Abstract
The house mouse (Mus musculus) is universally adopted as the mammalian laboratory model, and it is involved in most studies of large-scale comparative genomics. Paradoxically, this taxon is rarely the index species for evolutionary analyses of genome architecture owing to its highly rearranged karyotype. To unravel the origin and nature of this extensive repatterning genome, we performed a multidirectional chromosome painting study of representative species within the genus Mus. However, the latter includes four extant subgenera (Mus, Coelomys, Nannomys and Pyromys) between which the phylogenetic relationships remain elusive despite the numerous molecular studies. Comparative genomic maps were established using chromosome-specific painting probes of the laboratory mouse and Nannomys minutoides. Hence, by integrating closely related species within Mus, this study allowed us to: (i) unambiguously resolve for the first time the long-standing controversial phylogeny, (ii) trace the evolution of genome organization in the house mouse, (iii) track rearrangements that necessitated new centromere locations, i.e. formation of neocentromere or reactivation of latent centromeres, (iv) reveal an extremely high rate of karyotypic evolution, with a 10- to 30-fold acceleration which was coincidental with subgeneric cladogenesis and (v) highlight genomic areas of interest for high-resolution studies on neocentromere formation and synteny breakpoints.
Keywords: fluorescence in situ hybridization, phylogenomics, Mus, Nannomys, Coelomys, neocentromere
1. Introduction
In the last decade, considerable advances have been made in understanding mammalian genomic architecture through genome-sequencing initiatives and cross-species chromosome painting. Large-scale comparative mapping analyses have focused primarily on the three mammalian species for which the most complete genomic data are available: human, mouse and rat. The investigations have shown that (i) the random breakage model of genome evolution (Nadeau & Taylor 1984) is flawed, since extensive breakpoint reuse is apparent (Pevzner & Tesler 2003; Bailey et al. 2004; Zhao et al. 2004), (ii) centromeric shifts and/or neocentromere formation are relatively common (Ventura et al. 2001, 2003, 2004; Murphy et al. 2005) and (iii) rates of chromosomal repatterning show considerable variation among lineages (O'Brien et al. 1999; Murphy et al. 2005). However, bioinformatic approaches are limited by the availability of sequenced genomes. In contrast, cross-species chromosome painting, i.e. zoo-fluorescence in situ hybridization (FISH) analyses (flow-sorted fluorescent chromosome probes hybridized in situ to chromosomes of another target species) have provided genomic information, albeit at a far lower resolution, on species spanning most of the 20 modern orders of mammals. Most comparisons of genome organization have involved human chromosome-specific paints, while those using the house mouse as the index species have been relatively scarce despite the biomedical and genomic importance of this mammalian laboratory model, and have only recently started accumulating. The lack of interest in this rodent is attributed to its highly fragmented and rearranged karyotype compared to that of the human and other mammals (Stanyon et al. 1999; Nilsson et al. 2001; Gregory et al. 2002). Several zoo-FISH comparisons between Mus and other rodent genera have been undertaken (e.g. Yang et al. 2000; Cavagna et al. 2002; Rambau & Robinson 2003; Matsubara et al. 2004; Engelbrecht et al. 2006) and have shed some insight into our understanding of the origin and nature of the extensive repatterning in the house mouse genome. However, the investigations have not been extended to closely related species within the genus Mus. The only exception to this has been Mus platythrix, which falls within the subgenus Pyromys (Matsubara et al. 2003).
The genus Mus (Rodentia, Muridae and Murinae) is a highly speciose murid genus, which exhibits extensive chromosomal evolution (e.g. Britton-Davidian et al. 2000; Veyrunes et al. 2004; Piàlek et al. 2005). This genus encompasses at least 40 species divided into four subgenera: Mus sensu stricto, Nannomys, Coelomys and Pyromys (Musser & Carleton 1993). The Eurasian subgenus Mus is by far the most extensively studied, and consists of 11 species to which a new species from the island of Cyprus has recently been added (Cucchi et al. 2006). The three other subgenera are less well known. The subgenus Nannomys, the African pygmy mice, has a sub-Saharan distribution and comprises 19 recognized species. The two last subgenera are restricted to the Indian subcontinent and southeastern Asia: Pyromys with five species and Coelomys with four species (Musser & Carleton 1993). The Mus genus has been the focus of a plethora of phylogenetic studies (e.g. Bonhomme 1986, 1992; Jouvin-Marche et al. 1988; She et al. 1990; Catzeflis & Denys 1992; Boursot et al. 1993; Sourrouille et al. 1995; Lundrigan et al. 2002; Chevret et al. 2003, 2005; Suzuki et al. 2004; Veyrunes et al. 2005). However, while the monophyly of the genus and of each of the four subgenera are clearly established, the relationships between them are still unresolved, despite the large variety of molecular markers used. Even the sequencing of not less than six paternally, maternally and biparentally inherited genes failed to provide strong support for the intersubgeneric relationships (Lundrigan et al. 2002). This lack of resolution most likely reflects the rapid radiation of these four clades, which is thought to have occurred within 1 Myr (e.g. Chevret et al. 2005; Veyrunes et al. 2005). Thus, new genetic markers are required to resolve these phylogenetic uncertainties. Chromosomal rearrangements appear to be ideal candidates as they are considered to be rare genomic changes sensu Rokas & Holland (2000), and provide cladistic signatures with very low levels of homoplasy (e.g. Murphy et al. 2004; Wienberg 2004). In effect, zoo-FISH comparative chromosome painting constitutes a powerful and elegant method for both detecting chromosome homologies between species and resolving long-standing phylogenetic controversies such as within the Carnivora, Rodentia or Primate orders (de Oliveira et al. 2002; Nie et al. 2002; Muller et al. 2003; Li et al. 2004).
In the present study, a multidirectional chromosome painting analysis is performed between representative species of three subgenera of Mus (Nannomys, Coelomys and Mus). By including published data for the fourth subgenus (Pyromys; Matsubara et al. 2003), and using available rodent species as outgroups, a chromosomal phylogeny is reconstructed following three aims: (i) to test the performance of chromosomal rearrangements in resolving the phylogenetic relationships between the subgenera of Mus; (ii) to infer the ancestral karyotype of the genus Mus for use in future comparisons with other taxa and finally (iii) to gain insight into patterns and processes of genome organization and evolution leading to the house mouse karyotype.
2. Material and methods
(a) Animals, chromosome preparation and identification
In order to avoid nomenclatural ambiguities, we refer to subgenera to distinguish lineages (i.e. Nannomys, Coelomys, Mus and Pyromys) and not to the genus name (Mus).
