Abstract
The introduction of avian malaria (Plasmodium relictum) to Hawaii has provided a model system for studying the influence of exotic disease on naive host populations. Little is known, however, about the origin or the genetic variation of Hawaii's malaria and traditional classification methods have confounded attempts to place the parasite within a global ecological and evolutionary context. Using fragments of the parasite mitochondrial gene cytochrome b and the nuclear gene dihydrofolate reductase-thymidylate synthase obtained from a global survey of greater than 13 000 avian samples, we show that Hawaii's avian malaria, which can cause high mortality and is a major limiting factor for many species of native passerines, represents just one of the numerous lineages composing the morphological parasite species. The single parasite lineage detected in Hawaii exhibits a broad host distribution worldwide and is dominant on several other remote oceanic islands, including Bermuda and Moorea, French Polynesia. The rarity of this lineage in the continental New World and the restriction of closely related lineages to the Old World suggest limitations to the transmission of reproductively isolated parasite groups within the morphological species.
Keywords: avian malaria, Plasmodium relictum, Hawaii, phylogeography
1. Introduction
The introduction of avian malaria (Plasmodium relictum) to the remote Hawaiian Islands has been implicated in the widespread decline and the possible extinction of many species within the endemic avian radiation of honeycreepers (Warner 1968; van Riper et al. 1986). While mortality in introduced bird species is negligible, mortality in many endemic species can range from 50 to 90% (Jarvi et al. 2001), possibly reflecting their long isolation (ca 4 Myr; Fleischer & McIntosh 2001) from malarial parasites. Although the epidemiology of malaria in Hawaiian birds has been well studied, little is known about the diversity of parasite strains in Hawaii or their origin. Transmission of malaria was impossible until the human-mediated introduction of a competent vector (Culex quinquefasciatus) to Hawaii in 1826 (Warner 1968). Since then, resident Hawaiian birds may have been exposed to reservoirs of parasites harboured by the hundreds of exotic birds released in the late nineteenth and early twentieth centuries (Long 1981) and by the thousands of ducks and shorebirds which annually migrate to Hawaii from their breeding grounds in the Arctic.
Understanding the host range of P. relictum in Hawaii and elsewhere across the globe is essential for identifying its original host, understanding limits to its transmission and eventually understanding its extreme virulence in native honeycreepers relative to its effects in other hosts. Unfortunately, classical techniques for identifying parasites may confound this understanding. Plasmodium relictum has been reported to occur in a broad spectrum of hosts from all continents except Antarctica (Bennett et al. 1993); however, to date, identification has been based both on morphology, which can vary within strains of the same parasite species (Peirce 1979; van Riper 1991) and on biological characteristics, such as vector, endogenous development and host range (Garnham 1966). These characters may not accurately reflect phylogenetic relationships among parasites (Escalante et al. 1998).
The recent detection of extensive genetic diversity across avian malaria parasites (Ricklefs & Fallon 2002; Waldenström et al. 2002; Beadell et al. 2004) suggests that cryptic structure in parasite populations may underlie differences in host susceptibility, vector competence and parasite virulence. Here, we use molecular markers to characterize Hawaii's avian malaria and place this genotype in a global context in order to better understand its origin, present and historic impacts and limits to its transmission.
2. Material and methods
(a) Lineage identification
Although phylogenetic species limits have not been well defined in avian malaria parasites, a previous study indicated that mitochondrial lineages appear to represent reproductively isolated units (Bensch et al. 2004), and therefore, we characterized the Hawaiian strain and its global distribution using cytochrome b (cyt b). Determination of parasite haplotype frequencies in Europe and Nigeria (table 1) followed PCR methods described by Waldenström et al. (2004). Data for the Lesser Antilles and Venezuela were provided by S. Fallon. Detection and identification of all other parasite mitochondrial lineages generally followed the methods described by Beadell et al. (2004). Briefly, we screened DNA extracted from blood or tissue for parasites using primers F2/R2, 850F/1024R or 213F/372R (Beadell & Fleischer 2005). For positively infected samples, we then sequenced a larger fragment generated with primers 3760F/4292rw2 (533 bp), or with primers Fifi/4292rw2 (351 bp; Ishtiaq et al. 2006) or F2/4292rw2 (295 bp) in cases where degraded template precluded the amplification of the larger piece (see Appendix 1 in the electronic supplementary material). For lineages used in detailed phylogenetic analysis, we also obtained an additional 220 bp of sequence using primers L15368/H15730 (Fallon et al. 2003), which generated a 338 bp fragment overlapping previously generated fragments by 118 bp. Sequences were assembled, aligned and edited using Sequencher v. 4.1. For the purpose of describing the worldwide distribution of Hawaii's parasite lineage (GRW4; Bensch et al. 2000), we identified as GRW4 any sequence that identically matched the largest fragment of parasite cyt b isolated from a Hawaiian host (AY733090). In Korea, we detected the presence of GRW4 based on matching a single 91 bp fragment alone, but in all other locations, detection of GRW4 was based on the recovery of identical sequences of between 256 and 753 bp (see Appendix 1 in the electronic supplementary material). When defining all other lineages, we grouped together only identical sequences exhibiting matching sequence of 256 bp or more. Therefore, within the limits of the sequence examined, parasite lineages are defined by unique mitochondrial haplotypes.
Table 1.
