Abstract
A fundamental challenge in population genetics and molecular evolution is to understand the forces shaping the patterns of genetic diversity within and among species. Among them, mating systems are thought to have important influences on molecular diversity and genome evolution. Selfing is expected to reduce effective population size, Ne, and effective recombination rates, directly leading to reduced polymorphism and increased linkage disequilibrium compared with outcrossing. Increased isolation between populations also results directly from selfing or indirectly from evolutionary changes, such as small flowers and low pollen output, leading to greater differentiation of molecular markers than under outcrossing. The lower effective recombination rate increases the likelihood of hitch-hiking, further reducing within-deme diversity of selfers and thus increasing their genetic differentiation. There are also indirect effects on molecular evolutionary processes. Low Ne reduces the efficacy of selection; in selfers, selection should thus be less efficient in removing deleterious mutations. The rarity of heterozygous sites in selfers leads to infrequent action of biased conversion towards GC, which tends to increase sequences' GC content in the most highly recombining genome regions of outcrossers. To test these predictions in plants, we used a newly developed sequence polymorphism database to investigate the effects of mating system differences on sequence polymorphism and genome evolution in a wide set of plant species. We also took into account other life-history traits, including life form (whether annual or perennial herbs, and woody perennial) and the modes of pollination and seed dispersal, which are known to affect enzyme and DNA marker polymorphism. We show that among various life-history traits, mating systems have the greatest influence on patterns of polymorphism.
Keywords: mating system, selfing, sequence polymorphism, effective population size, selection, recombination
1. Introduction
Mating systems are probably a major factor controlling molecular diversity and genome evolution (reviewed by Charlesworth & Wright (2001)). Inbreeding directly reduces the effective population size, Ne, by reducing the number of independent gametes sampled for reproduction, and also reduces the effective recombination rate by increasing homozygosity (Nordborg 2000). The lower effective recombination rate allows the occurrence of hitch-hiking effects owing to the elimination of deleterious mutations (Charlesworth et al. 1993) or the spread of advantageous ones (Maynard-Smith & Haig 1974). These effects further reduce within-deme Ne and can span larger genomic regions in selfers than in outcrossers. These effects should be detectable in DNA sequences as lower neutral diversity within populations and increased population differentiation of selfers, together with more extensive linkage disequilibrium (LD). Bottlenecks may also often be more severe in inbreeders than in outcrossers (Schoen & Brown 1991) because a single seed can found a new population (Baker 1955), which can further reduce within-deme diversity. Extinction–recolonization dynamics and reduced gene flow through pollen also lead to strong population subdivision. Increased subdivision can compensate for locally reduced Ne, potentially leading to species-wide diversity that is not lower than that of outcrossing species, though extinction–recolonization dynamics is predicted usually to reduce Ne of inbreeding species and lead to lowered species-wide diversity (Ingvarsson 2002). Polymorphism in selfers should thus generally be low, particularly within populations, and therefore differentiation measures, such as Fst, should be higher than in the outcrossing species.
A second indirect type of effect is expected because the efficacy of selection is reduced when Ne is low. In populations of selfers, weakly deleterious mutations may be more likely to become fixed than in outcrossers, which can increase the rate of non-synonymous substitutions in a selfing lineage relative to the synonymous substitution rate (the Dn/Ds ratio). Within selfing populations, deleterious mutations may be found at higher frequencies and both the selection of slightly advantageous alleles and selection to optimize codon usage should be less effective. Homozygosity can also affect other processes that are important for molecular evolution. For example, when recombination occurs in a genome region containing sites heterozygous for GC and AT variants, correction of mismatches in heteroduplex DNA involves DNA repair, in which GC variants can be favoured over AT variants (Marais 2003). In highly homozygous species, this process will be rare. Homozygosity also affects transposable element dynamics. On the one hand, lack of heterozygosity for element insertions may affect the frequency of ectopic recombination, removing a force that can act against transposable element insertions (Charlesworth & Charlesworth 1995). On the other hand, homozygosity exposes insertions to selection, so that mutation–selection balance is changed towards elimination of deleterious insertions (Wright & Schoen 1999; Morgan 2001).
Even before the major analysis of allozyme diversity (Hamrick & Godt 1996), it was recognized that selfers have lower marker diversity than outcrossers and that their genetic variability is more structured. This has subsequently been confirmed for other marker types, including rapid amplification of polymorphic DNA (RAPDs) and microsatellites (Nybom 2004). Influences of other life-history traits on patterns of polymorphism have also been detected in plants, including life form (annual herbs, perennial herbs and woody perennial), seed dispersal (Hamrick & Godt 1996; Nybom 2004) and conservation status (Cole 2003). Sequence polymorphism data analyses to test these predictions and evaluate the importance of the various factors are, however, very scarce. It is important to obtain and analyse sequence data, because some predictions can be tested only using sequences (see §2) and nucleotide diversity estimates are the only diversity values that are predicted quantitatively by the theories outlined above. Several comparisons between species within the same genus with different mating systems have recently been made, and they revealed lower diversity in selfers than in outcrossers in plant species (Leavenworthia, Liu et al. 1998, 1999; Arabidopsis, Savolainen et al. 2000; including Lycopersicon, Baudry et al. 2001; Miscanthus, Chiang et al. 2003; Wright et al. 2003b) and in the nematode genus Caenorhabditis (Graustein et al. 2002; Cutter 2006).
The effect of mating system differences on selection efficacy and genome composition has occasionally been tested. Most such results come from Arabidopsis species. Marais et al. (2004) showed that in highly selfing species, no relationship is expected between local recombination rates and either the GC content or the codon usage. Indeed, no such effects were observed in Arabidopsis thaliana. It is also consistent with the highly selfing habit of A. thaliana that no clear relationship with recombination was found in the distribution of transposable elements, suggesting that the predominant form of selection affecting element abundance in the genome of this plant is selection against disruption of gene expression (Wright et al. 2003a). Unexpectedly, however, a comparison of A. thaliana with its outcrossing close relative Arabidopsis lyrata revealed no clear differences in either the efficacy of selection or in codon usage (Wright et al. 2002). This may be accounted for if only A. thaliana recently changed its breeding system to selfing, which seems likely to be the case (Shimizu et al. 2004; Charlesworth & Vekemans 2005).