The female Nannomys mattheyi, male Nannomys minutoides, female Coelomys pahari and male Mus musculus specimens used in this study originated, respectively, from Samaya in Mali, Stellenbosch in South Africa, India (precise locality unknown) and Clapiers in France. The chromosome preparations were made either from bone marrow of yeast-stimulated animals (N. mattheyi, C. pahari and M. musculus) or fibroblast cell-cultures established from skin biopsy following the standard procedures (N. minutoides). The identification of chromosomes was also accomplished by G- and DAPI banding concurrently with in situ hybridization.
(b) Flow sorting and chromosome-specific painting probes preparation
As cell cultures of N. mattheyi were not available, flow sorting was performed for N. minutoides. The chromosomes were prepared for sorting as described previously (Yang et al. 1997). The stained chromosome preparations were sorted on a dual laser cell sorter (FAC-Star Plus, Becton Dickinson). Flow-sorted chromosomes were used as templates for amplification by degenerate oligonucleotide-primed PCR (DOP-PCR) using 6 MW primers (Telenius et al. 1992). Primary DOP-PCR products were used as a source of template for the incorporation of biotin-16-dUTP (Boehringer).
(c) Fluorescence in situ hybridization
The complete set of commercial chromosome-specific painting probes from the house mouse M. musculus (Cambio) and those made from N. minutoides were hybridized across representative species of the other subgenera i.e. Nannomys, Coelomys and Mus, respectively. Hybridization and detection were carried out following the procedure described in Robinson et al. (2004). Biotin-labelled probes were visualized using Cy3-avidin (1 : 500 dilution, Amersham). Slides were then mounted in Vectashield mounting medium with DAPI (Vector Laboratories). Images were captured using the Genus software (Applied Imaging). The hybridization signals were assigned to specific chromosomal regions identified by DAPI staining.
(d) Phylogenetic analysis
The phylogenetic analysis was performed using the comparative chromosomal maps of Mus versus Nannomys and Coelomys, and published data on Pyromys (Matsubara et al. 2003). Three additional Murinae species were included as outgroups: Rattus rattus (Cavagna et al. 2002), Rhabdomys pumilio (Rambau & Robinson 2003) and Apodemus sylvaticus (Matsubara et al. 2004). A subsequent zoo-FISH analysis of A. sylvaticus by Stanyon et al. (2004) revealed several discrepancies between the two studies. We chose the former as it included one synteny in common with our analysis, which was overlooked in the latter. Contiguous chromosomal segment associations (syntenies) were used as characters to establish a binary data matrix (electronic supplementary material) following the procedure for encoding chromosomal data reviewed in Dobigny et al. (2004). We chose to exclude from the matrix the pericentromeric material homologous to Mus chromosome 14 on Pyromys chromosomes 5, 8 and 12, which may consist of duplications of 18S–28S ribosomal RNA genes (Matsubara et al. 2003; see also Thomas et al. 2003). In the absence of accurate information, we postulated that all characters had the same weight (i.e. same probability of appearance/fixation). The most parsimonious phylogenetic tree was obtained using an exhaustive search in Paup v. 4.0b10 (Swofford 1999). The robustness of each node was assessed by bootstrap estimates after 1000 iterations.
3. Results
(a) G-banded karyotypes
The G-banded karyotypes of the Nannomys species have already been described in Veyrunes et al. (2004): N. mattheyi has an all-acrocentric 2n=36 karyotype and N. minutoides has a diploid number of 2n=18 with all chromosomes being biarmed. The G-banding analyses (Veyrunes et al. 2004) indicate that the lower diploid number in the latter species resulted exclusively from Robertsonian (Rb) fusions; in particular, both sex chromosomes are involved in Rb fusions with pair 1 to form the chromosomes Rb(X.1) and Rb(Y.1). The G-banded karyotype of C. pahari represents the first ever published for this subgenus, and is composed of 48 acrocentric chromosomes.
(b) Flow-sorted karyotype of N. minutoides
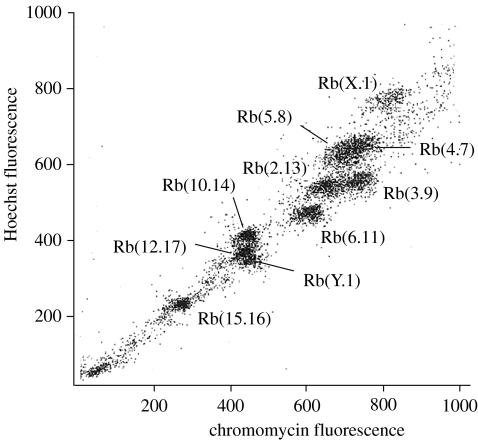
Figure 1 shows the flow karyotype of N. minutoides. Chromosome suspensions were sorted on base pair composition and chromosomal size. This resulted in 10 peaks which represent the eight autosomal pairs, and the Rb(X.1) and Rb(Y.1). Identification of the different peaks was achieved by hybridizing DOP-PCR generated probes onto DAPI-banded N. minutoides metaphases.
Figure 1.
Flow karyotype of a male Nannomys minutoides (2n=18) resolving 10 peaks, each containing an Rb fusion chromosome pair.
(c) Reciprocal chromosome painting between Mus and Nannomys
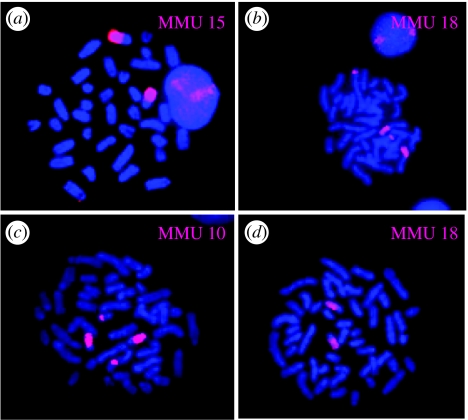
All the house mouse chromosome-specific probes successfully hybridized to the euchromatic regions of the pygmy mouse chromosomes (see figure 2 for examples). The 19 autosomal and X paints defined 27 segments of homology between Mus and Nannomys. Regions of homology are indicated on the G-banded karyotype of N. mattheyi (figure 3a). Fourteen Mus chromosomes (2–4, 6–8, 10–12, 14–16, 19 and X) were each retained as a single conserved block, five Mus probes (1, 9, 13, 17 and 18) each produced two signals, and finally the Mus chromosome 5 probe hybridized to three regions in the Nannomys karyotype.
Figure 2.
Examples of hybridization using mouse chromosome paints (MMU) to (a,b) Nannomys mattheyi and (c,d) Coelomys pahari metaphase spreads counterstained with DAPI.
