A.
To examine whether the 5′-flanking region of exon 1B was capable of functioning as a promoter, restriction fragments derived from the 5.8-kb BamHI genomic DNA fragment were fused upstream of a CAT reporter gene. As shown by the results in Fig. 5, the 2.2-kb X/X reporter construct carrying the 2.2-kb XbaI genomic fragment fused upstream of the CAT gene exhibited significant CAT activity in transfected U cells; the activity was not further increased by IFN treatment. In contrast, the same 2.2-kb XbaI genomic fragment in reverse orientation showed very low CAT activity. Reporter constructs prepared with genomic fragments flanking the 2.2-kb XbaI fragment, either the 1.3-kb X/B or the 2.3-kb B/X fragment, lacked promoter activity. As a negative control, the pCAT-Basic promoter-less plasmid without inserted human genomic DNA exhibited insignificant CAT activity (<2% conversion). As a positive control, the pCAT-Control (Promega) plasmid containing the simian virus 40 promoter and enhancer displayed high CAT activity levels.
A series of deletions of the 2.2-kb XbaI construct was generated by using internal AvaI restriction sites and was analyzed for CAT reporter expression. The internal 0.9-kb AvaI fragment, tested as the 0.9-kb A/A construct, lacked promoter activity. However, the 1.3-kb X/X (A/A del) construct obtained by deletion of the 0.9-kb AvaI fragment showed strong reporter activity. The activity of the 1.3-kb X/X (A/A del) construct was comparable in the absence and presence of IFN treatment. The 1.05-kb X/X (A/A del) plasmid, generated by further deletion of a 0.25-kb AvaI fragment from the 1.3-kb X/X (A/A del) construct, showed greatly reduced activity. Although the 1.3-kb X/X (A/A del) construct was not IFN-inducible, the pCAT-0.6-kb PI S/X reference plasmid that contains the 591-bp Sac/Xho genomic fragment flanking exon 1A into pCAT-Basic (16) showed promoter activity that was increased 4- to 5-fold by IFN treatment (Fig. 5). The pCAT-0.6-kb PI S/X construct includes an ISRE element (16). Finally, neither pCAT-Basic nor pCAT-Control showed IFN inducibility of the CAT reporter (data not shown).
DNA Sequence of the ADAR1 Constitutive Promoter Region.
The genomic DNA flanking exon 1B that possessed the necessary functional elements to support constitutive transcription was sequenced; 395 nucleotides of the sequence are shown in Fig. 6. Comparison of the sequence obtained for the ADAR1 P1–249 genomic DNA subclone to that for the 5′-RACE-1B cDNA revealed an exact match over the 107-nt region that constitutes exon 1B (Figs. 1 and 6). Intron 1B conformed to the GT–AG rule. Computer analysis of the promoter region sequence revealed three canonical CAAT boxes in the immediate vicinity of the transcription initiation sites determined by 5′-RACE and primer extension analyses. A consensus initiator positioning sequence Inr and a consensus TATA box were also present, but not at the standard positions (34, 37). Additional motifs identified (35) in the flanking region include PEA3 of the Ets-1 family, AP-1, AP-2, MRE, Cap-box, purine box, and cAMP CRE-like motif (Fig. 6). The promoter region upstream of exon 1B lacked elements corresponding to the interferon responsive ISRE and GAS elements, and the KCS element (30, 31); the interferon-inducible promoter upstream of exon 1A includes an ISRE- and KCS-like element (16).
Figure 6.
Genomic DNA sequence of the promoter Pc region of the human ADAR1 gene. The sequence of the promoter region as well as exon 1B and the 5′-sequence of intron 1B are shown. The nucleotide numbers of the genomic DNA sequence upstream of the AvaI site present in exon 1B are relative. The sequence designated as exon 1B corresponds to nucleotides 1 to 107 of the cDNA as defined by 5′-RACE and shown in Fig. 1. Potential transcription factor binding sites as described in the text are shown.