Despite the increasing availability of sequence data, diversity datasets of sufficient size for analysis are still scarce and broadly based analyses of the effect of mating systems on the pattern of plant sequence polymorphism are still lacking. We have analysed a large set of angiosperm species, using a sequence polymorphism database, Polymorphix (Bazin et al. 2005), to test the effect of mating systems on the following: (i) the amount of polymorphism; (ii) LD; (iii) the efficacy of selection at the molecular level; and (iv) the GC content.
2. Material and methods
(a) Dataset
Sequence polymorphism data were retrieved from the Polymorphix database (Bazin et al. 2005; http://pbil.univ-lyon1.fr/polymorphix/). This database contains alignments of sets of sequence from putative allele samples within species, obtained on the basis of BLAST similarity between sequences from EMBL/GenBank. When available, outgroups are also included in the alignment. The database uses the term ‘families’ for these alignments. For angiosperms, the database contains 2011 sequence families from a total of 887 species. However, owing to the high representation of sequences such as ITS or rRNA genes among the non-coding sequences, we used only alignments containing annotated coding regions, and we restricted our sample to alignments containing at least four sequences within a species. These alignments were cleaned up semi-automatically as follows. First, genes annotated as transposons, transposon-like and pseudogenes were discarded. We also excluded pathogen resistance genes and self-incompatibility genes, as these include gene families with at least some members likely to be highly polymorphic owing to balancing selection, making it impossible to determine whether the available sequences are true alleles or orthologous sequences in the outgroup species; thus, including them would tend to overestimate polymorphism. Second, for each alignment, we removed sequences containing stop codons within the expected coding regions. Finally, to avoid paralogues, we manually inspected all genes exhibiting silent-site (synonymous+intron) nucleotide diversity, πsilent, greater than 5%. When an alignment error was evident, it was corrected. When our inspection detected the presence of paralogues, they were removed. All the remaining alignments with excessively high diversity were discarded, even when we did not find any evident error. When several outgroups were available in the Polymorphix alignments, we chose the one with the lowest silent-site divergence. The final dataset contained 342 nuclear gene families (from a total of 137 species) and 67 chloroplast gene families (from a total of 58 species).
For each species, we recorded, from research papers or the botanical literature and databases, the mating system (selfing, mixed-mating and outcrossing) and the following other life-history traits: life form (annual or perennial herbs, and woody perennials); mode of pollination (wind and insect); dispersal (gravity, attached, ingested, wind/water); cultivated or not; and rare or not. The list of species used, with their characteristics, is available in the electronic supplementary material (ESM1).
(b) Polymorphism statistics
The diversity statistics were computed with a C++ program using the functions available in the Bio++ library (Dutheil et al. 2006, http://162.38.181.25/BioPP/). The program computes the following statistics: haplotype diversity and the two estimators of θ=4Neμ, Watterson's θ estimator (Watterson 1975), and the nucleotide diversity, π (Tajima 1983), for all site types and also for synonymous and non-synonymous positions in coding regions (πs and πn, respectively) and for non-coding sites that we combined with synonymous sites to compute silent-site diversity, πsilent. The populations of origin of the individuals sampled are not recorded in the database, so that the diversity statistics are species-wide estimates, and we did not study differentiation measures, such as Fst.
To estimate recombination, we computed the squared correlation, r2, for pairs of polymorphic sites from the genotype frequency contingency table, the slope of the regression of pairwise r2 against pairwise distances, excluding low-frequency alleles (less than 0.125) and Hudson's estimator of ρ=4Nec (Hudson 1987), where c is the recombination rate.
We computed f0=πn/πs to assess the efficacy of selection against weakly deleterious alleles. Nine genes with f0>1 were excluded. When outgroups were available, we also computed the neutrality index (NI) based on the McDonald–Kreitman table (McDonald & Kreitman 1991): NI=(Pn/Ps)/(Dn/Ds) (Rand & Kann 1996), where P stands for the number of polymorphic sites, D for the number of sites with fixed differences from the outgroup sequence, and n and s stand for the non-synonymous and synonymous positions, respectively. An NI<1 implies positive selection between the species and the outgroup. Weakly deleterious standing variation can partly mask positive selection. Therefore, as suggested by Fay et al. (2001), we also computed the NI after removing low-frequency variants (less than 0.125). In both cases, we applied a Jukes–Cantor correction to Dn and Ds (Nei & Gojobori 1986).
Finally, we computed three measures of GC content: the total GC content, GC content at the third position of codons (GC3) and GC in introns (GCintrons). Differences in GC3 could be owing either to biased gene conversion or selection on codon usage, but a difference in GCintrons suggests biased gene conversion. We also computed the proportion of sites at which GC versus AT polymorphisms are detected, at which the GC allele is more frequent than the AT allele. Given that a mutation has occurred, its fixation dynamics depends only on the forces presently acting. In other words, the dynamics is independent of whether or not the base composition is at equilibrium. Under neutrality, the frequency spectrum of variants should be symmetrical, whereas with biased conversion or selection on codon usage, it should be skewed (e.g. Galtier et al. 2001).