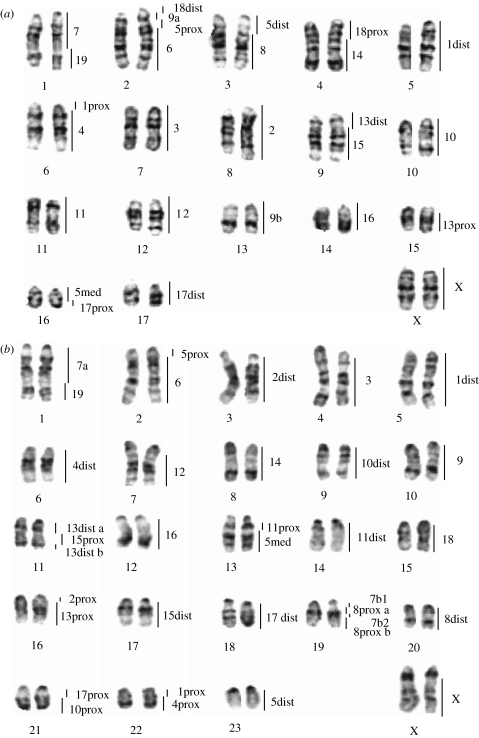
Figure 3.
G-banded karyotypes of (a) Nannomys mattheyi and (b) Coelomys pahari with the assignments of Mus musculus homologous segments (chromosome pair numbers on the right) revealed by the mouse probes. ‘dist’, ‘med’ and ‘prox’ refer to the distal, median and proximal segment of the chromosome, respectively, and a and b refer to unidentified subchromosomal segments.
Reciprocal chromosome painting (Nannomys paints onto the Mus karyotype) was used to define precisely the genome-wide homologies between these two subgenera. Twenty-six homologous segments were detected and summarized in table 1. In addition to confirming the painting results of the Mus probes, this procedure allowed us to assign subchromosomal homologies between the Mus and Nannomys chromosomes. The only exception to this was chromosome 9 of Mus, which is homologous to parts of chromosomes 2 and 13 of N. mattheyi, but are present as a single fused chromosome in the N. minutoides-derived painting probe, Rb(2.13).
Table 1.
Homologies between Mus musculus, Nannomys mattheyi and Coelomys pahari chromosomes, inferred from reciprocal and cross-species chromosome painting and G-banded patterns. (‘dist’, ‘med’ and ‘prox’ refer to the distal, median and proximal segment of the chromosome, respectively. ‘tot’ refers to entire chromosome; a, b refer to subsegments of chromosome.)
| Mus | Nannomys | Coelomys |
|---|---|---|
| 1prox | 6prox | 22prox |
| 1dist | 5tot | 5tot |
| 2prox | 8prox | 16prox |
| 2dist | 8dist | 3tot |
| 3tot | 7tot | 4tot |
| 4prox | 6med | 22dist |
| 4dist | 6dist | 6tot |
| 5prox | 2med b | 2prox |
| 5med | 16prox | 13dist |
| 5dist | 3prox | 23tot |
| 6tot | 2dist | 2dist |
| 7tot | 1prox | 1prox+19prox+med b |
| 8prox | 3med | 19med a+dist |
| 8med | 3dist | 20tot |
| 9tot | 13tot+2med a | 10tot |
| 10prox | 10prox | 21dist |
| 10dist | 10dist | 9tot |
| 11prox | 11prox | 13prox |
| 11dist | 11dist | 14tot |
| 12tot | 12tot | 7tot |
| 13prox | 15dist | 16dist |
| 13dist | 9prox | 11prox+dist |
| 14tot | 4dist | 8tot |
| 15prox | 9med | 11med |
| 15dist | 9dist | 17tot |
| 16tot | 14tot | 12tot |
| 17prox | 16dist | 21prox |
| 17dist | 17tot | 18tot |
| 18prox | 4prox | 15prox |
| 18dist | 2prox | 15dist |
| 19tot | 1dist | 1dist |
| Xtot | Xtot | Xtot |
(d) Chromosome painting of Mus and Nannomys probes onto Coelomys chromosomes
The 19 house mouse autosomal probes and the X paint delineated 35 homologous segments in the C. pahari genome (figure 3b and also figure 2 for examples). Nine Mus chromosomes (3, 6, 9, 12, 14, 16, 18, 19 and X) showed complete conservation of synteny (i.e. retained as single sites of hybridization), seven (1, 2, 4, 10, 11, 15 and 17) each painted two chromosomal regions and four (5, 7, 8 and 13) detected three segments in the Coelomys karyotype.
The hybridization of Nannomys probes onto Coelomys chromosomes (not shown) revealed 30 homologous segments in perfect concordance with the preceding results and allowed us to assign several subchromosomal homologies between Mus and Coelomys (table 1) via the Nannomys karyotype.
(e) Phylogenetic analysis
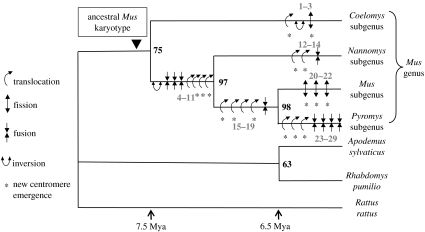
The reciprocal painting results, in combination with G-banding comparison, allowed us to identify most of the sub-regional homologies between the representative karyotypes of the three subgenera (table 1). The homologous adjacent segments (syntenic associations) identified between these species and those published for Pyromys and the outgroups were translated into 67 chromosomal characters (electronic supplementary material). The maximum parsimony analysis resulted in only one most parsimonious tree (74-steps long, consistency index=0.91, retention index=0.79, homoplasy index=0.09; figure 4). The first subgenus to diverge is Coelomys, followed by Nannomys, then Mus and Pyromys. There is no synapomorphy that supports the monophyly of the genus, resulting in the lowest bootstrap value (equal to 75) for this node. The two other nodes are much more robust (equal to 97 and 98) and are supported by several chromosomal changes, i.e. four translocations, three fusions and one inversion all provide strong support for the Nannomys/Mus/Pyromys cluster; four translocations and one fusion characterize the Mus/Pyromys clade (figure 4). Semantically, we considered ‘translocation’, the transfer of a fragment of one chromosome onto another chromosome, and ‘fusion and/or fission’, events involving two entire chromosomes.
Figure 4.