Sampling effort and frequency with which the Hawaiian lineage of Plasmodium (GRW4, lineage 15) was recovered from regions shown in figure 2. (Lineages defined as unique (differing by at least 1 bp) within a given region may be shared between regions.)
| no. | region | total host individuals sampled (n) | total species sampled (n) | host species with Plasmodium (n) | Plasmodium sequences recovered (n) | sequences matching GRW4 (n) | minimum unique lineages (n) |
|---|---|---|---|---|---|---|---|
| 1. | Hawaiian Archipelago | 320 | 17 | 8 | 79 | 78 | 2 |
| 2. | French Polynesiaa | 161 | 8 | 4 | 14 | 14 | 1 |
| 3. | USA | 161 | 21 | 10 | 61 | 0 | 12 |
| 4. | Bermuda | 142 | 14 | 7 | 42 | 39 | 3 |
| 5. | Antilles/Venezuela | 5553 | 169 | 47 | 303 | 2 | 17 |
| 6. | Guyana | 195 | 53 | 22 | 42 | 0 | 23 |
| 7. | Uruguay | 322 | 111 | 33 | 57 | 0 | 13 |
| 8. | Northern Europeb | 2835 | 26 | 19 | 305 | 131 | 36 |
| 9. | Southern Europec | 1151 | 9 | 8 | 206 | 4 | 16 |
| 10. | Nigeria | 827 | 71 | 33 | 101 | 7 | 30 |
| 11. | Western Africad | 656 | 105 | 62 | 174 | 0 | 44 |
| 12. | South Africa | 171 | 15 | 8 | 60 | 1 | 15 |
| 13. | Indian Ocean Islandse | 150 | 20 | 15 | 48 | 23 | 8 |
| 14. | India | 259 | 43 | 23 | 71 | 18 | 23 |
| 15. | Burma | 344 | 133 | 42 | 60 | 0 | 28 |
| 16. | Japan/Korea | 209 | 58 | 26 | 48 | 1 | 15 |
| 17. | Australia/Papua New Guinea | 454 | 106 | 30 | 56 | 3 | 22 |
French Polynesia: Moorea (Society Islands), Nuku Hiva (Marquesas).
Northern Europe: Belarus, Belgium, England, Germany, Lithuania, Sweden.
Southern Europe: France, Israel, Italy, Spain, Ukraine.
Western Africa: Annabon, Bioko, Cameroon, Gabon, Principe, Sao Tome.
Indian Ocean Islands: Anjouan, Fregate, Grand Comore, Madagascar, Mauritius, Mayotte, Moheli, Praslin, Reunion, Rodrigues.
(b) Phylogeny of Plasmodium spp.
To examine the concordance between parasite classification based on morphology and DNA, we assembled cyt b sequence from avian Plasmodium spp. previously identified by morphology and recognized as valid by Bennett et al. (1993). We obtained sequences from GenBank and from DNA extracted from blood smears, which were obtained from the International Reference Collection for Avian Haematozoa (IRCAH; Brisbane, Australia) or from M. Peirce. Classification of parasites in IRCAH smears was checked by C. Atkinson and M. Peirce and smears exhibiting multiple infections (upon visual inspection or after molecular analysis) were not used. We constructed phylogenetic trees using taxa for which we had recovered between 335 and 753 bp of sequence and rooted trees with sequences from Haemoproteus spp., parasites in the sister genus to avian Plasmodium (Perkins & Schall 2002). We estimated phylogenies using minimum evolution (ME; on K2P and GTR distances), maximum likelihood (ML), and maximum parsimony (MP) as implemented by PAUP* (Swofford 1999). For ML analysis, we chose the most likely model of base pair substitution (GTR+I+G) and parameters (pinv=0.5509, shape=0.6505) based on a likelihood ratio test employed by Modeltest v. 3.07 (Posada & Crandall 1998). Bootstrap support was estimated for each method using 1000 replicates.
(c) Phylogenetic analysis of lineages related to GRW4
We assembled a total of 166 unique Plasmodium cyt b sequences collected from our global survey and from GenBank. Owing to the large number of mitochondrial lineages, we initially constructed an ME tree using K2P distances and PAUP* to identify those lineages which shared the most recent common ancestry with GRW4 and which were most relevant in tracing the origin of this lineage. The resulting tree (data not shown) exhibited a clade containing GRW4 (lineages 15–37), a sister clade (lineages 1–14) and two clades immediately ancestral (lineages 38–51); together, these 51 lineages formed a monophyletic clade nested within the other 115 lineages. For subsequent analyses, therefore, we focused only on these 51 lineages plus several lineages with known morphological classification to help polarize the tree. Lineage 56 was included because a representative (AF254962) was originally classified as Plasmodium nucleophilum, however, its status is undergoing revision (G. Valkiunas, personal communication). Employing the longest fragment of cyt b available for each lineage (see Appendix 1 in the electronic supplementary material), we used ML to re-estimate a phylogeny using a model (GTR+I+G, pinv=0.5445, shape=0.7154) chosen by Modeltest. We estimated support for nodes based on 100 replicates. Owing to uncertainty about parasite species limits and since a dichotomously branching tree may not appropriately capture relationships among mitochondrial lineages within a species, we also generated a haplotype network using statistical parsimony as implemented in TCS1.21 (Clement et al. 2000). Lineages were joined at the 95% confidence criterion unless noted. Parasite lineages which could not be joined to GRW4 at the 90% level were excluded.
In order to generate a second, independent estimate of relationships among lineages, we followed the protocols by Bensch et al. (2004) to amplify and sequence a portion of the nuclear gene dihydrofolate reductase-thymidylate synthase (DHFR-TS; 236 bp) from samples for which we had already recovered mitochondrial sequence (see Appendix 2 in the electronic supplementary material). Since nuclear DNA occurs in much lower copy number than mitochondrial DNA, we recovered DHFR-TS from only a fraction of the samples for which we recovered mitochondrial lineages. In addition, we did not include DHFR-TS sequences from samples for which nuclear or mitochondrial sequence provided evidence of multiple infections (e.g. double peaks in the chromatogram or different sequences from different primer sets). Nuclear haplotype Q (AY033582) was derived from Plasmodium gallinaceum but not necessarily from the same strain of P. gallinaceum from which mitochondrial lineage 50 (AY099029) was derived. We used the methods described above to estimate an ML tree using the GTR+I+G (pinv=0.5620, shape=2.0791) model. Bootstrap support was based on 1000 replicates.