(c) Data analyses
The number of genes in the database is very unevenly distributed across species (see electronic supplementary material ESM1 for details). Most species are represented by one or a few genes, whereas for some ‘model species’, such as A. thaliana or maize, many genes are available. Since the last release of the database, other large diversity datasets have been published, including a very large one from A. thaliana (Nordborg et al. 2005). These datasets were not added to the database because they would further increase its skewness. We used species means as data points in statistical analyses. Each statistic was computed for each gene and then averaged by species, except for GC versus AT polymorphism, where all polymorphic sites in a species were analysed as a single dataset. We excluded alignments smaller than 100 bp, either in total or in the coding region. For the analysis of GC versus AT polymorphisms, we also excluded species with fewer than 10 such polymorphic sites. After averaging, the variables were normalized by arcsine transformation, except for NI and ρ, which were log-transformed.
A single large ANOVA, with all the different plant characters as factors (mating system, life form, pollination, etc.) would be ideal because some factors are likely to be correlated, such as mating system and life form, or mating system and rarity. However, for such an analysis, the dataset is too small and very unbalanced. Therefore, we did one-way and two-way ANOVAs with each factor or pair of factors. The full ANOVA is present only in electronic supplementary material (ESM4). To take phylogenetic relationships into account, we also repeated the ANOVAs for nuclear genes, including the species' plant families as a variable. For chloroplast genes, owing to the sparse data, we included only a higher taxonomic level: monocotyledons versus eudicotyledons. It would be preferable to correct the phylogenetic inertia at the genus level because mating system differences are frequent between closely related plants (e.g. Goodwillie 1999; Shimizu et al. 2004), but the dataset included few genera with contrasting mating systems; hence, this was not possible. Instead, we averaged the results of species within the same genus that all have the same life-history traits and repeated the analyses for the nuclear gene dataset. This reduced this dataset from 137 species to 108 genera. All the analyses were performed with type-III sum of squares using the generalized linear model procedure of the SAS software (SAS Institute, v. 8).
3. Results and discussion
In our dataset, only 10 species with mixed mating systems (intermediate levels of outcrossing) were available. Preliminary analyses showed that the diversity and other quantities estimated here behave like those of outcrossers. Theoretical arguments also suggest that for intermediate selfing rates, recombination is effective enough to lead to patterns similar to the expectations in panmictic populations (e.g. Charlesworth et al. 1993; Stephan 1995). Therefore, in the following sections, we grouped the mating systems into just two categories: selfers versus full or partial outcrossers.
(a) Effect of mating systems on patterns of polymorphism
Among the life-history traits tested in one-way ANOVAs, mating system has the greatest effect on the level of polymorphism, measured by either nucleotide or haplotype diversity (table 1). The differences between selfers and outcrossers are on an average less than twofold. Results are similar for Watterson's θ estimates (not shown). For nuclear genes, we found no effect of other traits except for a marginally significant effect of rarity status. These results are similar when we use the plant families or genera to correct phylogeny (table 1 shows the former results; the latter is shown in electronic supplementary material ESM2). Owing to multiple tests (six by variable), Bonferroni correction may be desirable. However, we have not explored multiple factors without a priori predictions, but have tested predictions that provide a priori expectations for most individual factors. Hence, this is not necessarily appropriate. In any case, a lower threshold for significance (p<0.0085, here) leaves the conclusion for mating system unchanged. The analyses suggest that a selfing mating system considerably reduces species-wide diversity. However, mating system differences explain only a low proportion of the total variance in diversity measures (13–20%; first R2 in table 1) and much less by rarity (R2=2–4%). These conclusions are similar to those of Hamrick & Godt (1996) for allozyme diversity. When we correct the effects of the plant family (second R2 in table 1), the proportion of the variance explained by the mating system differences becomes much lower (R2=6–8%).
Table 1.
Results of ANOVAs (family+one factor) for nuclear gene total nucleotide diversity (πtotal), silent-site nucleotide diversity (πsilent) and haplotype diversity. (To illustrate the impact of controlling for phylogeny, R2-values are given both with and without adding family effects. p-values and least-squares means are given for each ANOVA that includes the plant family effects. Significant values at 5% are boldfaced. Italicized values are no longer significant after Bonferroni correction (six tests: α=0.85%).)
| R2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| factor | n | p-values | without family effects | with family effects | least-squares estimates | ||||
| nuclear genes | |||||||||
| outcrossing | selfing | ||||||||
| mating system | πtotal | 105 | 0.002 | 0.156 | 0.083 | 0.011 | 0.006 | ||
| πsilent | 105 | 0.003 | 0.133 | 0.068 | 0.016 | 0.008 | |||
| haplotype diversity | 105 | 0.001 | 0.207 | 0.079 | 0.836 | 0.697 | |||
| annual herb | perennial herb | woody perennial | |||||||
| life form | πtotal | 131 | 0.640 | 0.042 | 0.002 | 0.008 | 0.008 | 0.011 | |
| πsilent | 131 | 0.589 | 0.054 | 0.010 | 0.015 | 0.012 | 0.013 | ||
| haplotype diversity | 131 | 0.895 | 0.011 | 0.001 | 0.782 | 0.800 | 0.802 | ||
| insects | wind | ||||||||
| pollination | πtotal | 132 | 0.887 | 0.006 | 0.001 | 0.009 | 0.008 | ||
| πsilent | 132 | 0.698 | 0.001 | 0.001 | 0.014 | 0.010 | |||
| haplotype diversity | 132 | 0.851 | 0.006 | 0.001 | 0.786 | 0.826 | |||
| gravity | wind | attached | ingested | ||||||
| dispersal | πtotal | 118 | 0.453 | 0.036 | 0.011 | 0.010 | 0.006 | 0.012 | 0.010 |
| πsilent | 118 | 0.383 | 0.031 | 0.013 | 0.014 | 0.008 | 0.019 | 0.015 | |
| haplotype diversity | 118 | 0.072 | 0.051 | 0.023 | 0.746 | 0.777 | 0.692 | 0.863 | |
| wild | cultivated | ||||||||
| cultivated status | πtotal | 136 | 0.739 | 0.002 | 0.001 | 0.009 | 0.009 | ||
| πsilent | 136 | 0.772 | 0.001 | 0.001 | 0.013 | 0.014 | |||
| haplotype diversity | 136 | 0.832 | 0.001 | 0.001 | 0.790 | 0.801 | |||
| widespread | rare | ||||||||
| rarity status | πtotal | 127 | 0.027 | 0.029 | 0.028 | 0.009 | 0.005 | ||
| πsilent | 127 | 0.009 | 0.042 | 0.044 | 0.014 | 0.006 | |||
| haplotype diversity | 127 | 0.021 | 0.020 | 0.030 | 0.800 | 0.642 | |||
| chloroplast genes | |||||||||
| outcrossing | selfing | ||||||||
| mating system | πtotal | 31 | 0.408 | 0.023 | 0.003 | 0.002 | |||
| πsilent | 31 | 0.605 | 0.009 | 0.004 | 0.003 | ||||
| haplotype diversity | 31 | 0.862 | 0.001 | 0.569 | 0.587 | ||||
With the small chloroplast gene dataset of only 31 species (table 1), we found no significant effect of any trait or combination of traits (details not shown), but selfers again exhibited lower diversity than outcrossers.