Most parsimonious phylogeny using PAUP, based on the 67 chromosomal characters (electronic supplementary material). Bootstrap values supporting each clade are indicated in bold on nodes. The chromosomal rearrangements, which have occurred within the genus Mus are mapped onto the tree and are numbered in grey. Each rearrangement is coded as follows: transl, translocation; inv, inversion; fiss, fission; fus, fusion followed by the character numbers of the table in the electronic supplementary material. Asterisk indicates the emergence of a new centromere. Rearrangements 1–3: transl* + inv 52–53; fiss* 3—rearrangements 4–11: inv 14; fus 16; fus 8; fus 58; transl 11; transl* 60; transl* 10; transl* 2—rearrangements 12–14: transl* 57; transl* 56; fus 59—rearrangements 15–19: transl* 1; transl* 18; transl 5; transl* 4; fus 13—rearrangements 20–22: fiss* 25; fiss* 26; fiss* 58—rearrangements 23–29: transl* 66; transl* 65; transl* 67; fus 64; fus 62; fus 61; fus 63. The divergence times mentioned follow Chevret et al. (2005) and Veyrunes et al. (2005).
4. Discussion
(a) Phylogenetic relationships
Despite the number of studies involved and the variety of molecular markers used (see §1), an unambiguous phylogenetic tree for the genus Mus has remained elusive. In such a context, subgeneric relationships within Mus were investigated using chromosomal rearrangements. They constitute alternative genetic markers with low levels of convergence, and are highly informative as being under-dominant mutations, they are fixed (or lost) rapidly in the populations, contrary to genes which may remain polymorphic for long periods of time.
Our phylogenomic analysis yielded a single most parsimonious tree in which the subgeneric relationships were resolved for the first time (figure 4). The nodes were well supported by the standard method of bootstrap, and more importantly, each one was supported by several unique non-ambiguously identified chromosomal rearrangements providing strong confidence to the topology retrieved. The first subgenus to diverge is Coelomys, followed by Nannomys at the base of a Mus–Pyromys clade. This topology was previously suggested by Lundrigan et al. (2002), but was not supported by high bootstrap values (except for one nuclear marker Tcp-1). Other studies have also clustered Mus with Pyromys, although with similarly weak support (e.g. Catzeflis & Denys 1992; Chevret et al. 2005). Curiously, the lowest bootstrap value in our study (equal to 75) defines the monophyly of the genus, a node consensually supported by a variety of molecular datasets (e.g. Lundrigan et al. 2002; Chevret et al. 2003, 2005; Suzuki et al. 2004; Veyrunes et al. 2005).
Palaeontological and molecular data indicate that the genus Mus originated in Asia, with the oldest true-Mus fossil reported from Pakistan from the Late Miocene (e.g. Suzuki et al. 2004; Chevret et al. 2005). The resolved cytogenetically based phylogenetic tree allowed us to order the dispersal events that took place during the evolution of the genus into three successive bursts of differentiation that occurred approximately 7 Myr ago (Chevret et al. 2005; Veyrunes et al. 2005), the first one involving Coelomys in southeastern Asia, followed by Nannomys with the colonization of Africa via the Middle East, then Mus in Eurasia and Pyromys in Southeast Asia and the Indian subcontinent.
(b) Genome comparison and ancestral Mus karyotype
Although the house mouse is perhaps the most widely studied mammal in terms of chromosomal evolution (e.g. Britton-Davidian et al. 2000; Capanna & Castiglia 2004; Piàlek et al. 2005), comparisons between subgenera within the genus Mus are very scarce. Thus, our cross-species multidirectional chromosome painting involving one representative species belonging to all four subgenera represents the first attempt to establish genome-wide comparative chromosome maps in the genus. Moreover, comparison of these data with those for five other murid genera (Yang et al. 2000; Cavagna et al. 2002; Rambau & Robinson 2003; Matsubara et al. 2004; Engelbrecht et al. 2006) has shed light on shared primitive and derived chromosomal syntenies in the murid lineage. The results reveal that drastic genome shuffles have occurred in the genus Mus. Several Mus autosomes are retained as complete chromosomes or chromosome blocks (e.g. 3, 12, 16 and 19), whereas others have undergone considerable disruption (e.g. 5 and 17). The small-sized chromosome 17 shows extensive fragmentation in all karyotypes, hybridizing to nine regions in the Chinese hamster chromosomes, eight in R. pumilio, four in Otomys irroratus, five in the black rat (Rattus rattus) and six in Apodemus. These results suggest that the synteny of mouse chromosome 17 evolved recently. Typically, the X chromosome is conserved across all taxa (e.g. Graves et al. 2002). Although intrachromosomal rearrangements usually escape detection by chromosome painting, the pattern shown by several syntenic associations allowed us to detect two inversion events. Thus, the 8/7/8/7 synteny on chromosome 19 of C. pahari provides evidence that a paracentric inversion occurred (figure 3b). In the same way, the combination 13a/15/13b on chromosome 11 in this same species, which is also present in the Apodemus karyotype (Matsubara et al. 2004, but not detected in Stanyon et al. 2004), suggests that it is the ancestral state, and was subsequently modified by an inversion (13a+b/15) in the lineage leading to three other Mus subgenera. The data allow us to reconstruct the likely ancestral karyotype of the genus Mus. This was done by mapping changes along the phylogenetic tree (figure 4) and inferring ancestral character states at the different nodes by listing shared syntenies between all ingroup species, or between at least one ingroup and an outgroup. Examples include the widespread associations 7/19, 10/17 and 13/15 found in species of all the investigated genera, the synteny 5/6 also observed in Rattus and Apodemus and 2/13 in Cricetulus and Rattus. Finally, the subgenera Coelomys and Nannomys share synteny 1/4 suggesting that it was present in the recent ancestor of the genus Mus. In summary, the ancestral Mus karyotype is thought to consist of 2n=46 acrocentric chromosomes (figure 5). It shares 13 autosomal pairs conserved in toto (block or synteny 7/19, 2, 3, 14, 10, 9, 11/5, 18, 2/13, 15, 8, 8, 17/10) with the 2n=54 ancestral murid karyotype proposed by Stanyon et al. (2004). The extensive repatterning of the house mouse karyotype has often led to its exclusion from most interspecific genomic comparisons (e.g. Richard et al. 2003; Stanyon et al. 2003). In contrast, the more representative Mus ancestral karyotype may be a helpful substitute for large-scale comparisons of genome organization.
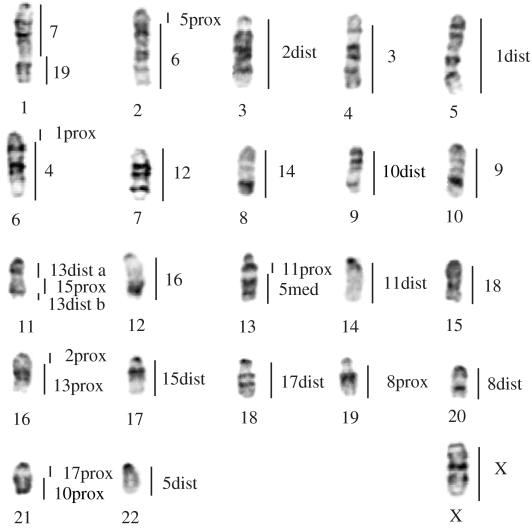
Figure 5.
