Abstract
We have identified two vesicular proteins, designated evectin (evt)-1 and -2. These proteins are ≈25 kDa in molecular mass, lack a cleaved N-terminal signal sequence, and appear to be inserted into membranes through a C-terminal hydrophobic anchor. They also carry a pleckstrin homology domain at their N termini, which potentially couples them to signal transduction pathways that result in the production of lipid second messengers. evt-1 is specific to the nervous system, where it is expressed in photoreceptors and myelinating glia, polarized cell types in which plasma membrane biosynthesis is prodigious and regulated; in contrast, evt-2 is widely expressed in both neural and nonneural tissues. In photoreceptors, evt-1 localizes to rhodopsin-bearing membranes of the post-Golgi, an important transport compartment for which specific molecular markers have heretofore been lacking. The structure and subcellular distribution of evt-1 strongly implicate this protein as a mediator of post-Golgi trafficking in cells that produce large membrane-rich organelles. Its restricted cellular distribution and genetic locus make it a candidate gene for the inherited human retinopathy autosomal dominant familial exudative vitreoretinopathy and suggest that it also may be a susceptibility gene for multiple sclerosis.
Many cells regulate the production of plasma membrane, the secretion of proteins, or the localization of lipids and proteins in response to environmental stimuli (1–4). Among the most dramatic examples of this form of regulation are those seen during neural development. A differentiating neuron, for example, may organize membrane biosynthesis and protein localization such that its growing axon extends over great distances toward the source of a chemoattractant (5). Similarly, oligodendrocytes of the vertebrate brain elaborate extraordinary quantities of specialized membrane (≈5,000 μm2 per day) in response to the cues that trigger their myelination of axons (6), and vertebrate photoreceptors achieve similar rates of membrane biosynthesis in the course of assembling and maintaining their outer segments (7). Current interest notwithstanding, the identification of molecules that may link extracellular signals to the dramatic changes in membrane biosynthesis and organization that underlie these developmental events remains a largely unrealized goal.
Membrane-associated proteins and lipids are sorted in the trans-Golgi network (TGN), a tubular assembly located adjacent to the cisternae of the trans-Golgi, and in a set of downstream membrane compartments known as the post-Golgi (8). The TGN serves as a central sorting station in which proteins and lipids destined for distinct subcellular regions are segregated into post-Golgi carriers. This sorting relies in part on cargo-intrinsic signals for basolateral versus apical routing, but the manner in which the activation of a cell surface receptor may be coupled to these cargo-intrinsic routing signals is also unknown.
We have identified two unusual proteins whose properties suggest that they may be coupling factors between extracellular signals and intracellular membrane biosynthesis and trafficking. These proteins carry a pleckstrin homology domain (PHD), a module that typically binds signaling phospholipids that are generated consequent to receptor activation; they have structural features characteristic of proteins involved in vesicular routing and fusion; they are apparently capable of shuttling between soluble and membrane-associated forms; and, in photoreceptors, they specifically associate with membranes of the post-Golgi compartment.
METHODS
Cloning of Evectins (evts).
A partial cDNA clone for evt-1 was isolated in a yeast one-hybrid screen designed to discover regulatory proteins that interact with the transcription factor SCIP. A GAL4 activation domain tagged cDNA library from forskolin-stimulated, cultured rat Schwann cells (9) was constructed in pGAD424 and was screened by using standard procedures (Yeast Protocols Handbook, CLONTECH). The partial evt-1 cDNA clone proved to be a false positive with respect to SCIP interaction. Full-length rat evt-1 cDNAs were obtained from a λZap Schwann cell cDNA library (10) by using the one-hybrid cDNA fragments as probes. Murine and human evt-1 cDNAs and murine evt-2 cDNAs were obtained as expressed sequence tag (EST) clones from the American Type Culture Collection.
Chromosomal Mapping.
Overlapping EST sequences for evt-1 and evt-2 were assembled and assigned to an EST cluster by using the EST-Assembler at TIGEMnet [http://gcg.tigem.it (11)]. The EST clusters were mapped by the EST Mapping Consortium by using radiation hybrids; see the Human Transcript Map database at http://www.ncbi.nlm.nih.gov.
Antibodies.
A rabbit antiserum was raised against a synthetic peptide derived from the α-helical, C-terminal region of the evt-1 PHD (residues 93–104) by using standard procedures (12). The evt-1 antiserum was affinity-purified against a glutathione S-transferase–evt-1 fusion protein containing residues 1–111 by using the Quickpure kit (Sterogen, Arcadia, CA).
Northern Blot Analyses.
Total cellular RNAs were isolated and analyzed by RNA blot as described (10). For evt-1 probes, we used a 500-bp ScaI-XhoI fragment from the 3′ end of the rat evt-1 cDNA clone. For evt-2 probes, we used a 1,100-bp PstI fragment from the murine EST clone W99864. Equal loading of lanes was assessed by methylene blue staining of 18S and 28S rRNAs.
Immunoblotting.
Protein samples were resuspended in loading buffer (3% β-mercaptoethanol/3% SDS/10% glycerol/1M urea/60 mM Tris, pH 6.8/0.1% Triton X-100/0.005% bromphenol blue). For Western blot analyses, proteins were separated by 12.5% SDS/PAGE (with 10 mM β-mercaptoethanol in the resolving gel), were transferred onto nitrocellulose filters, and were incubated with primary antibody for 1 hr. Immunoreactivity was detected by using the ECL method (Amersham).