To improve the resolution of hierarchical relationships among lineages, we used Bayesian analysis as implemented in MrBayes v. 3.1.1 (Ronquist & Huelsenbeck 2003) to estimate phylogenetic relationships among lineages for which we could combine both mitochondrial and nuclear markers. The parameters of the GTR+I+G model of DNA substitution were allowed to vary independently for each marker within the concatenated dataset. We performed two runs of 25 million generations, each with one cold and three heated chains and sampled the resulting trees every 1000 generations. Graphical plotting of ML scores suggested that stationarity was reached after approximately 100 000 generations; however, we discarded the first million generations as burn-in. Posterior probabilities of the nodes were estimated from the remaining 24 000 trees. Flat priors were assumed for all parameters.
Using a well-supported group of lineages within the Bayesian tree (clade A) and the program Mesquite v. 1.05 (Maddison & Maddison 2004), we calculated the likelihood that ancestral parasites were found in the New World. We employed the Mk1 model and considered ancestral state reconstruction to be significant when raw likelihood scores for the two possible states (in New World or not-in New World) differed by greater than 2 and the proportional likelihood of the best state was greater than or equal to 0.95.
3. Results and discussion
From 245 introduced and endemic-resident forest birds captured during various seasons between 1971 and 1998 on Hawaii, Maui, Molokai, Oahu and Kauai, we recovered only a single mitochondrial lineage of Plasmodium (lineage 15, n=75 sequenced infections, previously identified as GRW4; Bensch et al. 2000) and only one nuclear haplotype (DHFR-TS haplotype G). We detected a second lineage of parasite (lineage 43) in a single migratory golden plover (Pluvialis fulva) from the northwestern Hawaiian Islands; however, we found no evidence for transmission of this parasite to Hawaiian forest birds. This result corroborates reports of just a single morphological subspecies of parasite in Hawaii (P. relictum capistranoae; Laird & van Riper 1981; van Riper et al. 1986) and suggests that the recent expansion of native host populations into low-elevation forests over the last decade (Woodworth et al. 2005) has not been facilitated by the cryptic introduction of different parasite lineages of lower virulence.
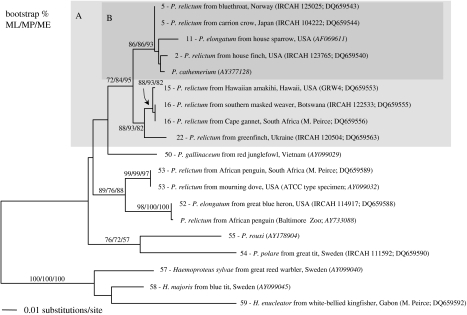
While Hawaiian mitochondrial lineages of P. relictum were monotypic, pairwise divergence of other parasites identified as P. relictum averaged 4.0% (range 0–7.6%), substantially greater than the intraspecific divergence observed in the human parasite P. falciparum across the entire mitochondrial genome (0.2%; Joy et al. 2003) and the divergence observed between sympatric haematozoan parasites restricted to different avian hosts (0.6%; Ricklefs et al. 2004). In addition, phylogenetic analysis of avian malaria parasites classified by morphology indicated that P. relictum does not form a monophyletic clade (figure 1). Deep nodes were generally not well supported, however, the close relationship of one isolate of Plasmodium cathemerium and two isolates of Plasmodium elongatum to P. relictum indicated that either morphology does not reflect evolutionary relationships or that previous classification has been in error. This conflict, as well as the disparity between the broad genetic diversity of parasites identified as P. relictum by morphology and the single type found in Hawaii, suggests that ecological data, such as host and geographical ranges, which have been compiled for parasites classically identified as P. relictum, are not necessarily applicable to the Hawaiian parasite.
Figure 1.
Phylogenetic relationships among morphologically identified species of Plasmodium estimated using ML, MP and ME with cyt b sequences. Numbers above branches indicate bootstrap support based on 1000 replicates. Numbers before species names correspond to mitochondrial lineage numbers in figure 3. Sequences were obtained directly from GenBank (accession number in italics) or from extracts of blood smears obtained from IRCAH and M. Peirce.
Pinpointing the original host is difficult given the broad host range of Plasmodium spp. in general (Bennett et al. 1993; Beadell et al. 2004; Fallon et al. 2005) and of the GRW4 lineage in particular. Worldwide, we recovered GRW4 from 39 species of birds, representing 13 families (see Appendix 1 in the electronic supplementary material). We found the lineage frequently in continental populations of common mynas (Acridotheres tristis) and house sparrows (Passer domesticus), both of which were introduced to Hawaii and also in great reed warblers (Acrocephalus arundinaceus). We did not detect GRW4 in a survey of 75 migratory shorebirds sampled from Hawaii, the French Frigate Shoals and Laysan Island; however, the lineage has been detected in a shorebird from Mauritania (Mendes et al. 2005).