Inferred haploid set of the ancestral karyotype of the genus Mus (2n=46) reconstructed with Coelomys, Nannomys and Mus G-banded chromosomes. Homology to Mus chromosomes is indicated to the right of each putative ancestral chromosome. ‘dist’, ‘med’ and ‘prox’ refer to the distal, median and proximal segment of the chromosome, respectively, and a and b refer to unidentified subchromosomal segments.
(c) Rates of genome reorganization in the genus Mus
Our analyses provide insight into rates of chromosomal evolution in the genus Mus. The chromosomal phylogeny identified 29 rearrangements that have been fixed during the diversification of the genus Mus. The subgenus Coelomys has a conserved karyotype, which differs from the ancestral one by only three rearrangements, whereas the others have undergone greater genome shuffles, with 11 rearrangements in Nannomys, 16 in Mus and 20 in Pyromys (figure 4). This analysis clearly shows that extensive genome repatterning is not unique to the house mouse karyotype, since only three rearrangements are autapomorphic, but is in fact a characteristic of the Mus lineage within the Muridae. In addition, the rearrangements are not randomly distributed along the branches (figure 4). The four subgenera of Mus diverged nearly simultaneously within 1 Myr during which nearly half of the rearrangements occurred, representing a rate of 13 mutations per million years. In contrast, as few as 3–7 were subsequently fixed in the terminal branches leading to the four subgenera during the last 6–7 Myr (Chevret et al. 2005; Veyrunes et al. 2005), yielding a rate range between 0.4 and 1.2 Myr−1. Thus, the pattern of karyotypic evolution exhibits a short phase of intensive diversification followed by a stage with a lower rate of chromosomal change. In Mus, this rate acceleration is concomitant with cladogenetic events, i.e. the separation of the four subgenera. Hence, we are tempted to correlate the karyotypic diversification with the speciation events on the basis that such an accumulation of rearrangements may lead to reproductive isolation (e.g. King 1993; Rieseberg 2001; Delneri et al. 2003; Olmo 2005). Such data provide additional support for higher rates of chromosomal reorganization in murids compared to other mammalian lineages, which generally display a low rate of chromosome exchange, of the order of 0.1–0.2 mutations per million years, although drastic karyotype reshuffling has also been evidenced in several lineages (O'Brien et al. 1999; Wienberg 2004). However, even within the murids, the evolution of genome structure in the genus Mus is remarkably extensive. For example, the 20 mouse paints revealed 37 homologous segments in R. rattus (Cavagna et al. 2002), which is only slightly higher than in Coelomys (equal to 35), but the divergence Mus/Rattus occurred 11–12 Myr ago, which is twice that estimated between the two subgenera of Mus (Chevret et al. 2005). Moreover, a chromosome painting survey in Apodemus, the only other Eurasian murine genus that matches Mus in terms of species diversity and geographic range, reveals the presence of only one translocation plus a few inversions among species belonging to the four major clades (Matsubara et al. 2004), even though their diversification was estimated to have occurred prior to the radiation of Mus (Michaux et al. 2002).
(d) Modes of genome reorganization in the genus Mus
The chromosomal changes that have occurred during the Mus radiation are mapped onto the branches of the phylogeny (figure 4), and therefore, are a posteriori polarized (e.g. fusion versus fission) using the outgroup criterion. Among the 29 rearrangements identified in the genus, the majority are translocations (14), followed by fusions (9) and fissions (4). Very few inversions were identified (2) which is likely due to the painting protocol used (discussed earlier) and would require more refined approaches to be identified (Zhao et al. 2004). Although we cannot assess the frequency of inversions in these species, the observed preponderance of translocations is in agreement with recent genome sequence comparisons between human, mouse and/or rat, which indicate that interchromosomal versus intrachromosomal rearrangements are much more frequent in the mouse lineage than in that of the human (Friedman & Hughes 2004) or the rat (Zhao et al. 2004). All fusion events identified are tandem and not Robertsonian (i.e. centromere–telomere instead of centromere–centromere fusion), whereas curiously, the Nannomys subgenus, and even more so the house mouse are taxa prone to the accumulation of Rb fusions (e.g. Veyrunes et al. 2004; Piàlek et al. 2005). This suggests that changes in the structure or nature of the centromere may have recently occurred in these lineages allowing a greater frequency of Rb fusions (Redi et al. 1990). Segmental translocations and particularly fissions necessitate the appearance of centromeres at new locations (figure 4). The process of emergence of new centromeres remains unclear, and may in fact involve different independent mechanisms, such as reactivation of ancestral latent centromeres, chromosome healing by telomere sequence seeding, or prior segmental duplications of pericentromeric or other sequences (Choo 1997; du Sart et al. 1997; Ventura et al. 2001, 2003, 2004; Amor et al. 2004; Nergadze et al. 2004). By tracking chromosomal segments throughout the phylogeny, the nature of these new centromeres can be ascertained. Thus, a minimum of two involve previous centromere locations, and 14 require possible de novo acquisition of a centromere (i.e. neocentromerization). Thus, neocentromere formation is apparently a recurrent event during the evolution of Mus, mirroring the situation in primates and marsupials (Ventura et al. 2004; Ferreri et al. 2005). One of the neocentromeres, which was identified, appeared following the break of synteny 5prox/6, the flanking regions of which have been studied by comparative cytogenomic mapping (Walentinsson et al. 2001; Thomas et al. 2003). Thomas et al. (2003) uncovered pericentromeric duplications at this breakpoint, the sequence divergence of which allowed them to date the event at 3–7 Myr ago. By including close relatives of the house mouse in our phylogenetic framework, we are able to more accurately time the occurrence of this event (i.e. break of the synteny 5prox/6 by the translocation of 5prox onto 5med (character 4; electronic supplementary material); it occurred before the split of the subgenera Mus and Pyromys dated at 7 Myr (Chevret et al. 2005; Veyrunes et al. 2005). The two rearrangements involving latent centromere location correspond to: (i) the synteny 1prox/4prox (character 55) which appeared along the branch leading to the genus Mus, and was subsequently broken along the branch leading to the Mus/Pyromys clade (character 1) and (ii) the synteny 5dist/8 (character 58) with fusion along the branch leading to the Nannomys/Mus/Pyromys cluster, and fission in the Mus lineage (figure 4). These events are homoplasic, both involving a synteny formation followed by a break further along the tree. These reversals (fusion then fission) suggest that ‘fossil’ (i.e. latent) centromeres may be a hotspot for breakpoints and centromere reactivation.