In Situ Hybridization.
Perfused tissues and organs were sectioned at 7–10 μm. 35S/33P in situ hybridization was carried out as described (13). Sense and antisense riboprobes for evt-1 and evt-2 were in vitro transcribed (Maxiscript, Ambion, Austin, TX). The evt-1 riboprobe was prepared from the 3′-most 500 bp of the rat cDNA. The evt-2 riboprobe was generated from the 5′-most 300 bp of the murine EST clone W99864, and the 700-bp PLP riboprobe was made from the 3′ end of the proteolipid protein cDNA.
Subcellular Fractionation of Frog Retinae.
Southern leopard frogs, Rana berlandieri, were dark-adapted for 2 hr before experiments. Isolated retinae were incubated with [35S]express protein labeling mixture (25 μCi/retina) at 22°C for 1 hr followed by 2 hr of cold chase. Retinal fractionation, purification of rod outer segments (ROSs) on step sucrose gradients, and preparation of post-nuclear supernatants enriched in photoreceptor biosynthetic membranes were all performed as described (14). The post-nuclear supernatants were fractionated on linear 20–39% (wt/wt) sucrose gradients at 100,000 × gav for 15 hr at 4°C, and 14 fractions were collected from the top of the gradient. Six subcellular fraction pools were created as described (15), were diluted with 10 mM Tris acetate (pH 7.4) and were centrifuged at 336,000 × gav for 30 min. Pellets were resuspended in 10 mM Tris acetate (pH 7.4), and supernatants were precipitated with 10% trichloroacetic acid and were analyzed by SDS/PAGE. Immunoblotting with affinity purified anti-evt-1 (diluted 1: 250) and anti-ADP ribosylation factor (anti-ARF3) (Transduction Laboratories, Lexington, KY; diluted 1: 500) was performed as described (15).
RESULTS
evts Are Vesicular Proteins Containing a Pleckstrin Homology Domain.
cDNAs for the principal protein described below were fortuitously isolated in a yeast one-hybrid screen whose objectives were unrelated to membrane biosynthesis or trafficking. We chose to pursue analysis of the original one-hybrid clones because (i) a Northern blot survey of tissue expression indicated that the mRNA corresponding to these clones was highly and restrictively expressed in myelinating glial cells—oligodendrocytes in the central nervous system (CNS) and Schwann cells in the peripheral nervous system (PNS); and (ii) preliminary sequencing predicted a protein with an intriguing structure. Therefore, we isolated full-length, 1,875-bp cDNAs from a rat Schwann cell library. In light of its structure, subcellular location, and probable bioactivity, we named the protein encoded by these cDNAs evectin-1 (evt-1), from the Latin evectus, meaning “carried” or “moved forward”.
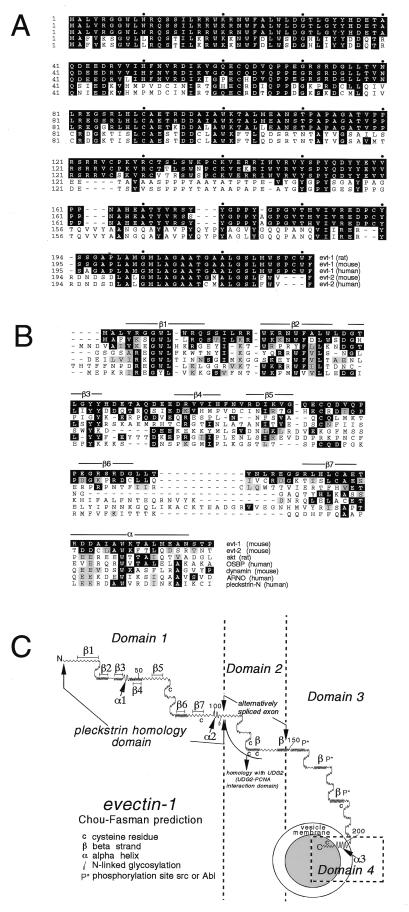
The deduced amino acid sequence of rat evt-1 (Fig. 1A) exhibits no substantial sequence similarity to any known protein. However, blast (National Center for Biotechnology Information) searches yield a variety of human and mouse ESTs for evt-1, and for a second, closely related protein, which we designated evectin-2 (Fig. 1A). evt-1 and -2 display ≈40% amino acid identity overall, although conservation between them is appreciably higher in specific subregions.
Figure 1.
(A) Amino acid sequences of rat, mouse, and human evt-1 and -2. cDNAs for rat evt-1 were cloned as described in the text. All other protein sequences were deduced from ESTs. Amino acid identities with rat evt-1 are indicated in black. (B) Comparison of the evt PHDs to the most closely related PHDs in public databases. The seven β strands and the C-terminal α helix of the evt-1 PHD are indicated. These elements were positioned based on (i) alignment of the evt-1 PHD with PHDs for which x-ray crystal or NMR structures have been determined and (ii) the evt-1 secondary structure prediction displayed in Fig. 1C. Amino acid identities with rat evt-1 are indicated in black, and conservative substitutions are indicated in gray. (C) Predicted secondary structure for rat evt-1, generated by the pepplot and plotstructure programs of the University of Wisconsin Genetics Computer Group (Madison, WI). The seven β strands of the PHD are indicated. Helix α2 corresponds to the conserved α helix that is located at the C-terminal end of all PHDs; helix α1 is conserved in some but not all PHDs. Helix α3 is hydrophobic and is predicted to span a membrane bilayer.