The effect of mating system was again always significant in two-way ANOVAs, which were possible for nuclear genes (table 2). Life form and dispersal also have significant effects when the species' mating systems were included, but not when the plant families are taken into account. The only character with a marginal effect on diversity, even when we control phylogeny, is rarity (table 2). In all other two-factor analyses, rarity is also the only significant effect when we control phylogeny (see electronic supplementary material (ESM3). We found no interaction between factors in any analysis. Finally, including the six factors in the same ANOVA, results are similar and mating system still has the strongest effect (see electronic supplementary material (ESM4)).
Table 2.
Results of ANOVAs each including the mating system and one or more other factors. (p-values are given for the mating system effect and for the other factor (life form, pollination, etc.) effects. The first rows correspond to two-way ANOVAs without interaction (not significant). The following rows correspond to three-way ANOVAs including family effects (without interaction). Significant values at 5% are boldfaced. Values italicized are no longer significant after Bonferronni correction (six tests: α=0.85%).)
| factors | n | R2 | p-values | ||
|---|---|---|---|---|---|
| mating system | other factors | ||||
| without family effects | life form | 105 | 0.242 | <0.0001 | 0.001 |
| pollination | 104 | 0.132 | 0.0002 | 0.5114 | |
| dispersal | 104 | 0.200 | <0.0001 | 0.040 | |
| cultivated status | 105 | 0.133 | 0.0001 | 0.827 | |
| rarity | 105 | 0.192 | 0.0002 | 0.025 | |
| with family effects | life form | 105 | 0.091 | 0.002 | 0.300 |
| pollination | 104 | 0.067 | 0.004 | 0.948 | |
| dispersal | 104 | 0.100 | 0.005 | 0.514 | |
| cultivated status | 105 | 0.696 | 0.003 | 0.931 | |
| rarity | 105 | 0.118 | 0.002 | 0.050 | |
As reviewed previously, stronger effects of mating system differences are expected within populations than at the species level (e.g. Liu et al. 1998; Charlesworth 2003), but there are currently very few sequence data available to analyse within-population diversity separately. Nybom (2004) analysed within-population diversity values, which can partly explain the different results. Hamrick & Godt's (1996) dataset is much larger than ours, partly because they did not compute mean species values across loci as we did. Hence, a species can be represented several times in their analysis. Thus, their dataset has more power to detect weak effects (explaining less than 10% of the variance). However, these previous studies (Hamrick & Godt 1996; Nybom 2004) did not remove phylogenetic effects, which we found eliminated the significance of the effects of life form and dispersal when the mating system was included in the model.
(b) Effects of mating systems on linkage disequilibrium
The mating system is the characteristic expected to have the strongest effect on LD patterns. Indeed, selfers have significantly higher LD values than outcrossers, as measured by r2-values within genes (table 3). The effects of mating system differences on the slope of the regression of r2 against distance between polymorphic sites also suggest lower effective recombination rates in selfers. For outcrossers the slope is negative, as expected with recombination (−1.98), whereas selfers yield a positive mean slope of 2.61. The results are similar when low-frequency alleles (less than 0.125) are included. However, these differences are not significant after controlling phylogeny (p=0.086 with the family effects added; table 3). Hudson's ρ estimate also suggests the same effect, but is again non-significant after controlling phylogeny: ρoutcrossers=6.34, while ρselfers=1.50 (p=0.097). Because we have clear predictions for the direction of the effect of mating systems, one-tailed t-tests are appropriate. After removing the effects of phylogeny, tests are close to the 5% threshold (slope of the regression of r2 against distance: p=0.055; Hudson's ρ estimate: p=0.049).
Table 3.
Results of ANOVAs for nuclear gene patterns of LD, including the family and the mating system effects. (LD statistics r2 (excluding low-frequency alleles less than 0.125), slope of the r2 statistics against physical distance and Hudson's estimate of ρ=4Nec (Hudson 1987), where Ne is the effective population size and c is the recombination rate. Significant values at 5% are boldfaced.)
| least-squares estimates | ||||||
|---|---|---|---|---|---|---|
| n | R2 | p-value (two-tailed) | p-value (one-tailed) | outcrossing | selfing | |
| r2 | 96 | 0.049 | 0.018 | 0.014 | 0.40 | 0.56 |
| slope r2/distance | 94 | 0.027 | 0.086 | 0.055 | −1.99 | 2.63 |
| Hudson's ρ estimate | 86 | 0.032 | 0.097 | 0.049 | 6.34 | 1.50 |
It should be noted that for many species included in our analysis, the data are single sequences from each of the multiple populations, which is the best sample structure for testing the LD since inclusion of multiple sequences per population tends to overestimate LD owing to recent relatedness of the individuals (Wakeley & Lessard 2003). However, owing to the very large variance of LD measures, it is not surprising that we lack power to detect significant effects on these statistics. Furthermore, LD reaches equilibrium more slowly than nucleotide diversity. If selfing often evolved recently, as is likely if it is evolutionarily unsuccessful in the long term (Schoen et al. 1997) owing to maladaptation (see §1, also §4), no strong difference between selfers and outcrossers might be found. Moreover, theoretical models and estimates of chiasma frequencies suggest that selfers may generally evolve increased recombination (Charlesworth et al. 1977, 1979; Roze & Lenormand 2005). If so, high crossing-over rates could partly compensate for the reduced effective recombination rate owing to homozygosity, so that LD might not be as much higher as is expected when the outcrossing rate is low.