For the first time, this study using cross-species chromosome painting allows to resolve the long-standing controversial phylogeny of the genus Mus. Conversely, this phylogenetic analysis provides a more accurate assessment of chromosome evolution in the genus. In addition, we highlight chromosomal genomic areas of interest for higher resolution studies (such as gene-mapping, FISH with BACs or in silico genome exploration) on sequence composition of neocentromeres and synteny breakpoints and their involvement in restructuring the mouse genome. The advantage of this phylogenetic framework, involving closely related species within Mus, is the shorter evolutionary timescale than the human– and/or rat–mouse split, allowing us to trace ancestral sequences at breakpoints, and date the rearrangements and associated segmental duplications more precisely.
Acknowledgments
We are grateful to B. Fu, E. Panetto and A. T. Pardini for technical assistance, and F. Bonhomme, M. Marquine, J. A. J. Nel, A. Orth and B. Sicard for collecting specimens. We thank F. Bonhomme for his comments on the manuscript. This study was supported by a CNRS–NRF collaboration (no. 13293 and 15439, 2002–2004), and CNRS-UM II grants to UMR 5554. This is publication ISEM no. 2006-054.
Footnotes
Present address: Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, UK.
Supplementary Material
References
- Amor D.J, Bentley K, Ryan J, Perry J, Wong L, Slater H, Choo K.H.A. Human centromere repositioning “in progress”. Proc. Natl Acad. Sci. USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. doi:10.1073/pnas.0308637101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A, Baertsch R, James Kent W, Haussler D, Eichler E.E. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004;5:R23. doi: 10.1186/gb-2004-5-4-r23. doi:10.1186/gb-2004-5-4-r23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme F. Evolutionary relationships in the genus Mus. Curr. Top. Microbiol. Immun. 1986;127:19–34. doi: 10.1007/978-3-642-71304-0_3. [DOI] [PubMed] [Google Scholar]
- Bonhomme F. Genetic diversity and evolution in the genus Mus. In: Goldowitz W.D, Winner R.E, editors. Techniques for the genetic analysis of brain and behavior. Elsevier Sciences Publishers, BV; North Holland, NY: 1992. [Google Scholar]
- Boursot P, Auffray J.C, Britton-Davidian J, Bonhomme F. The evolution of house mice. Annu. Rev. Ecol. Syst. 1993;24:119–152. doi:10.1146/annurev.es.24.110193.001003 [Google Scholar]
- Britton-Davidian J, Catalan J, Ramalhinho M.G, Ganem G, Auffray J.C, Capela R, Biscoito M, Searle J, Mathias M.L. Rapid chromosomal evolution in island mice. Nature. 2000;403:158. doi: 10.1038/35003116. doi:10.1038/35003116 [DOI] [PubMed] [Google Scholar]
- Capanna E, Castiglia R. Chromosomes and speciation in Mus musculus domesticus. Cytogenet. Genome Res. 2004;105:375–384. doi: 10.1159/000078210. doi:10.1159/000078210 [DOI] [PubMed] [Google Scholar]
- Catzeflis F, Denys C. The African Nannomys (Muridae): an early offshoot from the Mus lineage—evidence from scnDNA hybridization experiments and compared morphology. Israel J. Zool. 1992;38:219–231. [Google Scholar]
- Cavagna P, Stone G, Stanyon R. Black rat (Rattus rattus) genomic variability characterized by chromosome painting. Mamm. Genome. 2002;13:157–163. doi: 10.1007/BF02684021. [DOI] [PubMed] [Google Scholar]
- Chevret P, Jenkins P, Catzeflis F. Evolutionnary systematics of the Indian mouse Mus famulus: molecular (DNA/DNA hybrization and 12S rRNA sequences) and morphological evidence. Zool. J. Linn. Soc. 2003;137:385–401. doi:10.1046/j.1096-3642.2003.00050.x [Google Scholar]
- Chevret P, Veyrunes F, Britton-Davidian J. Molecular phylogeny of the genus Mus (Rodentia: Murinae) based on mitochondrial and nuclear data. Biol. J. Linn. Soc. 2005;84:417–427. doi:10.1111/j.1095-8312.2005.00444.x [Google Scholar]
- Choo K.H.A. Centromere DNA dynamics: latent centromeres and neocentromere formation. Am. J. Hum. Genet. 1997;61:1225–1233. doi: 10.1086/301657. doi:10.1086/301657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucchi T, Orth A, Auffray J.-C, Renaud S, Fabre L, Catalan J, Hadjisterkotis E, Bonhomme F, Vigne J.-D. A new endemic species of the genus Mus (Rodentia, Mammalia) on the island of Cyprus. Zootaxa. 2006;1241:1–36. [Google Scholar]
- de Oliveira E.H.C, Neusser M, Figueiredo W.B, Nagamachi C, Pieczarka J.C, Sbalqueiro I.J, Wienberg J, Muller S. The phylogeny of howler monkeys (Alouatta, Platyrrhini): reconstruction by multicolor cross-species chromosome painting. Chromosome Res. 2002;10:669–683. doi: 10.1023/a:1021520529952. doi:10.1023/A:1021520529952 [DOI] [PubMed] [Google Scholar]
- Delneri D, Colson I, Grammenoudi S, Roberts I.N, Louis E.J, Oliver S.G. Engineering evolution to study speciation in yeasts. Nature. 2003;422:68–72. doi: 10.1038/nature01418. doi:10.1038/nature01418 [DOI] [PubMed] [Google Scholar]
- Dobigny G, Ducroz J.F, Robinson T.J, Volobouev V. Cytogenetics and cladistics. Syst. Biol. 2004;53:470–484. doi: 10.1080/10635150490445698. doi:10.1080/10635150490445698 [DOI] [PubMed] [Google Scholar]
- du Sart D, et al. A functional neo centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nat. Genet. 1997;16:144–153. doi: 10.1038/ng0697-144. doi:10.1038/ng0697-144 [DOI] [PubMed] [Google Scholar]
- Engelbrecht A, Dobigny G, Robinson T.J. Further insights into the ancestral murine karyotype: the contribution of the Otomys–Mus comparison using chromosome painting. Cytogenet. Genome Res. 2006;112:126–130. doi: 10.1159/000087524. doi:10.1159/000087524 [DOI] [PubMed] [Google Scholar]
- Ferreri G.C, Liscinsky D.M, Mack J.A, Eldridge M.D.B, O'Neill R.J. Retention of latent centromeres in the mammalian genome. J. Hered. 2005;96:217–224. doi: 10.1093/jhered/esi029. doi:10.1093/jhered/esi029 [DOI] [PubMed] [Google Scholar]