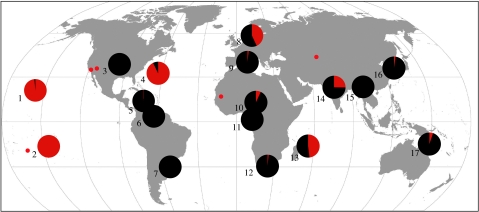
The apparent lack of host-specificity of GRW4 is reflected by its broad geographic distribution. In addition to Hawaii, we detected GRW4 throughout the Old World, where it was particularly common relative to other malaria lineages in Europe (but only in adults of migratory species), India and several Indian Ocean islands (figure 2; table 1). Evidence from our surveys and extensive sampling of thousands of North American birds by R. Ricklefs and co-workers (R. E. Ricklefs, personal communication), however, suggests that the lineage is rare in birds from mainland North and South America. To our knowledge, the only two mainland hosts in which GRW4 has been detected are a house sparrow from California (Schrenzel et al. 2003) and a house finch (Carpodacus mexicanus) from Arizona (M. Kimura, personal communication). The only other New World record of this lineage was derived from two individuals in the Lesser Antilles (Fallon et al. 2005). Given the wide host range of GRW4 and its presence in at least some New World host families (Emberizidae and Mimidae), our failure to detect GRW4 more widely in the New World likely reflects its rarity there and not simply an artefact attributable to differences in the composition of hosts sampled in different regions.
Figure 2.
Map depicting the global distribution of the single mitochondrial lineage of malaria parasite (GRW4) found in resident Hawaiian passerines. Pie charts indicate the proportion of all sequenced Plasmodium infections in a given region that were identical to GRW4 (red). Details concerning locations and sampling effort are in table 1. Red dots indicate additional locations in which GRW4 has been reported previously (Schrenzel et al. 2003; Mendes et al. 2005; Ishtiaq et al. 2006; M. Kimura, personal communication) or in which GRW4 was recovered from a relatively small group of samples (Kazakhstan).
In contrast to its rarity relative to other lineages elsewhere in the New World, GRW4 was the only lineage detected in resident passerines of Bermuda, an oceanic island of volcanic origin lying 1000 km off the coast of North America. Colonization of Bermuda by GRW4 is likely to have occurred only recently since mosquitoes were reported absent from Bermuda by the Spanish sailor Diego Ramirez, who was shipwrecked on the then uninhabited island in 1603 (account published in Wilkinson 1950). Given the subfossil evidence of unique endemic-resident passerines existing in Bermuda prior to human colonization (Olson et al. 2005), it is possible that as in Hawaii, the arrival of a competent vector (C. quinquefasciatus is currently present) and an Old World lineage of Plasmodium may have contributed to the extinction of another island avifauna.
In the Pacific, the Hawaiian form of malaria was also the only lineage of malaria parasite detected in passerines of French Polynesia, though we detected a second lineage exclusively in jungle fowl (Gallus gallus). In a survey of birds from Moorea, we found GRW4 at low frequency in several introduced species including red-browed firetails (Neochmia temporalis; 2 out of 34 individuals), silvereyes (Zosterops lateralis; 1 out of 60 individuals) and common mynas (A. tristis; 2 out of 10 individuals). We did not sample any of the native passerines on Moorea because populations of those species, namely the Tahiti reed warbler (Acrocephalus caffer longirostris), Pacific swallow (Hirundo tahitica) and Polynesian swiftlet (Aerodramus leucophaeus), if extant, were extremely small. However, among a small sample of endemic Marquesan reed warblers (Acrocephalus mendanae) collected in 1987 on Nuku Hiva, we detected GRW4 in 9 out of 11 individuals. Since populations of Marquesan reed warblers remain fairly robust (Holyoak & Thibault 1984; J.-C. Thibault, personal communication), this finding presents the possibility that, unlike its effect on Hawaiian honeycreepers, GRW4 may not pose a threat to these endemic French Polynesian passerines, which are relatively recently diverged from a mainland ancestor (ca 1–2 Ma; Fleischer et al. unpublished data) and of Old World descent. Conversely, for older Polynesian endemics such as the Pomarea flycatchers, which have probably evolved for a longer time in isolation (ca 3.6 Ma; Cibois et al. 2004), the introduction of GRW4 may represent a previously unrecognized factor driving the decline of these species, most of which are threatened or endangered (BirdLife International 2000). The detection of GRW4 in French Polynesia and the Cook Islands (Ishtiaq et al. 2006), and of a closely related parasite in the Marianas (lineage 32), warrants further investigation into the effects of avian malaria on isolated avifaunas outside of Hawaii.