Both evt-1 and -2 have a structure that can be readily subdivided into four domains. Neither protein sequence carries a cleaved, N-terminal signal peptide that would allow for insertion into membranes via a conventional endoplasmic reticulum-to-Golgi routing. Instead, both proteins immediately begin with an ≈110-aa region that exhibits all of the distinctive structural features of PHDs that, first recognized in pleckstrin, have now been identified in a wide variety of proteins (16). The PHD-mediated binding of phosphoinositides often has been found to promote the translocation of these proteins to their sites of action at the plasma membrane or other cellular membranes (17). Although the evt-1 and -2 PHDs are not closely related to those of any previously identified PHD (Fig. 1B), they are nonetheless predicted to fold into the 7 β strand sandwich (β1–β7 in Fig. 1C), closed C-terminally by a conserved α helix (α2 in Fig. 1C), which is characteristic of all solved PHD structures (Fig. 1C). They also carry all of the critically conserved amino acid motifs that are diagnostic of PHDs (Fig. 1B) and exhibit the correct predicted spacing of the conserved features of PHD secondary structure (16).
The evt-1 and -2 PHDs are most closely related to those of (i) the kinase Akt (Fig. 1B), which regulates the apoptotic death pathway (18) and which also has a PHD located at its extreme N terminus; (ii) the oxysterol binding protein, which translocates to the Golgi apparatus on binding oxygenated derivatives of cholesterol (19); (iii) the GTPase dynamin, an essential regulator of vesicular trafficking during clathrin-mediated endocytosis (20); and (iv) ARNO, a guanine nucleotide exchange factor for the ARF family of G proteins (21).
Downstream of the PHD, the evts contain a 35-residue alternatively spliced domain (Domain 2 in Fig. 1C). In evt-1, this domain is similar to the DNA-editing enzyme uracil-DNA glycoylase 2. The amino acids that are identical between evt-1 and uracil-DNA glycoylase 2 are the same residues that are critical for binding of uracil-DNA glycoylase 2 to proliferating cell nuclear antigen (22). Domain 2 is therefore likely to be a protein–protein interaction module. Downstream of Domain 2 is a set of predicted β strands that carry potential phosphorylation sites for src or Abl family kinases (Domain 3 in Fig. 1C). This domain displays no significant similarity to any known protein.
The extreme C termini of the evts (Domain 4, helix α3 in Fig. 1C) are highly conserved hydrophobic α helices (Fig. 1 A and C) that are predicted to span a lipid bilayer. Such C-terminal helices are a general feature of small, membrane-associated proteins that lack cleaved N-terminal signal peptides and allow these proteins to be post-translationally inserted into membranes (23). Proteins that carry a C-terminal membrane anchor include the apoptosis regulator Bcl-2, the polyoma virus middle T antigen, and a large number of the SNARE proteins that mediate vesicle trafficking, docking, and fusion, such as synaptobrevin (VAMP) and the syntaxins of the mammalian nervous system (23). In all cases in which orientation has been closely examined, such proteins are anchored in the membrane such that they are cytoplasmically displayed.
evt-1 Expression Is Specific to the Nervous System.
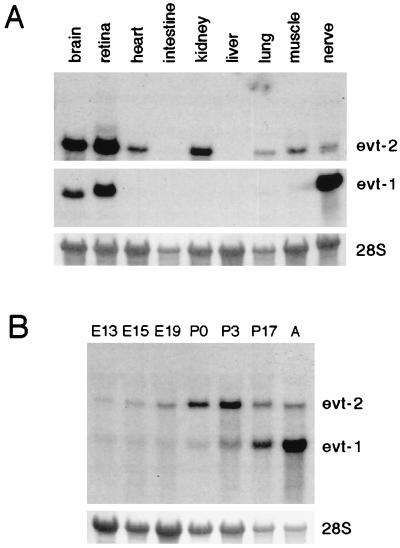
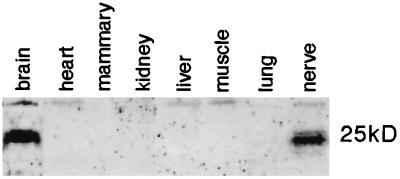
A Northern blot survey of RNAs isolated from adult mammalian tissues revealed a restricted pattern of evt-1 expression. Its 1.9-kilobase mRNA is expressed in rat brain, peripheral nerve, and retina but is undetectable in skeletal muscle, small intestine, spleen, liver, lung, kidney, or heart (Fig. 2A). In both the brain and the sciatic nerve, evt-1 is up-regulated through the first and second postnatal weeks, during the onset of axonal myelination by oligodendrocytes (CNS) and Schwann cells (PNS) (Fig. 2B). In contrast to myelination genes, however, expression of evt-1 mRNA is maintained at relatively high levels in the adult CNS and PNS. An affinity-purified evt-1 antibody specifically recognizes a protein of ≈25 kDa on Western blots in tissues that express the evt-1 mRNA (Fig. 3).