(c) Effects of mating systems on the efficacy of selection
Our dataset is not perfect for testing differences in the efficacy of selection, because we could analyse only a heterogeneous set of genes, different from one species to another (an ideal dataset would compare the diversity of genes in selfers with that of orthologues from outcrossing sister species, with a third outgroup to allow the lineages in which changes occurred to be inferred). There is, however, a loose correlation between taxonomy and genes available, so that taking phylogeny into account partially controls gene heterogeneity. As a result, we found significant results only when we statistically removed the effects of the plant family. For the entire dataset, we find no significant effect of the mating system on the πn/πs ratio (f01 in table 4). However, several genes had no non-synonymous variants (πn=0). These are either uninformative low-diversity genes (mainly in selfers) or highly constrained genes for which no effective population size effect is expected, and which should not be included. After removing these genes, selfers tend to have higher f0 ratios than outcrossers, suggesting that selection is less effective in selfers at removing weakly deleterious alleles.
Table 4.
Results of ANOVAs for the ratio f0 for plant family and mating system effects. (The analysis of f0 was computed for all genes (f01) or excluding zero values (f02). The analysis of the NI was computed either including (NI1) or excluding (NI2) low-frequency alleles less than 0.125 (see main text). Significant values at 5% are boldfaced.)
| least-squares estimates | |||||
|---|---|---|---|---|---|
| n | p-value | R2 | outcrossing | selfing | |
| f01 | 87 | 0.414 | 0.003 | 0.273 | 0.328 |
| f02 | 71 | 0.006 | 0.080 | 0.286 | 0.445 |
| NI1 | 44 | 0.020 | 0.094 | 1.032 | 2.353 |
| NI2 | 39 | 0.061 | 0.058 | 0.742 | 1.653 |
We also found a significant mating system effect on the NI (NI1 in table 4). An NI<1 suggests adaptive evolution in a gene, with an excess of non-synonymous substitutions between a species and its outgroup. However, weakly deleterious alleles segregating at low frequencies within a species (increased Pn) can mask the signal of positive selection (Fay et al. 2001). Taking the raw data, the mean NI value in outcrossers is close to 1 (1.03), but it is higher in selfers (2.37). However, when low-frequency alleles are removed (NI2 in table 4), the mean NI for outcrossers becomes 0.742, while for selfers it remains high (1.65), though the difference is non-significant (p=0.061; because the data are unbalanced for these statistics, both one- and two-tailed t-tests on the residuals of ANOVAs, after removing the effects of phylogeny give similar p-values). Although this result must be viewed with caution, it suggests that positive selection may be more efficient in outcrossers than in selfers.
Differences in the efficacy of selection owing to differences in the effective population size have been documented, mainly in animals (Ohta 1993a,b; Eyre-Walker et al. 2002; Keightley et al. 2005). In plants, based on the comparisons with Drosophila melanogaster and using a model in which all mutations in a gene have the same selective effects, Bustamente et al. (2002) inferred that A. thaliana has mainly fixed slightly detrimental mutations owing to its predominantly selfing mating system. However, comparisons with the closely related outcrossing species A. lyrata revealed no clear differences (Wright et al. 2002). Despite the deficiencies of our present dataset, we found a slight but significant signal. By lowering adaptation, inbreeding may have a cost over long evolutionary time-scales. However, advantageous mutations with large effect whose fixation is facilitated by inbreeding (Charlesworth 1992), may compensate for this. Thus, even if inbreeding has a cost, selfing species may not necessarily be evolutionary dead-ends (Takebayashi & Morrell 2001).
(d) Effect of mating systems on base composition
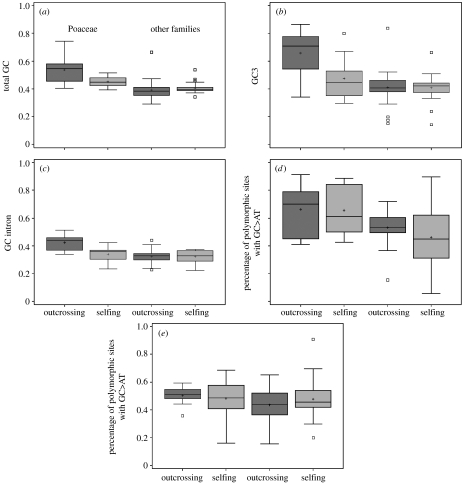
Heterogeneity in GC content was initially found in vertebrate genomes and named ‘isochores’ (for a review, see Eyre-Walker & Hurst 2001). It is well known that the Poaceae have an unusual genomic base composition compared with other plant families, and their genomes exhibit great heterogeneity in GC content (Carels & Bernardi 2000; Wong et al. 2002; Wang et al. 2004). Accordingly, in our dataset, the main phylogenetic effect is expected to be between Poaceae and other families. Therefore, in our analyses of base composition, we replaced the plant family effect with these two categories. The results are presented in figure 1 and table 5. We found a strong effect of Poaceae on base composition (total GC, GC3 and GCintrons), consistent with the previous results showing that Poaceae genes are more GC rich and vary in GC content more than those of other plants. We also found a significant mating system effect, but only in the Poaceae: outcrossers have higher GC content than selfers in this family for all three GC measures. Polymorphism patterns are less clear. We found a significant excess of high-frequency GC3 alleles in Poaceae, but no mating system effect. For GCintrons, we found no effects of the mating system and the plant family (Poaceae versus others). The effect on the pattern of GC versus AT polymorphism in third codon positions is mainly owing to GC-rich genes. Between genes, GC3 is strongly correlated with the proportion of third position sites at which GC alleles have the highest frequency (r=0.452, p<0.0001). When the previous analyses are done on the residuals of the regression of GC3 polymorphism on GC3, the Poaceae no longer have a significant excess of high-frequency GC3 alleles.