- Friedman R, Hughes A.L. Two patterns of genome organization in mammals: the chromosomal distribution of duplicate genes in human and mouse. Mol. Biol. Evol. 2004;21:1008–1013. doi: 10.1093/molbev/msh076. doi:10.1093/molbev/msh076 [DOI] [PubMed] [Google Scholar]
- Graves J.A.M, Gecz J, Hameister H. Evolution of the human X—a smart and sexy chromosome that controls speciation and development. Cytogenet. Genome Res. 2002;99:141–145. doi: 10.1159/000071585. doi:10.1159/000071585 [DOI] [PubMed] [Google Scholar]
- Gregory S.G, et al. A physical map of the mouse genome. Nature. 2002;418:743–750. doi: 10.1038/nature00957. doi:10.1038/nature00957 [DOI] [PubMed] [Google Scholar]
- Jouvin-Marche E, Cuddihy A, Butler S, Hansen J.N, Fitch W.M, Rudikoff S. Modern evolution of a single-copy gene: the immunoglobulin Ck in wild mice. Mol. Biol. Evol. 1988;5:500–511. doi: 10.1093/oxfordjournals.molbev.a040514. [DOI] [PubMed] [Google Scholar]
- King M. Cambridge University Press; Cambridge, UK: 1993. Species evolution. The role of chromosome change. [Google Scholar]
- Li T, O'Brien P.C.M, Biltueva L, Fu B, Wang J, Nie W, Ferguson-Smith M.A, Graphodatsky A.S, Yang F. Evolution of genome organizations of squirrels (Sciuridae) revealed by cross-species chromosome painting. Chromosome Res. 2004;12:317–335. doi: 10.1023/B:CHRO.0000034131.73620.48. doi:10.1023/B:CHRO.0000034131.73620.48 [DOI] [PubMed] [Google Scholar]
- Lundrigan B.L, Jansa S.A, Tucker P.K. Phylogenetic relationships in the genus Mus, based on paternally, maternally, and biparentally inherited characters. Syst. Biol. 2002;51:410–431. doi: 10.1080/10635150290069878. doi:10.1080/10635150290069878 [DOI] [PubMed] [Google Scholar]
- Matsubara K, Nishida-Umehara C, Kuroiwa A, Tsuchiya K, Matsuda Y. Identification of chromosome rearrangements between the laboratory mouse (Mus musculus) and the Indian spiny mouse (Mus platythrix) by comparative FISH analysis. Chromosome Res. 2003;11:57–64. doi: 10.1023/a:1022010116287. doi:10.1023/A:1022010116287 [DOI] [PubMed] [Google Scholar]
- Matsubara K, Nishida-Umehara C, Tsuchiya K, Nukaya D, Matsuda Y. Karyotypic evolution of Apodemus (Muridae, Rodentia) inferred from comparative FISH analyses. Chromosome Res. 2004;12:383–395. doi: 10.1023/B:CHRO.0000034103.05528.83. doi:10.1023/B:CHRO.0000034103.05528.83 [DOI] [PubMed] [Google Scholar]
- Michaux J.R, Chevret P, Filippucci M.G, Macholan M. Phylogeny of the genus Apodemus with a special emphasis on the subgenus Sylvaemus using the nuclear IRBP gene and two mitochondrial markers: cytochrome b and 12S rRNA. Mol. Phylogenet. Evol. 2002;23:123–136. doi: 10.1016/S1055-7903(02)00007-6. doi:10.1016/S1055-7903(02)00007-6 [DOI] [PubMed] [Google Scholar]
- Muller S, Hollatz M, Wienberg J. Chromosomal phylogeny and evolution of gibbons (Hylobatidae) Hum. Genet. 2003;113:493–501. doi: 10.1007/s00439-003-0997-2. [DOI] [PubMed] [Google Scholar]
- Murphy W.J, Pevzner P.A, O'Brien S.J. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. doi:10.1016/j.tig.2004.09.005 [DOI] [PubMed] [Google Scholar]
- Murphy W.J, et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. doi:10.1126/science.1111387 [DOI] [PubMed] [Google Scholar]
- Musser G.G, Carleton M.D. Family Muridae. In: Wilson D.E, Reeder D.M, editors. Mammal species of the world. A taxonomic and geographic reference. Smithsonian Institution Press; Washington, DC; London, UK: 1993. pp. 501–755. [Google Scholar]
- Nadeau J.H, Taylor B.A. Lengths of chromosomal segments conserved since divergence of man and mouse. Proc. Natl Acad. Sci. USA. 1984;81:814–818. doi: 10.1073/pnas.81.3.814. doi:10.1073/pnas.81.3.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nergadze S.G, Rocchi M, Azzalin C.M, Mondello C, Giulotto E. Insertion of telomeric repeats at intrachromosomal break sites during Primate evolution. Genome Res. 2004;14:1704–1710. doi: 10.1101/gr.2778904. doi:10.1101/gr.2778904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie W, Wang J, O'Brien P.C.M, Fu B, Ying T, Ferguson-Smith M.A, Yang F. The genome phylogeny of domestic cat, red panda and five mustelid species revealed by comparative chromosome painting and G-banding. Chromosome Res. 2002;10:209–222. doi: 10.1023/a:1015292005631. doi:10.1023/A:1015292005631 [DOI] [PubMed] [Google Scholar]
- Nilsson S, Helou K, Walentinsson A, Szpirer C, Nerman O, Stahl F. Rat–Mouse and Rat–Human comparative maps based on gene homology and high-resolution Zoo-FISH. Genomics. 2001;74:287–298. doi: 10.1006/geno.2001.6550. doi:10.1006/geno.2001.6550 [DOI] [PubMed] [Google Scholar]
- O'Brien S.J, et al. The promise of comparative genomics in mammals. Science. 1999;286:458–481. doi: 10.1126/science.286.5439.458. doi:10.1126/science.286.5439.458 [DOI] [PubMed] [Google Scholar]
- Olmo E. Rate of chromosome changes and speciation in reptiles. Genetica. 2005;125:185–203. doi: 10.1007/s10709-005-8008-2. doi:10.1007/s10709-005-8008-2 [DOI] [PubMed] [Google Scholar]
- Pevzner P, Tesler G. Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc. Natl Acad. Sci. USA. 2003;100:7672–7677. doi: 10.1073/pnas.1330369100. doi:10.1073/pnas.1330369100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pialek J, Hauffe H.C, Searle J.B. Chromosomal variation in the house mouse. Biol. J. Linn. Soc. 2005;84:535–563. doi:10.1111/j.1095-8312.2005.00454.x [Google Scholar]
- Rambau R.V, Robinson T.J. Chromosome painting in the African four-striped mouse Rhabdomys pumilio: detection of possible murid specific contiguous segment combinations. Chromosome Res. 2003;11:91–98. doi: 10.1023/a:1022887629707. doi:10.1023/A:1022887629707 [DOI] [PubMed] [Google Scholar]