DISCUSSION
Two important points emerge from our results reported herein on the transcription-control regions of the ADAR1 gene encoding the RNA-specific adenosine deaminase. First, human ADAR1 transcripts possess alternative exon 1 structures that predict the synthesis of two differently sized forms of ADAR1, one 1,226 amino acids and the other 931 amino acids. Second, the alternative exon 1A and exon 1B structures of the ADAR1 transcripts initiate from different promoters, one interferon inducible (PI) and the other constitutively active (PC). These findings now provide a molecular explanation for the earlier observation that two forms of ADAR1 protein occur in mammalian cells, one inducible by interferon and the other constitutively expressed (9, 14). The schematic diagram shown in Fig. 7A provides a summary of the relative organization of the 5′-region of the human ADAR1 gene. The ADAR1 gene structure was previously determined to include 15 exons that span ≈30-kb (14) on chromosome 1q (15). With the identification of the alternative exon 1B positioned about ≈9 kb upstream of the previously identified exon 1, now designated exon 1A, the size of the human ADAR1 gene increases to ≈39 kb and spans 16 exons.
Figure 7.
Schematic summary of the organization of the human ADAR1 gene possessing alternative exon 1 structures of transcripts that initiate from the Pc and PI promoters. (A) Organization of the promoter region of the ADAR1 gene. Pc, the constitutively active promoter that initiates transcription with exon 1B; PI, the interferon-inducible promoter that initiates transcription with exon 1A. (B) Schematic representation of the ORFs of the ADAR1 transcripts. Transcripts with the alternative exon 1A possess a 1,226 amino acid-deduced ORF that begins at AUG1 (methionine 1) present in exon 1A and ends at an UAG in exon 15 (16). Transcripts with the alternative exon 1B possess a 931 amino acid-deduced ORF that begins at AUG296 (methionine 2) present in the unusually large (1,586 nt) exon 2 and terminate at the UAG in exon 15. R, dsRNA-binding domain.
Human ADAR1 gene expression is complex. At least two mature similarly sized ≈6.7-kb ADAR1 mRNAs are produced from the ADAR1 gene that can be distinguished from each other by the nature of the exon 1 at their 5′-ends and by their IFN inducibility. Although the level of ADAR1 expression is increased by IFN treatment (9, 17), significant basal amounts of ADAR1 transcripts are routinely observed in human cell lines and organs in the absence of IFN treatment (9–11). A single major ≈6.7-kb ADAR1 mRNA is detectable in human cell lines in the absence of exogenous cytokine treatment as well as in various organs with hybridization probes corresponding to different regions of the ADAR1 cDNA (9–11, 16). Our findings establish that the basal level of mature ADAR1 RNA corresponds at least in part to the newly identified exon 1B-containing transcripts that initiate from the constitutively active PC promoter (Figs. 3 and 4). By contrast, the increased level of the similarly sized ≈6.7-kb ADAR1 RNA observed in IFN-treated cells corresponds to the exon 1A-containing transcripts that initiate from the IFN-inducible PI promoter (Figs. 3 and 4). The PI promoter of ADAR1 possesses ISRE- and KCS-like elements associated with IFN responsiveness (16).
ADAR1 transcripts containing exon 1A possess an ORF of 1,226 amino acids (Fig. 7B Upper). This ORF sequence is in agreement with the previously characterized human ADAR1 cDNA clones that were isolated in screens for IFN-regulated cDNAs (11, 22). Interestingly, the sequence of the exon 1A form of ADAR1 was also obtained by screening cDNA libraries prepared by using RNA from human natural killer cells (10) or HeLa cells (11) not known to be treated exogenously with IFN, possibly indicating PI promoter activity in response to autocrine IFN action or some other inducer present under the culture conditions preceding RNA isolation. Exon 1A includes the initiator methionine codon, AUG1, whose flanking −3 and +4 nucleotides GcaAUGA are purines, characteristic of a strong translational start codon (36). The next methionine in the 1,226 amino acid ORF is nearly 300 amino acid residues downstream, AUG296, and occurs within the unusually large 1,586 nt exon 2 (14). The newly identified exon 1B lacks AUG codons and when spliced to exon 2, a longer 5′-untranslated region is predicted than with exon 1A-containing transcripts. The long ORF of the ADAR1 transcripts possessing exon 1B linked to exon 2 is predicted to begin at AUG296 of reading frame 1; only two methionine codons are present in the ≈1 kb of sequence upstream of AUG296, and both are in frame 2 and predict short ORFs, 24 and 31 residues. The nucleotides flanking AUG296 at −3 and +4, GagAUGG, predict a strong initiation codon (36) beginning the 931 amino acid ORF (Fig. 7B Lower). These findings provide an explanation for the differently sized forms of ADAR1 protein that have been detected by Western immunoblot analysis or purified from a variety of sources (2, 8, 9, 14).