Figure 2.
Expression of evt mRNAs. (A) Northern blot of total RNAs isolated from the indicated adult mouse tissues, probed for evt-1 mRNA [a 1.9-kilobase mRNA (Middle)], and then stripped and reprobed for evt-2 mRNA [a 3.5-kilobase mRNA (Top)]. (Bottom) 28S rRNA levels visualized with methylene blue. (B) A developmental Northern blot of total RNAs isolated from embryonic day (E) 13 head and from E15 and E19, postnatal days (P) 0, P3, and P17, and adult (A) rat brain, simultaneously probed for both evt-1 and evt-2 mRNA. (Bottom) 28S rRNA levels visualized with methylene blue.
Figure 3.
Tissue expression of evt-1. Shown is a Western blot of total proteins isolated from the indicated tissues, from an SDS gel electrophoresed under strong reducing and denaturing conditions (see Methods). Under these conditions, evt-1 runs as a closely spaced doublet at 25 kDa. The presence of the doublet is consistent with differential phosphorylation or other post-translational modification, but these have not yet been investigated.
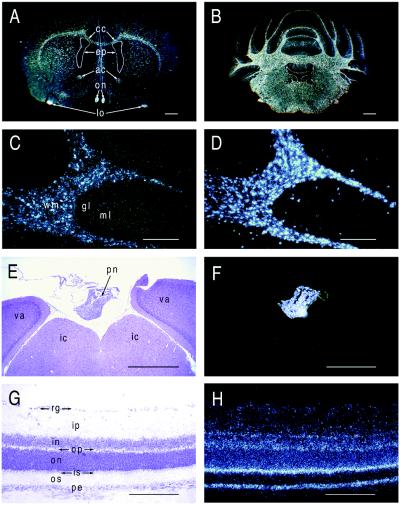
In situ hybridization analyses demonstrate that evt-1 mRNA is prominently expressed in oligodendrocytes, the cells that synthesize CNS myelin (Fig. 4). Every myelinated tract of the adult rat CNS is labeled by hybridization with an evt-1 probe (Fig. 4 A and B). In the cerebellum and elsewhere, the evt-1 signal colocalizes with mRNA for proteolipid protein, the most abundant protein of CNS myelin (Fig. 4 C and D). evt-1 mRNA also is expressed by the myelinating Schwann cells of peripheral nerves (data not shown).
Figure 4.
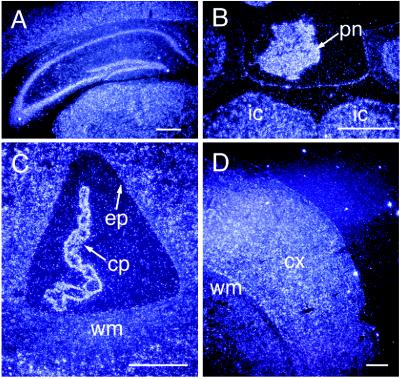
evt-1 expression in the adult CNS. (A) Dark-field in situ hybridization image of evt-1 mRNA in a coronal section of the adult rat brain. cc, corpus callosum; ep, ependyma; ac, anterior commissure; on, optic nerve; lo, lateral olfactory tract. (B) Dark-field image of evt-1 mRNA in a coronal section of the adult rat cerebellum. Labeling is confined to the white matter and ependyma. (C) High power image of evt-1 mRNA in the cerebellum. wm, white matter; gl, granule cell layer; ml, molecular layer. (D) A section adjacent to that in C, hybridized with a probe for proteolipid protein mRNA, a myelin-specific transcript. (E) Bright-field image of the pineal gland (pn), situated above the inferior colliculus (ic) and the visual area of the cortex (va). (F) Dark field image of evt-1 mRNA in a section adjacent to that in E, illustrating strong expression in the pineal. (G) Bright field image of adult rat retinal laminae. rg, retinal ganglion cells; ip, inner plexiform layer; in, inner nuclear layer; op, outer plexiform layer; on, outer nuclear layer; is, inner segment of photoreceptors; os, outer segment of photoreceptors; pe, pigment epithelium. (H) Dark field image of evt-1 mRNA in a section adjacent to that in G, illustrating strong expression in the inner segments of photoreceptors and in the pigment epithelium. [Bars = 1 mm (A and B), 0.4 mm (C and D), 0.8 mm (E and F), and 0.5 mm (G and H).]
In addition to myelinating glia in the CNS and PNS, the evt-1 gene is expressed in only three other CNS regions: the retina, the ependyma, and the pineal. evt-1 mRNA is readily detected in retinal tissue by Northern blot analysis (Fig. 2A) and in the inner segment region of retinal photoreceptors by in situ hybridization, with slightly lower levels in the pigment epithelium (Fig. 4 G and H). High evt-1 mRNA levels are also present in (i) the ependymal cells that line the ventricles of the brain but not in cells of the choroid plexus, with which the ependyma is physically contiguous (Fig. 4A); and (ii) the pineal gland (Fig. 4 E and F).
evt-2 Expression Is Widespread.