- Redi C.A, Garagna S, Zuccotti M. Robertsonian chromosome formation and fixation: the genomic scenario. Biol. J. Linn. Soc. 1990;41:235–255. [Google Scholar]
- Richard F, Messaoudi C, Bonnet-Garnier A, Lombard M, Dutrillaux B. Highly conserved chromosomes in an Asian squirrel (Menetes berdmorei, Rodentia: Sciuridae) as demonstrated by ZOO-FISH with human probes. Chromosome Res. 2003;11:597–603. doi: 10.1023/a:1024905018685. doi:10.1023/A:1024905018685 [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. doi:10.1016/S0169-5347(01)02187-5 [DOI] [PubMed] [Google Scholar]
- Robinson T.J, Fu B, Ferguson-Smith M.A, Yang F. Cross-species chromosome painting in the golden mole and elephant-shrew: support for the mammalian clades Afrotheria and Afroinsectiphillia but not Afroinsectivora. Proc. R. Soc. B. 2004;271:1477–1484. doi: 10.1098/rspb.2004.2754. doi:10.1098/rspb.2004.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Holland P.W.H. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. doi:10.1016/S0169-5347(00)01967-4 [DOI] [PubMed] [Google Scholar]
- She J.X, Bonhomme F, Boursot P, Thaler L, Catzeflis F. Molecular phylogenies in the genus Mus: Comparative analysis of electrophoretic, scnDNA hybridization, and mtDNA RFLP data. Biol. J. Linn. Soc. 1990;41:83–103. [Google Scholar]
- Sourrouille P, Hanni C, Ruedi M, Catzeflis F.M. Molecular systematics of Mus crociduroides, an endemic mouse of Sumatra (Muridae: Rodentia) Mammalia. 1995;59:91–102. [Google Scholar]
- Stanyon R, Yang F, Cavagna P, O'Brien P.C.M, Bagga M, Ferguson-Smith M.A, Wienberg J. Reciprocal chromosome painting shows that genomic rearrangement between rat and mouse proceeds ten times faster than between humans and cats. Cytogenet. Cell Genet. 1999;84:150–155. doi: 10.1159/000015244. doi:10.1159/000015244 [DOI] [PubMed] [Google Scholar]
- Stanyon R, Stone G, Garcia M, Froenicke L. Reciprocal chromosome painting shows that squirrels, unlike rodents, have a highly conserved genome organization. Genomics. 2003;82:245–249. doi: 10.1016/s0888-7543(03)00109-5. doi:10.1016/S0888-7543(03)00109-5 [DOI] [PubMed] [Google Scholar]
- Stanyon R, Yang F, Morescalchi A.M, Galleni L. Chromosome painting in the long-tailed field mouse provides insights into the ancestral murid karyotype. Cytogenet. Genome Res. 2004;105:406–411. doi: 10.1159/000078213. doi:10.1159/000078213 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Shimada T, Terashima M, Tsuchiya K, Aplin K. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 2004;33:626–646. doi: 10.1016/j.ympev.2004.08.003. doi:10.1016/j.ympev.2004.08.003 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Massachusetts, MA: 1999. PAUP*. Phylogenetic Analysis Using Parsimony (*and other Methods), version 4. 0b. [Google Scholar]
- Telenius H, Pelmear A.H, Tunnacliffe A, Carter N.P, Behmel A, Ferguson-Smith M.A, Nordenskjold M, Pfragner R, Ponder B.A.J. Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosomes Cancer. 1992;4:257–263. doi: 10.1002/gcc.2870040311. [DOI] [PubMed] [Google Scholar]
- Thomas J.W, et al. Pericentromeric duplications in the laboratory mouse. Genome Res. 2003;13:55–63. doi: 10.1101/gr.791403. doi:10.1101/gr.791403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Archidiacono N, Rocchi M. Centromere emergence in evolution. Genome Res. 2001;11:595–599. doi: 10.1101/gr.152101. doi:10.1101/gr.152101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, et al. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 2003;13:2059–2068. doi: 10.1101/gr.1155103. doi:10.1101/gr.1155103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, et al. Recurrent sites for new centromere seeding. Genome Res. 2004;14:1696–1703. doi: 10.1101/gr.2608804. doi:10.1101/gr.2608804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrunes F, Britton-Davidian J, Robinson T.J, Calvet E, Denys C, Chevret P. Molecular phylogeny of the African pygmy mice, subgenus Nannomys (Rodentia, Murinae, Mus): implications for chromosomal evolution. Mol. Phylogenet. Evol. 2005;36:358–369. doi: 10.1016/j.ympev.2005.02.011. doi:10.1016/j.ympev.2005.02.011 [DOI] [PubMed] [Google Scholar]
- Veyrunes F, Catalan J, Sicard B, Robinson T.J, Duplantier J.M, Granjon L, Dobigny G, Britton-Davidian J. Autosome and sex chromosome diversity among the African pygmy mice, subgenus Nannomys (Muridae; Mus) Chromosome Res. 2004;12:369–382. doi: 10.1023/B:CHRO.0000034098.09885.e6. doi:10.1023/B:CHRO.0000034098.09885.e6 [DOI] [PubMed] [Google Scholar]
- Walentinsson A, Helou K, Levan G. A dual-color FISH gene map of the proximal region of rat chromosome 4 and comparative analysis in human and mouse. Mamm. Genome. 2001;12:900–908. doi: 10.1007/s00335-001-2090-2. doi:10.1007/s00335-001-2090-2 [DOI] [PubMed] [Google Scholar]
- Wienberg J. The evolution of eutherian chromosomes. Curr. Opin. Genet. Dev. 2004;14:657–666. doi: 10.1016/j.gde.2004.10.001. doi:10.1016/j.gde.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Yang F, O'Brien P.C.M, Wienberg J, Ferguson-Smith M.A. A reappraisal of the tandem fusion theory of karyotype evolution in the Indian muntjac using chromosome painting. Chromosome Res. 1997;5:109–117. doi: 10.1023/a:1018466107822. doi:10.1023/A:1018466107822 [DOI] [PubMed] [Google Scholar]
- Yang F, O'Brien P.C, Ferguson-Smith M.A. Comparative chromosome map of the laboratory mouse and Chinese hamster defined by reciprocal chromosome painting. Chromosome Res. 2000;8:219–227. doi: 10.1023/a:1009200912436. doi:10.1023/A:1009200912436 [DOI] [PubMed] [Google Scholar]
- Zhao S, et al. Human, Mouse, and Rat genome large-scale rearrangements: stability versus speciation. Genome Res. 2004;14:1851–1860. doi: 10.1101/gr.2663304. doi:10.1101/gr.2663304 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.