Alternative splicing is an important strategy for generating multiple protein isoforms possessing different biological activities from a single genomic locus (37). Our results establish that alternative ADAR1 gene promoters, one constitutively active and the other interferon inducible, create alternative exon 1 structures on the 15 exon ADAR1 transcripts that consequently possess different 5′-untranslated regions and different ORFs, one of 1,226 residues and the other of 931 residues in the same frame. These findings suggest that the differences in apparent molecular weight observed for ADAR1 proteins are caused at least in part by the origin of exon 1 rather than by differential proteolysis of the larger ≈150-kDa protein to yield the ≈110-kDa protein. Consistent with these conclusions, antisera prepared against three nonoverlapping regions of the ADAR1 cDNA expressed in Escherichia coli including the N-terminal, central, and C-terminal regions of the 1,226 residue ORF all detect the IFN-inducible ≈150-kDa protein, whereas the constitutively expressed ≈110-kDa protein is detected only by the antisera prepared against the central and C-terminal regions but not the N-terminal region of the ORF (9). Although the alternative 5′-UTR may conceivably affect ADAR1 mRNA stability or translational activity, the C-terminal 931 residues of the 1,226 residue ORF includes the three dsRNA-binding domain motifs and the catalytic region. However, the shortened 931 amino acid ADAR1 protein lacks the Z-DNA binding domain Zα (33). The biological significance of this deletion of Z-DNA binding activity is unknown.
It is now apparent from the results described herein that the ADAR1 RNA-specific adenosine deaminases constitute a family of enzymes specified by alternative exon 1 splicing of transcripts initiated from at least two promoters. In eukaryotes, alternative promoters are known to be associated with gene expression that is developmentally regulated or tissue specific (37). This is exemplified by the Pax genes, regulators in the developing nervous system (38, 39), and by the Apobec1 gene encoding the apolipoprotein B RNA-editing protein (40). Cytokines including interferons modulate the expression of a number of cellular genes; both the basal level of expression and the fold induction after cytokine treatment vary markedly (19). It is conceivable that alternative promoters may be used by other cytokine-regulated genes in addition to ADAR1. For ADAR1, it is now of utmost importance to define the tissue-specific utilization of the PC- constitutive and PI-inducible promoters that drive transcription of the ADAR1 gene and the possible effect of cytokines other than interferons as well as growth factors on their activities in uninfected and virus-infected cells and tissues.
Acknowledgments
This work was supported in part by Research Grant AI-12520 from the National Institute of Allergy and Infectious Diseases, U.S. Public Health Service.
Footnotes
Abbreviations. ADAR1, the RNA-specific adenosine deaminase; dsRNA, double-stranded RNA; UTR, untranslated region; ISRE, IFN-stimulated response element; RACE, rapid amplification of cDNA ends; RT-PCR, reverse transcription—PCR; CAT, chloramphenicol acetyltransferase.