In contrast to evt-1, the ≈3.5-kilobase evt-2 mRNA was detected in many of the adult tissues that we examined by Northern blot analysis (Fig. 2A). In further distinction to evt-1, evt-2 mRNA was readily detected in the embryonic brain (Fig. 2B). In situ hybridization analyses for evt-2 mRNA in the adult brain revealed a complementarity of expression with evt-1 in many regions (Fig. 5). Although evt-1 is confined to white matter tracts and is excluded from neuronal populations, there is widespread expression of evt-2 mRNA in neurons throughout the brain, and this mRNA is excluded from the white matter. Similarly, evt-2 mRNA is expressed at high levels in the choroid plexus and not at all in the contiguous layer of ependymal cells. Although this complementarity of expression holds generally, there are obvious exceptions in the pineal, the retina, and peripheral nerve, where both mRNAs are well expressed.
Figure 5.
evt-2 expression in the adult CNS. (A) Dark-field in situ hybridization image of evt-2 mRNA in a coronal section of the adult mouse hippocampus, illustrating widespread neuronal expression in all hippocampal fields and in overlying cortex. (B) Expression of evt-2 mRNA in the adult mouse pineal gland (pn) and in neurons of the underlying inferior colliculus (ic). Compare with Fig. 4F. (C) Dark-field image of evt-2 mRNA in the lateral ventricle, illustrating strong expression in the choroid plexus (cp) but not in the ependyma (ep) or white matter tracts (wm). Compare with Fig. 4A. (D) Widespread expression of evt-2 mRNA in a lateral region of adult mouse neocortex (cx) but not in underlying white matter. (Bars = 1 mm.)
Genetic Loci of the evt Genes.
The human evt-1 and evt-2 genes have been mapped through the human EST database assembly and mapping consortium (see Methods). evt-1 maps to an interval within human chromosome 11q13 at 80–84 centimorgans from the centromere, between the DNA markers D11S916 and D11S911. Within this same interval, near marker D11S533, is the gene for an inherited retinal degeneration disorder designated autosomal dominant familial exudative vitreoretinopathy (adFEVR) (24–26). The human evt-2 gene maps to chromosome 4q1, 50–56 centimorgans from the centromere, between D4S1587 and D4S405.
evt-1 Associates with Membranes of the Post-Golgi Compartment.
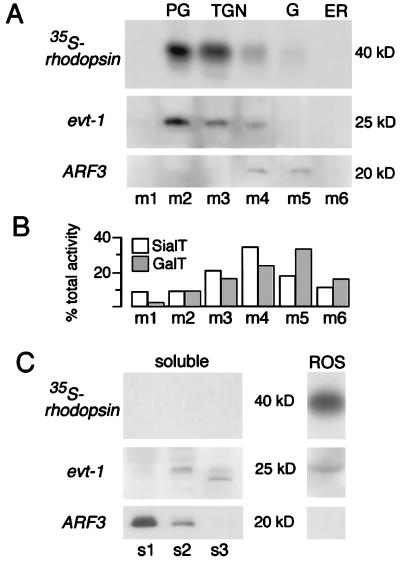
The high level expression of evt-1 by retinal photoreceptors led us to carry out a density gradient fractionation of these cells. We performed this analysis for the photoreceptors of the frog (R. berlandieri) retina because (i) we found that our evt-1 antibody cross-reacted with frog evt-1; and (ii) the outer segment membranes of frog photoreceptors are synthesized and shed at an exceptionally high steady-state rate, which allows for the metabolic labeling by pulse–chase of multiple rhodopsin-bearing membrane compartments (7). In particular, it permits the visualization of post-Golgi membrane vesicles, which, because of their transient nature and low abundance, are difficult to detect in other systems (14). Our fractionation of frog photoreceptors revealed a remarkable evt-1 localization (Fig. 6). Within membrane fractions, the protein was found to be the most specific marker for the post-Golgi compartment thus far identified (fraction pool m2 in Fig. 6A). Indeed, the distribution of evt-1 was nearly indistinguishable from that of 35S-rhodopsin after a 1-hr 35S-methionine pulse followed by a 2-hr cold chase of isolated retinae (Fig. 6 A and C), a labeling protocol that results in maximal visualization of rhodopsin as it transits out of the TGN and then through the post-Golgi on its way to the ROS (27). A modest evt-1 signal was present in fractions containing the TGN (fraction pools m3 and m4), but no evt-1 was detected in endoplasmic reticulum fractions or in the Golgi proper (fraction pool m5). A portion (30–40%) of membrane-bound evt-1 is present in the ROS (Fig. 6), with 30–35% in post-Golgi membranes and the remainder distributed in other membrane fractions, primarily the TGN. For comparison, Fig. 6B illustrates the previously published differential distributions, on identically run sucrose gradients of frog retinal membranes, of the enzymatic activities for sialyltransferase and galactosyltransferase, which are established markers for the TGN and the Golgi, respectively (14, 28).
Figure 6.