Figure 1.
Box plots of (a) total GC content, (b) GC3, (c) intron GC content, (d) proportion of AT versus GC polymorphic sites in third positions of codons for which GC alleles are at the higher frequency, and (e) the same quantity in introns. The two left boxes correspond to Poaceae and the two right ones to the other families. Statistical tests corresponding to these box plots are presented in table 5.
Table 5.
P-values of the two-way ANOVAs of GC content, including the clade (Poaceae versus other families) and mating system effects and interactions. (The analysis was computed for total GC content (GC), GC3, GC in introns, third codon position AT versus GC polymorphism (PolGC3) and intron AT versus GC polymorphism (PolGCintrons) (see main text). The corresponding box plots are presented in figure 1. Significant values at 5% are boldfaced.)
| Poaceae/others | mating system | interaction | |
|---|---|---|---|
| GC | <0.0001 | 0.005 | 0.0002 |
| GC3 | <0.0001 | 0.004 | 0.004 |
| GCintrons | <0.0001 | 0.001 | 0.001 |
| PolGC3 | 0.001 | 0.446 | 0.535 |
| PolGCintrons | 0.343 | 0.764 | 0.327 |
Much evidence supports the hypothesis that biased gene conversion can explain isochores, including the observation in mammals that GC-rich regions are associated with high recombination rates (Galtier et al. 2001; Galtier 2004; Meunier & Duret 2004). Our results suggest that GC heterogeneity in Poaceae genomes could be a consequence of biased gene conversion, as we find that outcrossing species are enriched in GC. The excess of high-frequency GC alleles in Poaceae, especially in GC-rich genes, is also consistent with this mechanism. More effective selection on codon usage in outcrossers than in selfers cannot be excluded but it cannot explain GC enrichment of introns in outcrossing Poaceae. There is also no reason why selection on codon usage should be more effective in the Poaceae, given that no difference is observed for either f0 (Poaceae=0.280, other species=0.284, p=0.956) or NI (Poaceae=1.69, other species=1.72, p=0.914). Through biased gene conversion, outcrossing may strongly affect genomes' base composition. This may also influence amino acid composition, as it has been shown that compositional bias may affect protein evolution in plants (Wang et al. 2004). If the biased gene conversion hypothesis is correct, it remains to be explained why this mechanism is effective in Poaceae, but not detected in other angiosperm families.
4. Conclusions
Our analysis is the first wide compilation of sequence polymorphism data in plants. We found that, as expected from population genetics theory, mating systems have significant effects on diversity and LD, selection efficacy and base composition. Species-wide diversity, as studied here, is expected to be less sensitive to mating systems than within-population diversity. Nevertheless, our results strongly suggest that mating systems should be taken into account when interpreting sequence diversity patterns. However, mating systems explain only a low proportion of the observed variation among species. This is partly owing to the taxonomic heterogeneity of our dataset. Despite the increasing amount of sequence data, they are still rather too scarce to make large comparisons among non-model species.
Finally, mating systems can evolve rapidly (Goodwillie 1999; Shimizu et al. 2004) and we know very little about the time when each species' mating system was established, and how long it has remained the same. If many selfing species changed recently from outcrossing, the theoretically predicted patterns will not be found. Understanding the dynamics of the transition to selfing, and its consequences for polymorphism patterns and genome evolution, is still incomplete.
Acknowledgments
We thank the numerous colleagues who provided us with information to compile the life-history traits dataset, especially H. Nybom and T. Lenormand. We thank N. Galtier and the anonymous reviewers for their helpful comments on the manuscript and K. Belkhir and J. Dutheil for their help with C++ programming. This work was initiated during a visit by S.G. at the Institute of Evolutionary Biology in Edinburgh funded by the Royal Society.
Supplementary Material
List of species used for nuclear (a) and chloroplast (b) genes with their characteristics. References are only given for mating system characterisation. References are not necessary the first ones describing the mating system. For instance we used previous compilations as Barrett & Eckert (1990) or Charlesworth (1985). Species were classified as mixed species either when intermediate selfing rates (between 0.2 and 0.8) were estimated or when indicated by authors (example: cleistogamous flowers + mainly outcrossing chasmogamous ones).
Results are given for one-way ANOVAs (first group), two-way ANOVAS including mating system (second group), two-way ANOVAs including family effects (third group), and three-way ANOVAs including mating system and family effects (last group).