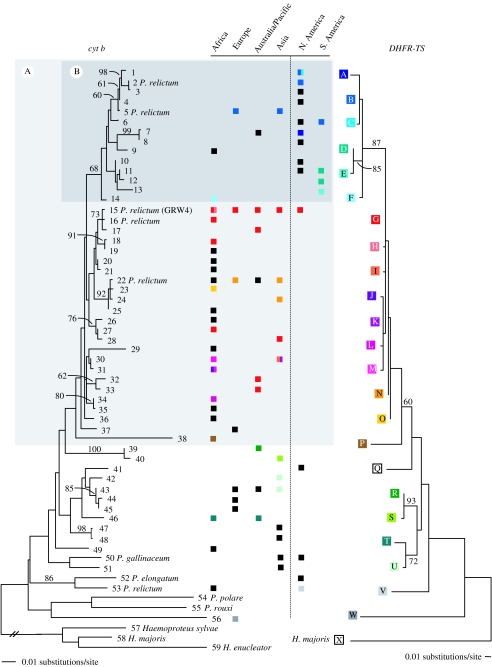
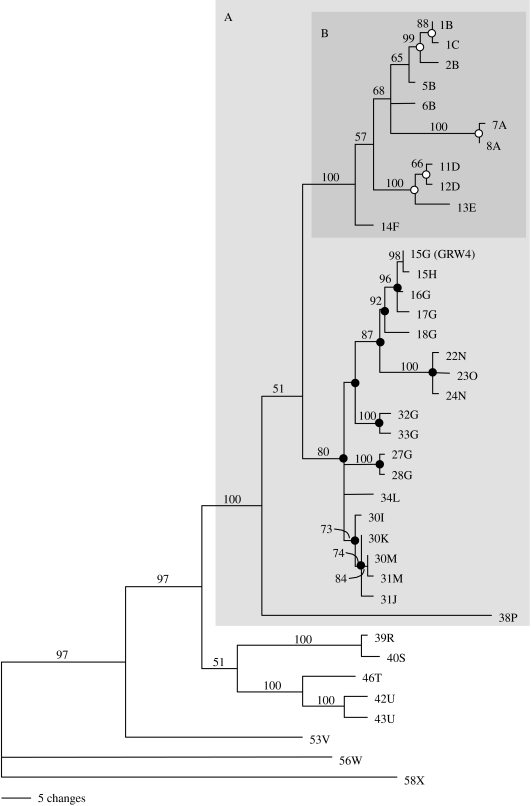
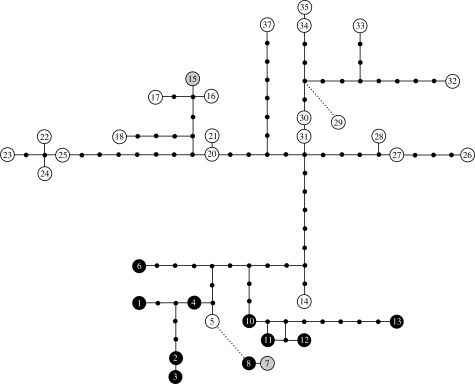
While GRW4 itself exhibited a broad geographical distribution, the distribution of related parasite lineages provided evidence of an Old World ancestry for GRW4. Phylogenetic reconstructions of mitochondrial parasite lineages and associated nuclear haplotypes yielded broadly concordant topologies (figure 3). Although there was little support for deep nodes when loci were analysed separately, Bayesian analysis of data from both mitochondrial and nuclear loci combined recovered similar clustering of parasite lineages and provided support for the monophyly of parasite genotypes 1B to 38P (clade A, figure 4). Within this group, but with the exception of 15G (GRW4), all the parasite genotypes detected in the New World fell within a well-supported clade (clade B, genotypes 1B–14F) that was either sister to or derived from the remainder of genotypes in clade A. Among the remainder of clade A, all genotypes except 15G (GRW4) were recovered exclusively from the Old World and likelihood estimation of ancestral origins confirmed that the immediate ancestors of GRW4 probably derived from the Old World (figure 4). Among the 24 parasite mitochondrial lineages comprising this group, 18 were recovered from hosts in Africa (figure 3). A haplotype network (figure 5), which may more appropriately describe non-bifurcating relationships among mitochondrial lineages derived from a single species, similarly indicated broad geographical substructure within clade A and close association of GRW4 (lineage 15) with Old World lineages recovered from Africa (16) and New Guinea (17). The derived position of GRW4 relative to other lineages from the Old World further suggests that its range has only recently expanded to include parts of the New World.
Figure 3.
Phylogenetic trees of parasite mitochondrial lineages (cyt b; left; numbered) and associated nuclear haplotypes (DHFR-TS; right; lettered), constructed using maximum likelihood (GTR+I+G for both markers). The distribution of mitochondrial lineages across global regions is indicated with squares, colour-coded to help identify the associated DHFR-TS sequence (when available, otherwise black). Background shading reflects the limits of two clades with good support in analysis of combined data (figure 4). Mitochondrial lineages which derived from at least one parasite identified as P. relictum by morphology are indicated on the left. Bootstrap support values (greater than or equal to 60) are indicated above branches.
Figure 4.
Majority rule consensus tree of avian malaria parasite lineages generated by Bayesian analysis of combined mitochondrial (cyt b) and nuclear (DHFR-TS) sequence. Parasite genotypes are identified by their respective cyt b lineage (number) and DHFR-TS haplotype (letter), which are depicted separately in figure 3. Clade credibility values are indicated above branches. Background shading identifies two well-supported clades (A and B) referenced in the text. Dots within clade A indicate nodes for which New World (open circle) or non-New World (black) origin could be confidently assigned based on ancestral trait reconstruction performed with Mesquite.
Figure 5.
Statistical parsimony network of Plasmodium mitochondrial lineages related to the Hawaiian strain (lineage 15). Sampled haplotypes are numbered as in figure 3 and inferred haplotypes are indicated by black dots. Shading indicates whether the lineage was detected in the Old World (white), New World (black) or in both regions (grey). Lineages 9, 19 and 36 were not included due to missing sequence. Lineages 7, 8 and 29 were joined at the 90% connection limit (13 substitutions).