Subcellular colocalization of evt-1 with radiolabeled rhodopsin in transit through the post-Golgi compartment and into the ROS. (A) After pulse–chase labeling and subcellular fractionation on sucrose density gradients, radiolabeled membrane proteins isolated from three retinae were separated by SDS/PAGE and were autoradiographed. The majority of radiolabeled rhodopsin is recovered in post-Golgi fraction pool m2. A duplicate gel was blotted and probed sequentially with antibodies to evt-1 and ARF3. (B) The differential distributions of sialyltransferase and galactosyltransferase activities, markers for the TGN and the trans-Golgi, respectively, in membrane pools from identically run sucrose gradients, as reported (14, 15, 28). (C) Soluble proteins recovered in high speed supernatants obtained from three retinae were trichloroacetic acid-precipitated and were separated by SDS/PAGE. Top indicates complete absence of radiolabeled rhodopsin from the cytosolic fractions. evt-1 and ARF3 (Middle and Bottom) are detected by immunoblotting. Purified ROS were isolated from only one half of one retina because the high rhodopsin content of the ROS precludes loading more protein.
A fraction of photoreceptor evt-1 (≈10%) is soluble and appears in supernatants from a subset of sucrose gradient spins (Fig. 6C). Only very low levels of the protein are detected in pool s1, which represents true soluble proteins. Instead, evt-1 appears predominantly in pools s2 and s3 (the supernatants from the pellets that yield fractions m2 and m3 of Fig. 6A), which contain proteins that are either part of large cytosolic complexes or are loosely associated with membranes that sediment in these fractions and then are eluted after membranes are diluted from sucrose and sedimented at high speed. The evt-1 membrane and supernatant profiles are particularly informative when compared with those of a soluble protein that lacks a hydrophobic membrane anchor but that nonetheless reversibly associates with membranes. Among such proteins are ARF proteins, which are important in membrane trafficking and vesicular processing (21). The membrane-bound form of one of these proteins (ARF3) is primarily associated with the Golgi complex and the TGN and is not detected in evt-1+ post-Golgi membranes (fraction pools m4 and m5 in Fig. 6A). Note that soluble ARF3 appears primarily in fraction s1 whereas soluble evt-1 appears in supernatant fractions s2 and s3 (Fig. 6C). Taken together, the above data indicate that evt-1 is specifically recruited to cellular membranes in the late TGN and, most prominently, in the post-Golgi compartment of vertebrate photoreceptors.
DISCUSSION
Roles for the evts in Membrane Trafficking.
The evts define a family of vesicular proteins: They are structurally distinctive, and they specifically associate with and provide the first molecular marker for the post-Golgi compartment in photoreceptors. What roles are these proteins likely to play within cells? It is perhaps best to first consider evt-1 because the limited number of evt-1+ cells are related to one another through cell biology and/or developmental origin. Photoreceptors, oligodendrocytes, and myelinating Schwann cells are all highly polarized cells that synthesize an exceptionally large, apically configured organelle composed almost entirely of extended sheets of tightly compacted and specialized plasma membrane. The compacted disks of the photoreceptor ROS and the compacted laminae of myelin look remarkably similar, and there are no other cell types in the body that maintain the extraordinarily high levels of membrane biosynthesis and maintenance that are characteristic of these cells. And both myelinating glia and photoreceptors produce specialized lipids and proteins (e.g., the proteolipid protein in myelin and rhodopsin in rods) that must be specifically transported into their major organelles. Retinal pigment epithelial cells, which are also evt-1+, similarly phagocytize and catabolize large quantities of rod disk membranes as these membranes are shed from the tips of rod outer segments. Although pinealocytes and ependymal cells do not synthesize large membranous organelles, they nonetheless regulate the secretion of proteins in response to extracellular cues, a well known example being the circadian release of melatonin by pinealocytes. These cells also express photoreceptor-restricted genes, including opsins (29), whereas vertebrate retinae express several pineal-associated proteins, including melatonin. evt-1 clearly falls into this group of retinal-plus-pineal proteins. The cells of the ependyma form a ciliated epithelium that insulates the brain parenchyma from the cerebrospinal fluid that fills the ventricles. In the region of the third ventricle, this layer of cells gives rise to the pinealocytes of the pineal gland. The four sets of evt-1+ cells in the nervous system are thus related to each other both by their unusual membrane biology and by development.
Within these cells, evt-1 appears to be distributed between two pools: a soluble reservoir and a membrane-bound fraction that is associated predominantly with the post-Golgi. This suggests that the recruitment of evt-1 to post-Golgi membranes may be an intracellular readout of extracellular signals that trigger rapid membrane biosynthesis or catabolism. The PHDs of the evts are likely to play a key role in any such recruitment because these lipid-binding domains have been found to translocate other proteins from soluble pools to cellular membranes (17).
It is possible that evt-1 specifically integrates into photoreceptor post-Golgi membranes via a charged glycolipid or a cholesterol derivative whose synthesis is completed in the trans-Golgi. The most extensively studied set of lipids to which a subset of PHD-containing proteins bind are the phosphoinositides. Such proteins are often links in receptor-initiated signaling cascades. Consistent with the relative paucity of evt-1 expression that we have observed at the plasma membrane, preliminary analyses suggest that the evt-1 PHD does not bind PIP2 with high affinity (M. Lemmon, personal communication). Binding of other phospholipids, or of cholesterol or its metabolic derivatives, has not been excluded. The possibility of cholesterol binding by the evt-1 PHD is of particular interest, given the abundance of cholesterol in the myelin membrane (6) and the likely abundance of sterols in the post-Golgi membranes of photoreceptors (30) as well as the observed similarity of the evt PHDs to that of the oxysterol binding protein (Fig. 1B). Once associated with the post-Golgi, we envision that both evt-1 and -2 function as transport modules for the directed import or assembly of proteins or lipids into membranous organelles.