References
- Baker H.G. Self-compatibility and establishment after ‘long-distance’ dispersal. Evolution. 1955;9:347–348. doi:10.2307/2405656 [Google Scholar]
- Baudry E, Kerdelhue C, Innan H, Stephan W. Species and recombination effects on DNA variability in the tomato genus. Genetics. 2001;158:1725–1735. doi: 10.1093/genetics/158.4.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin E, Duret L, Penel S, Galtier N. Polymorphix: a sequence polymorphism database. Nucleic Acids Res. 2005;33:D481–D484. doi: 10.1093/nar/gki076. doi:10.1093/nar/gki076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamente C.D, Nielsen R, Sawyer S.A, Olsen K.M, Purugganan M.D, Hartl D.L. The cost of inbreeding in Arabidopsis. Nature. 2002;416:531–534. doi: 10.1038/416531a. doi:10.1038/416531a [DOI] [PubMed] [Google Scholar]
- Carels N, Bernardi G. Two classes of genes in plants. Genetics. 2000;154:1819–1825. doi: 10.1093/genetics/154.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Evolutionary rates in partially self-fertilizing species. Am. Nat. 1992;140:126–148. doi: 10.1086/285406. doi:10.1086/285406 [DOI] [PubMed] [Google Scholar]
- Charlesworth D. Effects of inbreeding on the genetic diversity of populations. Phil. Trans. R. Soc. B. 2003;358:1051–1070. doi: 10.1098/rstb.2003.1296. doi:10.1098/rstb.2003.1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Transposable elements in inbreeding and outbreeding populations. Genetics. 1995;140:415–417. doi: 10.1093/genetics/140.1.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Vekemans X. How and when did Arabidopsis thaliana become highly self-fertilising? Bioessays. 2005;27:472–476. doi: 10.1002/bies.20231. doi:10.1002/bies.20231 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Wright S.I. Breeding systems and genome evolution. Curr. Opin. Genet. Dev. 2001;11:685–690. doi: 10.1016/s0959-437x(00)00254-9. doi:10.1016/S0959-437X(00)00254-9 [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Strobeck C. Effects of selfing on selection for recombination. Genetics. 1977;68:213–226. doi: 10.1093/genetics/86.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B, Strobeck C. Selection for recombination in self-fertilizing species. Genetics. 1979;93:237–244. doi: 10.1093/genetics/93.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Morgan M.T, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y.-C, Schaal B.A, Chou C.-H, Huang S, Chiang T.-Y. Contrasting selection mode at the ADH1 locus in outcrossing Miscanthus sinensis vs. inbreeding Miscanthus condensatus (Poaceae) Am. J. Bot. 2003;90:561–570. doi: 10.3732/ajb.90.4.561. [DOI] [PubMed] [Google Scholar]
- Cole C.T. Genetic variation in rare and common plants. Annu. Rev. Ecol. Syst. 2003;34:213–237. doi:10.1146/annurev.ecolsys.34.030102.151717 [Google Scholar]
- Cutter A.D. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics. 2006;172:171–184. doi: 10.1534/genetics.105.048207. doi:10.1534/genetics.105.048207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutheil J, Gaillard S, Bazin E, Glémin S, Ranwez V, Galtier N, Belkhir K. Bio++: a set of C++ libraries for sequence analysis, phylogenetics, molecular evolution and population genetics. BMC Bioinform. 2006;7:188. doi: 10.1186/1471-2105-7-188. doi:10.1186/1471-2105-7-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Hurst L.D. The evolution of isochores. Nat. Rev. Genet. 2001;2:549–555. doi: 10.1038/35080577. doi:10.1038/35080577 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley P.D, Smith N.G.C, Gafffney D. Quantifying the slightly deleterious mutation model of molecular evolution. Mol. Biol. Evol. 2002;19:2142–2149. doi: 10.1093/oxfordjournals.molbev.a004039. [DOI] [PubMed] [Google Scholar]
- Fay J.C, Wyckoff G.J, Wu C.I. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N. Recombination, GC-content and the human pseudoautosomal boundary paradox. Trends Genet. 2004;20:347–349. doi: 10.1016/j.tig.2004.06.001. doi:10.1016/j.tig.2004.06.001 [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwillie C. Multiple origins of self-compatibility in Linanthus section Leptosiphon (Polemoniaceae): phylogenetic evidence from internal-transcribed-spacer sequence data. Evolution. 1999;53:1387–1395. doi: 10.1111/j.1558-5646.1999.tb05403.x. doi:10.2307/2640885 [DOI] [PubMed] [Google Scholar]
- Graustein A, Gaspar J.M, Walters J.M, Palopoli M.F. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics. 2002;161:99–107. doi: 10.1093/genetics/161.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamrick J.L, Godt M.J.W. Effects of life history traits on genetic diversity in plant species. Phil. Trans. R. Soc. B. 1996;351:1291–1298. [Google Scholar]
- Hudson R.R. Estimating the recombination parameter of a finite population model without selection. Genet. Res. 1987;50:245–250. doi: 10.1017/s0016672300023776. [DOI] [PubMed] [Google Scholar]
- Ingvarsson P.K. A metapopulation perspective on genetic diversity and differentiation in partially self-fertilizing plants. Evolution. 2002;56:2368–2373. doi: 10.1111/j.0014-3820.2002.tb00162.x. doi:10.1554/0014-3820(2002)056[2368:AMPOGD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Keightley P.D, Lercher M.J, Eyre-Walker A. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. doi:10.1371/journal.pbio.0030042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Zhang L, Charlesworth D. Genetic diversity in Leavenworthia populations with different inbreeding levels. Proc. R. Soc. B. 1998;265:293–301. doi: 10.1098/rspb.1998.0295. doi:10.1098/rspb.1998.0295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Charlesworth D, Kreitman M. The effect of mating system differences on nucleotide diversity at the phosphoglucose isomerase locus in plant genus Leavenworthia. Genetics. 1999;151:343–357. doi: 10.1093/genetics/151.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 2003;19:330–338. doi: 10.1016/S0168-9525(03)00116-1. doi:10.1016/S0168-9525(03)00116-1 [DOI] [PubMed] [Google Scholar]