The mitochondrial lineage GRW4 was associated with nuclear haplotype G everywhere except on several Indian Ocean islands where GRW4 was instead associated with haplotype H (figure 3). We identified several additional cases of a single mitochondrial lineage (e.g. 1, 30 and 31) associated with multiple nuclear haplotypes as well as cases of a single DHFR-TS sequence associated with multiple divergent mitochondrial lineages (e.g. haplotype G and lineages 15, 16, 17, 18, 27, 28, 32, 33; figure 3). If every mitochondrial lineage of Plasmodium represents a sexually isolated unit (Bensch et al. 2004), then these results may reflect either incomplete lineage sorting among genes in otherwise reproductively isolated species or insufficient variation in our markers. On the other hand, the sharing of mitochondrial or nuclear haplotypes among different parasite lineages may simply represent intraspecific genetic variation. The resolution of our data precludes investigation of this on a fine scale, however, several cases in which nuclear and mitochondrial haplotypes exhibit similar clustering (e.g. lineages 11, 12, 13 and nuclear haplotypes D, E), combined with an apparent lack of genetic exchange with closely related parasites, provide an indication that species limits may be very narrow. Within the resolution of our data, the complete linkage disequilibrium of mitochondrial and nuclear markers found among parasites in clade B (average cyt b p-dist 1.5%) relative to other parasites in clade A (avg. p-dist 1.9%) and among parasites in clade A (avg. p-dist 2.2%) relative to the next most related lineages suggests that these groups, at least, are reproductively isolated. Except for two parasites, described as P. elongatum and P. cathemerium, all other morphologically described parasites with associated mitochondrial sequences falling within clade A were identified as P. relictum (lineages 2, 5, 15, 16 and 22; figure 3). Given the above results, the morphological taxon P. relictum appears to be composed of at least two, and probably several more, reproductively isolated groups.
The geographical structuring of parasite lineages within clade A is surprising in light of the massive commercial and migratory movement of birds worldwide. Parasites are often lost when their hosts are introduced to novel regions (Torchin et al. 2003; Colautti et al. 2004) and competence of novel hosts, host migration patterns, and competing strains of parasite may retard the exchange of parasites between hemispheres. Nonetheless, the prominence of GRW4 on several remote oceanic islands and its wide host distribution suggest that these are not primary factors limiting the range of GRW4. Instead, differential vector–parasite compatibility may be limiting the transmission of GRW4 and driving the genetic isolation of populations of P. relictum. Vector incompatibility may prevent the transmission of GRW4 in northern Europe (Waldenström et al. 2002) and appears to be responsible for the isolation of New World and Old World forms of Plasmodium vivax (Li et al. 2001), the dominant form of malaria in humans.
We found further evidence for transmission limits in Bermuda, where we recovered three lineages of Plasmodium, but GRW4 was the only lineage of Plasmodium detected in blood from resident Bermuda passerines (n=42 sequenced infections) sampled between 2002 and 2004. Among resident birds, we detected GRW4 in both introduced Old World hosts (house sparrows (P. domesticus) and European starlings (Sturnus vulgaris)) and New World hosts (grey catbirds (Dumetella carolinensis) and Eastern bluebirds (Sialia sialis), but never in white-eyed vireos (Vireo griseus; n=16) or great kiskadees (Pitangus sulphuratus; n=33)). As in Hawaii, Bermuda provides a wintering ground for numerous North American migrants, some of which may exhibit transmissible erythrocytic-stage malaria infections. Assuming that winter parasitemias are not low enough to prevent transmission, the absence of all lineages except GRW4 in Bermuda residents suggests that either resident Bermuda birds are not competent hosts for most North American Plasmodium lineages or that the local vector is refractory to these lineages. The former is unlikely to be true for all resident passerines since many of these species colonized or were introduced to the island from the New World only within the last several hundred years (C. E. McIntosh et al., unpublished work). In addition, we found one of the non-GRW4 lineages (lineage 1) in a migratory ovenbird (Seiurus aurocapillus) in Bermuda and in house sparrows from continental North America, but never in resident house sparrows from Bermuda (n=15 sequenced infections). The other non-GRW4 lineage in Bermuda was recovered from two migratory yellow-throated warblers (Dendroica dominica). This sequence matched a parasite lineage recovered in several other North American species and was only distantly related to lineages in clades A. Given the distinct parasite lineages in resident and migratory species, it appears that refractoriness of the local strain of C. quinquefasciatus to parasites carried by migrants may be important in structuring the parasite community in Bermuda.
If transmission of GRW4 was initially limited to the Old World, as seems possible given the distribution of lineages most closely related to it, the spread and admixture of Old World populations of C. quinquefasciatus with genetically differentiated New World populations (Fonseca et al. 2006) may facilitate the expansion of GRW4 into new locations. Future experimental infections of New World mosquitoes with isolates of P. relictum from different regions could shed light on the mechanism underlying the present rarity of GRW4 in the New World. Given the diversity of lineages encompassed by the morphological taxon P. relictum, future assessment of the ecological and evolutionary impacts of GRW4 and other avian malarias will require a molecular characterization of the pathogen in question. This will be particularly valuable, for example, when identifying an independent source of GRW4 with which to assess coevolutionary models of virulence change in Hawaii. Previous hypotheses of virulence change (van Riper 1991; Atkinson et al. 1995) have been based in part on comparisons of pathogenicity between the Hawaiian parasite and a North American strain that was presumed to be its closest counterpart (van Riper 1991). Our results, which provide evidence of cryptic population structure within P. relictum and an Old World origin for the Hawaiian parasite, should provide a more robust foundation for understanding the evolution of virulence and the dynamics of host–parasite–vector interactions in Hawaii's model system.
Acknowledgments
For sampling of field specimens and access to tissues, we are extremely indebted to R. Adlard, J. Austin, M. Braun, R. Cheke, C. Dove, J. Dumbacher, B. Flint, C. Gebhard, C. Huddleston, P. Jones, P. Marra, J.-Y. Meyer, C. Milensky, K. Murata, S. Olson, M. Ono, B. Peer, T. Pratt, J. Rappole, P. Raust, C. Rehkemper, B. Schmidt, D. Kumar Sharma and curators and collection managers J. Fjeldsa (Zoological Museum University of Copenhagen), R. Brumfeld (LSU Museum of Natural Science Collection of Genetic Resources) and S. Birks (Burke Museum). We thank S. Fallon and R. Ricklefs for valuable summaries of their North American parasite surveys and for comments on previous versions of this manuscript. S. Olson and D. Wingate provided unique insight into Bermudean natural history. C. Dorsey, K. Durrant, C. McIntosh and J. Reed provided assistance in the laboratory. This project was funded by the NIH (GM063258) and supported in part by the USGS Wildlife and Invasive Species Programs. Any use of trade, product or firm names in this publication is for descriptive purposes only and does not imply endorsement by the US Government.