Contributions of evt-1 to Human Disease.
evt-1 is a tenable candidate gene for adFEVR, an inherited disorder characterized by inflammation of retinal blood vessels, neovascularization and vascular drop-out, generalized hyperpermeability of retinal vessels, and consequent retinal degeneration (24). The name of the disorder derives from the ill-defined white exudate that is commonly observed to surround the blood vessels at the back of the retina—those closest to the evt-1+ pigment epithelium. We estimate that the interval that contains evt-1, between the chromosome 11 DNA markers D11S916 and D11S911, is ≈1.2 megabases in length. Two different multipoint linkage studies, using three families, have positioned the gene for adFEVR near D11S533, which is located within this same interval (25, 26). This, together with the fact that the retinal photoreceptors and pigment epithelia adjacent to the retinal vasculature are among the very few evt-1+ cell types in the body, suggests that the human evt-1 gene should be examined for mutations in adFEVR patients.
Patients with multiple sclerosis (MS), an autoimmune disease that results in demyelination of CNS axons and oligodendrocyte death, also commonly present with an accompanying retinal disorder that is similar to adFEVR. Termed retinal venous sheathing, this disorder also is characterized by deposits of a confluent white exudate along retinal blood vessels and by retinal vascular inflammation and hyperpermeability (31). As for the demyelinating lesions of MS, retinal venous sheathing is frequently observed to resolve and then recur. Some authors have concluded that nearly all MS patients experience retinal venous sheathing during their lifetime (32) and that MS should therefore be viewed as a disease both of myelinating glia and of cells in the retina because the retina proper lacks both myelin and oligodendrocytes.
Given that (i) evt-1 and adFEVR map to positions that are currently indistinguishable on chromosome 11; (ii) adFEVR and the retinal venous sheathing of MS are similar retinal disorders; and (iii) CNS expression of the evt-1 gene is largely confined to the two CNS tissues that are directly compromised in MS, we suggest that evt-1 may be a gene that contributes to the pathogenesis of MS. It is possible, for example, that particular alleles of the human evt-1 gene might be more or less prone to provoke or participate as targets of the autoreactive cellular immune responses that underlie MS. Of epidemiological interest is the curious tendency of MS to occur at a greater frequency in extreme northern latitudes in the northern hemisphere and extreme southern latitudes in the southern hemisphere (33). These geographical locations are subject to the most substantial variations in day length during the course of the year, which notably influences the bioactivity of the evt-1+ cells of the pineal. Thus, three of the four evt-1+ CNS regions are also to greater or lesser extents tied to MS. There are at least two points that must be borne in mind vis-à-vis the possibility that certain alleles of evt-1 may be MS risk factors. First, although there is clearly a strong genetic component to MS, the disease is nonetheless a multigenic disorder with an obvious environmental contribution (34). Second, evt-1 is expressed by myelinating Schwann cells in the PNS (which are also evt-2+), and these cells are almost never involved in demyelination in MS. Nonetheless, the above observations are consistent with the hypothesis that certain alleles of the human evt-1 gene may be predisposing to MS, and this possibility now can be tested.
Acknowledgments
We thank Dan Ortuño and Darcie Baynes for excellent technical assistance, Trevor Kilpatrick for drawing our attention to the connection between the retina and multiple sclerosis, and James Goldman for suggesting the Lemke/Deretic collaboration. This work was supported by grants from the National Institutes of Health (NS23896 and NS34803 to G.L. and EY12421 and EY6891 to D.D.), a Career Development Award from Research to Prevent Blindness (to D.D.), postdoctoral fellowships from the Deutsche Forschungsgemeinschaft and the Klingenstein Foundation (to R.K.), and the Medical Scientist Training Program at the University of California at San Diego (to A.N.).
ABBREVIATIONS
- adFEVR
autosomal dominant familial exudative vitreoretinopathy
- ARF
ADP ribosylation factor
- CNS
central nervous system
- EST
expressed sequence tag
- evt
evectin
- MS
multiple sclerosis
- PHD
pleckstrin homology domain
- PNS
peripheral nervous system
- ROS
rod outer segment
- TGN
trans-Golgi network
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF118562).