References
- 1.Simpson L, Emeson R B. Annu Rev Neurosci. 1996;19:27–52. doi: 10.1146/annurev.ne.19.030196.000331. [DOI] [PubMed] [Google Scholar]
- 2.Rueter S M, Emeson R B. Modifications and Editing of RNA. Washington, D.C.: Am. Soc. Microbiol.; 1998. pp. 343–361. [Google Scholar]
- 3.Seeburg P H, Higuchi M, Sprengel R. Brain Res Rev. 1998;26:217–229. doi: 10.1016/s0165-0173(97)00062-3. [DOI] [PubMed] [Google Scholar]
- 4.Bass B L. Trends Biochem Sci. 1997;22:157–162. doi: 10.1016/s0968-0004(97)01035-9. [DOI] [PubMed] [Google Scholar]
- 5.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Nature (London) 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 6.Cattaneo R. Curr Opin Genet Dev. 1994;4:895–900. doi: 10.1016/0959-437x(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 7.Kumar M, Carmichael G G. Proc Natl Acad Sci USA. 1997;94:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bass B L, Nishikura K, Keller W, Seeburg P H, Emeson R B, O’Connell M A, Samuel C E, Herbert A. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson J B, Samuel C E. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connell M A, Krause S, Higuchi M, Hsuan J J, Totty N F, Jenny A, Keller W. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herbert A, Alfken J, Kim Y-G, Mian I S, Nishikura K, Rich A. Proc Natl Acad Sci USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson J B, Thomis D C, Hans S L, Samuel C E. Virology. 1995;210:508–511. doi: 10.1006/viro.1995.1370. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, George C X, Patterson J B, Samuel C E. J Biol Chem. 1997;272:4419–4428. doi: 10.1074/jbc.272.7.4419. [DOI] [PubMed] [Google Scholar]
- 15.Weier H-U G, George C X, Greulich K M, Samuel C E. Genomics. 1995;30:372–375. doi: 10.1006/geno.1995.0034. [DOI] [PubMed] [Google Scholar]
- 16.George C X, Samuel C E. Gene. 1999;229:203–213. doi: 10.1016/s0378-1119(99)00017-7. [DOI] [PubMed] [Google Scholar]
- 17.Wathelet M G, Szpirer J, Nols C B, Clauss I M, De Wit L, Islam M Q, Levan G, Horisberger M A, Content J, Szpirer C, et al. Somatic Cell Mol Genet. 1988;14:415–426. doi: 10.1007/BF01534709. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Samuel C E. J Biol Chem. 1999;274:5070–5077. doi: 10.1074/jbc.274.8.5070. [DOI] [PubMed] [Google Scholar]
- 19.Der S D, Zhou A, Williams B R G, Silverman R H. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner R W, Yoo C, Wrabetz L, Kamholz J, Buchhalter J, Hassan N F, Khalili K, Kim S U, Perussia B, McMorris F A, et al. Mol Cell Biol. 1990;10:5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scadden A D J, Smith C W J. EMBO J. 1997;16:2140–2149. doi: 10.1093/emboj/16.8.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shepherd N S, Pfrogner B D, Coulby J N, Ackerman S L, Vaidyanathan G, Sauer R H, Balkenhol T C, Sternberg N. Proc Natl Acad Sci USA. 1994;91:2629–2633. doi: 10.1073/pnas.91.7.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning, a Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Tanaka H, Samuel C E. Proc Natl Acad Sci USA. 1994;91:7995–7999. doi: 10.1073/pnas.91.17.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frohman M A, Dush M, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomis D C, Floyd-Smith G, Samuel C E. J Biol Chem. 1992;267:10723–10728. [PubMed] [Google Scholar]
- 29.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 30.Kuhen K L, Samuel C E. Virology. 1997;227:119–130. doi: 10.1006/viro.1996.8306. [DOI] [PubMed] [Google Scholar]
- 31.Kuhen K L, Vessey J W, Samuel C E. J Virol. 1998;72:9934–9939. doi: 10.1128/jvi.72.12.9934-9939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Zeng Y, Murray J M, Nishikura K. J Mol Biol. 1995;254:184–195. doi: 10.1006/jmbi.1995.0610. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Herbert A, Rich A, Samuel C E. Methods Companion Methods Enzymol. 1998;15:199–205. doi: 10.1006/meth.1998.0624. [DOI] [PubMed] [Google Scholar]
- 34.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faisst S, Meyer S. Nucleic Acids Res. 1992;20:3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lodish H, Baltimore D, Berk A, Zipursky S L, Matsudaira P, Darnell J. Molecular Cell Biology. 3rd Ed. New York: Freeman; 1995. [Google Scholar]
- 38.Okladnova O, Syagailo Y V, Mossner R, Riederer P, Lesch I-P. Mol Brain Res. 1998;60:177–192. doi: 10.1016/s0169-328x(98)00167-3. [DOI] [PubMed] [Google Scholar]
- 39.Chalepakis G, Stoykova A, Wijnholds J, Tremblay P, Gruss P. J Neurobiol. 1993;24:1367–1384. doi: 10.1002/neu.480241009. [DOI] [PubMed] [Google Scholar]
- 40.Nakamuta M, Oka K, Krushkal J, Kobayashi K, Yamamoto M, Li W-H, Chan L. J Biol Chem. 1995;270:13042–13056. doi: 10.1074/jbc.270.22.13042. [DOI] [PubMed] [Google Scholar]