Supplementary Material
Appendix 1: avian hosts, geographical origin, frequency of detection and GenBank accession numbers for parasite mitochondrial (cyt b) lineages. GenBank numbers for sequences obtained from previously published data are in italics. Appendix 2: avian hosts, geographical origin, frequency of detection, associated mitochondrial lineage and GenBank accession numbers for DHFR-TS haplotypes shown in figure 3. DHFR-TS did not amplify from all samples for which a mitochondrial lineage w
References
- Atkinson C.T, Woods K.L, Dusek R.J, Sileo L.S, Iko W.M. Wildlife disease and conservation in Hawaii: pathogenicity of avian malaria (Plasmodium relictum) in experimentally infected iiwi (Vestiaria coccinea) Parasitology. 1995;111:S59–S69. doi: 10.1017/s003118200007582x. [DOI] [PubMed] [Google Scholar]
- Beadell J, Fleischer R. A restriction enzyme-based assay to distinguish between avian hemosporidians. J. Parasitol. 2005;91:683–685. doi: 10.1645/GE-3412RN. doi:10.1645/GE-3412RN [DOI] [PubMed] [Google Scholar]
- Beadell J.S, Gering E, Austin J, Dumbacher J.P, Peirce M, Pratt T.K, Atkinson C.T, Fleischer R.C. Prevalence and differential host-specificity of two avian blood parasite genera in the Australo-Papuan region. Mol. Ecol. 2004;13:3829–3844. doi: 10.1111/j.1365-294X.2004.02363.x. doi:10.1111/j.1365-294X.2004.02363.x [DOI] [PubMed] [Google Scholar]
- Bennett G.F, Bishop M.A, Peirce M.A. Checklist of the avian species of Plasmodium Marchiafava & Celli, 1885 (Apicomplexa) and their distribution by avian family and Wallacean life zones. Syst. Parasitol. 1993;26:171–179. doi:10.1007/BF00009724 [Google Scholar]
- Bensch S, Stjernman M, Hasselquist D, Ostman O, Hansson B, Westerdahl H, Pinheiro R.T. Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc. R. Soc. B. 2000;267:1583–1589. doi: 10.1098/rspb.2000.1181. doi:10.1098/rspb.2000.1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch S, Pérez-Tris J, Waldenström J, Hellgren O. Linkage between nuclear and mitochondrial DNA sequences in avian malaria parasites: multiple cases of cryptic speciation? Evolution. 2004;58:1617–1621. doi: 10.1111/j.0014-3820.2004.tb01742.x. doi:10.1554/04-026 [DOI] [PubMed] [Google Scholar]
- BirdLife International. Lynx Edicions; BirdLife International; Barcelona, Spain; Cambridge, UK: 2000. Threatened birds of the world. [Google Scholar]
- Cibois A, Thibault J.-C, Pasquet E. Biogeography of eastern Polynesian monarchs (Pomarea): an endemic genus close to extinction. Condor. 2004;106:837–851. doi:10.1650/7491 [Google Scholar]
- Clement M, Posada D, Crandall K.A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. doi:10.1046/j.1365-294x.2000.01020.x [DOI] [PubMed] [Google Scholar]
- Colautti R.I, Ricciardi A, Grigorovich I.A, MacIsaac H.J. Is invasion success explained by the enemy release hypothesis? Ecol. Lett. 2004;7:721–733. doi:10.1111/j.1461-0248.2004.00616.x [Google Scholar]
- Escalante A.A, Freeland D.E, Collins W.E, Lal A.A. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc. Natl Acad. Sci. USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. doi:10.1073/pnas.95.14.8124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon S.M, Bermingham E, Ricklefs R.E. Island and taxon effects in parasitism revisited: avian malaria in the Lesser Antilles. Evolution. 2003;57:606–615. doi: 10.1111/j.0014-3820.2003.tb01552.x. doi:10.1554/0014-3820(2003)057[0606:IATEIP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Fallon S.M, Bermingham E, Ricklefs R.E. Host specialization and geographic localization of avian malaria parasites: a regional analysis in the Lesser Antilles. Am. Nat. 2005;165:466–480. doi: 10.1086/428430. doi:10.1086/428430 [DOI] [PubMed] [Google Scholar]
- Fleischer R.C, McIntosh C.E. Molecular systematics and biogeography of the Hawaiian avifauna. Stud. Avian Biol. 2001;22:51–60. [Google Scholar]
- Fonseca D, Smith J, Wilkerson R, Fleischer R. Pathways of expansion and multiple introductions illustrated by large genetic differentiation among worldwide populations of the southern house mosquito. Am. J. Trop. Med. Hyg. 2006;74:284–289. [PubMed] [Google Scholar]
- Garnham P.C.C. Blackwell Scientific Publications; Oxford, UK: 1966. Malaria parasites and other Haemosporidia. [Google Scholar]
- Holyoak D.T, Thibault J.-C. Contribution à l'étude des oiseaux de Polynésie orientale. Mémoires Museum national Histoire naturelle, Paris (sér. A) Zoologie. 1984;127:1–209. [Google Scholar]
- Ishtiaq F, Beadell J, Baker A.J, Rahmani A.R, Jhala Y.V, Fleischer R. Prevalence and evolutionary relationships of haematozoan parasites in native versus introduced populations of common myna Acridotheres tristis. Proc. R. Soc. B. 2006;273:587–594. doi: 10.1098/rspb.2005.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvi S.I, Atkinson C.T, Fleischer R.C. Immunogenetics and resistance to avian malaria in Hawaiian honeycreepers (Drepanidinae) Stud. Avian Biol. 2001;22:254–263. [Google Scholar]