References
- 1.Luini A, De Matteis M A. Trends Cell Biol. 1993;3:290–292. doi: 10.1016/0962-8924(93)90002-i. [DOI] [PubMed] [Google Scholar]
- 2.Bretscher M S. Cell. 1996;85:465–467. doi: 10.1016/s0092-8674(00)81246-5. [DOI] [PubMed] [Google Scholar]
- 3.Mellman I. Annu Rev Cell Dev Biol. 1996;12:575–562. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 4.Cockcroft S. BioEssays. 1998;20:423–432. doi: 10.1002/(SICI)1521-1878(199805)20:5<423::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 5.Metin C, Deleglise D, Serafini T, Kennedy T E, Tessier-Lavigne M. Development (Cambridge, UK) 1997;124:5063–5074. doi: 10.1242/dev.124.24.5063. [DOI] [PubMed] [Google Scholar]
- 6.Pfeiffer S E, Warrington A E, Bansal R. Trends Cell Biol. 1993;3:191–197. doi: 10.1016/0962-8924(93)90213-k. [DOI] [PubMed] [Google Scholar]
- 7.Besharse J C. In: The Retina: A Model for Cell Biological Studies. Adler R, Farber D, editors. New York: Academic; 1986. pp. 297–352. [Google Scholar]
- 8.Keller P, Simons K. J Cell Sci. 1997;110:3001–3009. doi: 10.1242/jcs.110.24.3001. [DOI] [PubMed] [Google Scholar]
- 9.Lemke G E, Brockes J P. J Neurosci. 1984;4:75–83. doi: 10.1523/JNEUROSCI.04-01-00075.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monuki E S, Weinmaster G, Kuhn R, Lemke G. Neuron. 1989;2:783–793. doi: 10.1016/0896-6273(89)90247-x. [DOI] [PubMed] [Google Scholar]
- 11.Banfi S, Guffanti A, Borsani G. Trends Genet. 1998;14:80–81. doi: 10.1016/s0168-9525(97)01354-1. [DOI] [PubMed] [Google Scholar]
- 12.Harlow E, Lane D P. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 13.Kilpatrick T J, Brown A, Lai C, Gassmann M, Goulding M, Lemke G. Mol Cell Neurosci. 1996;7:62–74. doi: 10.1006/mcne.1996.0005. [DOI] [PubMed] [Google Scholar]
- 14.Deretic D, Papermaster D S. J Cell Biol. 1991;113:1281–1293. doi: 10.1083/jcb.113.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deretic D, Puleo-Scheppke B, Trippe C. J Biol Chem. 1996;271:2279–2286. doi: 10.1074/jbc.271.4.2279. [DOI] [PubMed] [Google Scholar]
- 16.Lemmon M A, Ferguson K M. Curr Top Microbiol Immunol. 1998;228:39–74. doi: 10.1007/978-3-642-80481-6_3. [DOI] [PubMed] [Google Scholar]
- 17.Irvine R. Curr Biol. 1998;8:R557–R559. doi: 10.1016/s0960-9822(07)00360-0. [DOI] [PubMed] [Google Scholar]
- 18.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 19.Levine T P, Munro S. Curr Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- 20.Schmid S L, McNiven M A, De Camilli P. Curr Opin Cell Biol. 1998;10:504–512. doi: 10.1016/s0955-0674(98)80066-5. [DOI] [PubMed] [Google Scholar]
- 21.Boman A L, Kahn R A. Trends Biochem Sci. 1995;20:147–150. doi: 10.1016/s0968-0004(00)88991-4. [DOI] [PubMed] [Google Scholar]
- 22.Muller-Weeks S J, Caradonna S. Exp Cell Res. 1996;226:346–355. doi: 10.1006/excr.1996.0235. [DOI] [PubMed] [Google Scholar]
- 23.Kutay U, Hartmann E, Rapoport T A. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 24.Criswick V G, Schepens C L. Am J Ophthalmol. 1969;68:578–594. doi: 10.1016/0002-9394(69)91237-9. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Muller B, Fuhrmann C, van Nouhuys C E, Laqua H, Humphries P, Schwinger E, Gal A. Am J Hum Genet. 1992;51:749–754. [PMC free article] [PubMed] [Google Scholar]
- 26.Price S M, Periam N, Humphries A, Woodruff G, Trembath R C. Ophthalmic Genet. 1996;17:53–57. doi: 10.3109/13816819609057871. [DOI] [PubMed] [Google Scholar]
- 27.Deretic D. Electrophoresis. 1997;18:2537–2541. doi: 10.1002/elps.1150181408. [DOI] [PubMed] [Google Scholar]
- 28.Deretic D, Papermaster D S. J Cell Sci. 1993;106:803–813. doi: 10.1242/jcs.106.3.803. [DOI] [PubMed] [Google Scholar]
- 29.Blackshaw S, Snyder S H. J Neurosci. 1997;17:8074–8082. doi: 10.1523/JNEUROSCI.17-21-08074.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez de Turco E B, Deretic D, Bazan N G, Papermaster D S. J Biol Chem. 1997;272:10491–10497. doi: 10.1074/jbc.272.16.10491. [DOI] [PubMed] [Google Scholar]
- 31.Wray S H. In: Multiple Sclerosis: Clinical and Pathogenetic Basis. Raine C S, McFarland H F, Tourtellotte W W, editors. London: Chapman & Hall; 1997. pp. 21–30. [Google Scholar]
- 32.Engell T, Anderson P K. Acta Neurol Scand. 1982;65:601. doi: 10.1111/j.1600-0404.1982.tb03113.x. [DOI] [PubMed] [Google Scholar]
- 33.Kurtzke J F. In: Multiple Sclerosis: Clinical and Pathogenetic Basis. Raine C S, McFarland H F, Tourtellotte W W, editors. London: Chapman & Hall; 1997. pp. 91–139. [Google Scholar]
- 34.Weinshenker B G. Neurol Clin. 1996;14:291–308. doi: 10.1016/s0733-8619(05)70257-7. [DOI] [PubMed] [Google Scholar]