- Marais G, Charlesworth B, Wright S.I. Recombination and base composition: the case of the highly self-fertilizing plant Arabidopsis thaliana. Genome Biol. 2004;5:R45. doi: 10.1186/gb-2004-5-7-r45. doi:10.1186/gb-2004-5-7-r45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard-Smith J, Haig D. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- McDonald J.H, Kreitman M. Adaptative protein evolution at the ADH locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. doi:10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- Meunier J, Duret L. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. doi:10.1093/molbev/msh070 [DOI] [PubMed] [Google Scholar]
- Morgan M.T. Transposable element number in mixed mating populations. Genet. Res. 2001;77:261–275. doi: 10.1017/s0016672301005067. doi:10.1017/S0016672301005067 [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nordborg M. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics. 2000;154:923–929. doi: 10.1093/genetics/154.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. doi:10.1371/journal.pbio.0030196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybom H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004;13:1143–1155. doi: 10.1111/j.1365-294X.2004.02141.x. doi:10.1111/j.1365-294X.2004.02141.x [DOI] [PubMed] [Google Scholar]
- Ohta T. Amino acid substitution at the ADH locus of Drosophila is facilitated by small population size. Proc. Natl Acad. Sci. USA. 1993a;90:4548–4551. doi: 10.1073/pnas.90.10.4548. doi:10.1073/pnas.90.10.4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. An examination of the generation-time effect on molecular evolution. Proc. Natl Acad. Sci. USA. 1993b;90:10 676–10 680. doi: 10.1073/pnas.90.22.10676. doi:10.1073/pnas.90.22.10676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand D.M, Kann L.M. Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol. Biol. Evol. 1996;13:735–748. doi: 10.1093/oxfordjournals.molbev.a025634. [DOI] [PubMed] [Google Scholar]
- Roze D, Lenormand T. Self-fertilization and the evolution of recombination. Genetics. 2005;170:841–857. doi: 10.1534/genetics.104.036384. doi:10.1534/genetics.104.036384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute. SAS Institute, Inc; Cary, NC: 1999. SAS/STAT User's guide. [Google Scholar]
- Savolainen O, Langley C.H, Lazzaro B.P, Fr H. Contrasting patterns of nucleotide polymorphism at the alcohol dehydrogenase locus in the outcrossing Arabidopsis lyrata and the selfing Arabidopsis thaliana. Mol. Biol. Evol. 2000;17:645–655. doi: 10.1093/oxfordjournals.molbev.a026343. [DOI] [PubMed] [Google Scholar]
- Schoen D.J, Brown A.H.D. Intraspecific variation in population gene diversity and effective population size correlates with the mating system in plants. Proc. Natl Acad. Sci. USA. 1991;88:4494–4497. doi: 10.1073/pnas.88.10.4494. doi:10.1073/pnas.88.10.4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen D.J, L'Heureux A.M, Marsolais J, Johnston M.O. Evolutionary history of the mating system in Amsinckia (Boraginaceae) Evolution. 1997;51:1090–1099. doi: 10.1111/j.1558-5646.1997.tb03956.x. doi:10.2307/2411038 [DOI] [PubMed] [Google Scholar]
- Shimizu K.K, et al. Darwinian selection on a selfing locus. Science. 2004;306:2081–2084. doi: 10.1126/science.1103776. doi:10.1126/science.1103776 [DOI] [PubMed] [Google Scholar]
- Stephan W. An improved method for estimating the rate of fixation of favorable mutations based on DNA polymorphism data. Mol. Biol. Evol. 1995;12:959–962. doi: 10.1093/oxfordjournals.molbev.a040274. [DOI] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi N, Morrell P.L. Is self-fertilization an evolutionary deed end? Revisiting an old hypothesis with genetic theories and a macroevolutionary approach. Am. J. Bot. 2001;88:1143–1150. [PubMed] [Google Scholar]
- Wakeley J, Lessard S. Theory of the effects of population structure and sampling on patterns of linkage disequilibrium applied to genomic data from humans. Genetics. 2003;164:1043–1053. doi: 10.1093/genetics/164.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.C, Singer G.A, Hickey D.A. Mutational bias affects protein evolution in flowering plants. Mol. Biol. Evol. 2004;21:90–96. doi: 10.1093/molbev/msh003. doi:10.1093/molbev/msh003 [DOI] [PubMed] [Google Scholar]
- Watterson G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. doi:10.1016/0040-5809(75)90020-9 [DOI] [PubMed] [Google Scholar]
- Wong G.K, Wang J, Tao L, Tan J, Zhang J, Passey D.A, Yu J. Compositional gradients in Gramineae genes. Genome Res. 2002;12:851–856. doi: 10.1101/gr.189102. doi:10.1101/gr.189102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S.I, Schoen D.J. Transposon dynamics and the breeding system. Genetica. 1999;107:139–148. doi:10.1023/A:1003953126700 [PubMed] [Google Scholar]
- Wright S.I, Lauga B, Charlesworth D. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 2002;19:1407–1420. doi: 10.1093/oxfordjournals.molbev.a004204. [DOI] [PubMed] [Google Scholar]
- Wright S.I, Agrawal N, Bureau T.E. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 2003a;13:1897–1903. doi: 10.1101/gr.1281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S.I, Lauga B, Charlesworth D. Subdivision and haplotype structure in natural populations of Arabidopsis lyrata. Mol. Ecol. 2003b;12:1247–1263. doi: 10.1046/j.1365-294x.2003.01743.x. doi:10.1046/j.1365-294X.2003.01743.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of species used for nuclear (a) and chloroplast (b) genes with their characteristics. References are only given for mating system characterisation. References are not necessary the first ones describing the mating system. For instance we used previous compilations as Barrett & Eckert (1990) or Charlesworth (1985). Species were classified as mixed species either when intermediate selfing rates (between 0.2 and 0.8) were estimated or when indicated by authors (example: cleistogamous flowers + mainly outcrossing chasmogamous ones).
Results are given for one-way ANOVAs (first group), two-way ANOVAS including mating system (second group), two-way ANOVAs including family effects (third group), and three-way ANOVAs including mating system and family effects (last group).