- Joy D.A, et al. Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300:318–321. doi: 10.1126/science.1081449. doi:10.1126/science.1081449 [DOI] [PubMed] [Google Scholar]
- Laird M, van Riper C., III . Questionable reports of Plasmodium from birds in Hawaii, with the recognition of P. relictum ssp. capistranoae (Russell, 1932) as the avian malaria parasite there. In: Canning E.V, editor. Parasitological topics. Allen Press; Lawrence, KS: 1981. pp. 151–165. [Google Scholar]
- Li J, Collins W.E, Wirtz R.A, Rathore D, Lal A, McCutchan T.R. Geographic subdivision of the range of the malaria parasite, Plasmodium vivax. Emerg. Infect. Dis. 2001;7:35–42. doi: 10.3201/eid0701.010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J. Universe Books; New York, NY: 1981. Introduced birds of the world; p. 528. [Google Scholar]
- Maddison, W. P., Maddison, D. R. 2004 Mesquite: a modular system for evolutionary analysis. Version 1.05 http://mesquiteproject.org
- Mendes L, Piersma T, Lecoq M, Spaans B, Ricklefs R.E. Disease-limited distributions? Contrasts in the prevalence of avian malaria in shorebird species using marine and freshwater habitats. Oikos. 2005;109:396–404. doi:10.1111/j.0030-1299.2005.13509.x [Google Scholar]
- Olson, S. L., Wingate, D. B., Hearty, P. J. & Grady, F. V. 2005 Prodromus of vertebrate paleontology and geochronology of Bermuda. In Insular vertebrate evolution: the Palaeontological Approach (ed. J. Alcover & P. Bover). Proc Int Symp Insular Vertebrate Evolution:the Paleontological Approach Monografies de la Societat d'Historia Natural de les Balears, 12, 219–232.
- Peirce M.A. Some additional observations on haematozoa of birds in the Mascarene Islands. Bull. Brit. Ornithol. Club. 1979;99:68–71. [Google Scholar]
- Perkins S.L, Schall J.J. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J. Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Ricklefs R.E, Fallon S.M. Diversification and host switching in avian malaria parasites. Proc. R. Soc. B. 2002;269:885–892. doi: 10.1098/rspb.2001.1940. doi:10.1098/rspb.2001.1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs R.E, Fallon S.M, Bermingham E. Evolutionary relationships, cospeciation, and host switching in avian malaria parasites. Syst. Biol. 2004;53:111–119. doi: 10.1080/10635150490264987. doi:10.1080/10635150490264987 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Schrenzel M.D, Maalouf G.A, Keener L.L, Gaffney P.M. Molecular characterization of malarial parasites in captive passerine birds. J. Parasitol. 2003;89:1025–1033. doi: 10.1645/GE-3163. doi:10.1645/GE-3163 [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 1999. PAUP*: phylogenetic analysis using parsimony (*and other methods) [Google Scholar]
- Torchin M.E, Lafferty K.D, Dobson A.P, McKenzie V.J, Kuris A.M. Introduced species and their missing parasites. Nature. 2003;421:628–630. doi: 10.1038/nature01346. doi:10.1038/nature01346 [DOI] [PubMed] [Google Scholar]
- van Riper C., III The impact of introduced vectors and avian malaria on insular passeriform bird populations in Hawaii. Bull. Soc. Vector Ecol. 1991;16:59–83. [Google Scholar]
- van Riper C, III, van Riper S.G, Goff M.L, Laird M. The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecol. Monogr. 1986;56:327–344. doi:10.2307/1942550 [Google Scholar]
- Waldenström J, Bensch S, Hasselquist D, Ostman O. A new nested polymerase chain reaction method very efficient in detecting Plasmodium and Haemoproteus infections from avian blood. J. Parasitol. 2004;90:191–194. doi: 10.1645/GE-3221RN. [DOI] [PubMed] [Google Scholar]
- Waldenström J, Bensch S, Kiboi S, Hasselquist D, Ottosson U. Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol. Ecol. 2002;11:1545–1554. doi: 10.1046/j.1365-294x.2002.01523.x. doi:10.1046/j.1365-294X.2002.01523.x [DOI] [PubMed] [Google Scholar]
- Warner R.E. The role of introduced diseases in the extinction of the endemic Hawaiian avifauna. Condor. 1968;70:101–120. [Google Scholar]
- Wilkinson H. Spanish intentions for Bermuda 1603–1615. Bermuda Hist. Q. 1950;7:50–89. [Google Scholar]
- Woodworth B, et al. Host population persistence in the face of introduced vector-borne diseases: Hawaii amakihi and avian malaria. Proc. Natl Acad. Sci. USA. 2005;102:1531–1536. doi: 10.1073/pnas.0409454102. doi:10.1073/pnas.0409454102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 1: avian hosts, geographical origin, frequency of detection and GenBank accession numbers for parasite mitochondrial (cyt b) lineages. GenBank numbers for sequences obtained from previously published data are in italics. Appendix 2: avian hosts, geographical origin, frequency of detection, associated mitochondrial lineage and GenBank accession numbers for DHFR-TS haplotypes shown in figure 3. DHFR-TS did not amplify from all samples for which a mitochondrial lineage w