Abstract
We have analyzed two Arabidopsis strains differing in the mean seed size and seed number they produced. The accession Cape Verde Islands (Cvi) yielded on average about 40% fewer seeds than the laboratory strain Landsberg erecta (Ler), but Cvi seeds were almost twice as heavy. Maternal and nonmaternal genetic factors were involved in the seed size variation, and interactions between both types of factors presumably occurred. The Ler/Cvi seed size difference increased through seed development from ovule maturation until seed desiccation, suggesting that multiple processes of seed development were affected. In addition, it involved changes in the final cell number and cell size of the seed coat and the embryo. Cell number variation was controlled mainly by maternal factors, whereas nonmaternal allelic variation mostly affected cell size. By using a recombinant inbred line population derived from Ler and Cvi, we mapped quantitative trait loci (QTLs) affecting 12 life history traits related to seed size, fruit size, seed number, and plant resources. Five of the seed size QTLs colocated with QTLs for other traits, suggesting that they control seed size via maternal components affecting ovule number and/or carpel development, ovule development, or reproductive resource allocation in the mother plant. The six remaining putative seed size QTLs did not show a significant effect on any other trait, suggesting that this allelic variation may be involved specifically in seed development processes.
The size of the seeds produced by a plant has been classically considered an important adaptive character (1, 2). Interspecific seed size variation has been associated with different habitat characteristics (3), and intraspecific variation has been correlated with different fitness components of seedling and adult plants (4, 5). However, as pointed out by Silvertown (2), “the adaptiveness of seed size is one of the clearest examples of the general difficulties inherent in the evolutionary interpretation of a character considered in isolation from the rest” and is still an open question. In particular, seed size is associated with the number of seeds produced by a plant, as suggested by the negative correlations (tradeoffs) frequently found between both characters. Ecological and physiological theories have assumed that this is due to the limited resources of the mother plant (1). Given the modular structure of plants, the total number of seeds they produced can be partitioned into different fitness components (life history traits), namely, the number of seeds per fruit, the number of fruits per inflorescence, and the number of inflorescences. Tradeoffs have been detected in crop plants between seed size and seed number components, but positive correlations have been also found for wild species (1, 2, 6). How many of these correlations are caused by antagonistic pleiotropy (and represent energetic or developmental constraints on the evolution of seed size) and how much might be coadaptation is unknown.
To understand the evolutionary forces driving the seed size phenotypic variation in natural populations, we need to identify the genes responsible for this variation. Unfortunately, because of its quantitative nature, the genetic analysis of this trait has been restricted mostly to the estimation of heritabilities and to comparisons of reciprocal crosses. High heritabilities have been reported for crop species grown under controlled experimental conditions, in contrast to the lower values often found in the wild (2). The analysis of reciprocal crosses in several species have generally shown strong maternal effects, indicating that seed size is influenced by the phenotype of the mother plant, by the maternal genotype of the seeds, and/or by the interactions between the mother plant and the offspring (7).
With the advent of molecular markers and the development of quantitative trait locus (QTL) mapping procedures, the genetic and correlative analyses of quantitative traits have become feasible (8). QTL mapping studies allow the identification of genomic regions controlling a quantitative trait. In addition, the analysis of multiple traits in the same experimental mapping population enables the detection of loci with pleiotropic effects on various characters (9). Several seed size QTL analyses have been performed in domesticated species (10–17). Paterson et al. (14) showed a correspondence of QTLs for seed size, flowering time, and shattering in sorghum, rice, and maize, suggesting that domestication of these species arose from mutations at orthologous loci and that few genes with large effects determine the phenotypes for these traits. However, the results from domesticated species cannot be applied directly to natural populations because artificial selection might have overcome pleiotropy that would be deleterious under natural conditions (18).
In the past few years, the molecular and cellular mechanisms of seed development and seed size have begun to be elucidated. Anatomically and genetically, seeds are complex structures consisting of three parts with different genetic composition: the embryo, the endosperm, and the seed coat or testa. Seeds are formed on the mother plant inside the fruit, and the development of seeds and fruits is tightly coordinated and triggered by fertilization. The analysis of seed mutants in various species shows that seed growth and development is controlled by the interactions among the three seed components and the maternal plant tissues (19–25). Physiologically, seeds behave as sinks where assimilates from maternal sources (leaves) are allocated. Various large seeded plant species such as maize, wheat, pea, and Vicia faba have been used to determine the relationships involved in assimilate partitioning between the mother plant and its offspring. Thus, several genes encoding sugar and amino acid transporters that participate in floem and testa unloading, various key carbohydrate enzymes and genes involved in the regulation of cell division and seed maturation, and hormones like gibberellins have been identified that affect resource allocation (26). This has led to the establishment of a working model for the metabolic control of seed growth (26).
In the present work, we have analyzed natural genetic variation for seed size in the annual weed Arabidopsis thaliana. Inbred genotypes isolated from different natural populations of Arabidopsis are usually referred to as ecotypes in a broad sense. However, because this term is applied to this plant independent of the ecological characteristics of the populations, we will refer to them as accessions. Arabidopsis is a species with considerable seed size variation among populations, as shown by the 3.5-fold difference among the mean seed weights of different accessions described by Krannitz et al. (4). These genetic differences in seed size have been correlated with survival of the seedlings (4). Furthermore, crossing of accessions revealed significant reciprocal effects on seed size over several generations, suggesting that the cytoplasmic genotype might be involved in this variation (27). Here, we have analyzed two Arabidopsis genotypes that show an almost 2-fold difference in their mean seed weight: the pure line Landsberg erecta (Ler), obtained as a mutant (er) from an accession of Northern Europe (28, 29), and Cvi, an accession from the tropical Cape Verde Islands (30). To determine in which seed developmental phases the phenotypic variation originates and whether it is caused by changes in cell size and/or cell number, we have characterized microscopically the seeds of these two lines and the seeds obtained from their reciprocal crosses. To dissect the seed size allelic variation present in Ler and Cvi, we determined the maternal and nonmaternal contributions to seed size and established the number of loci segregating in an experimental population of recombinant inbred lines (RILs) derived from these two genotypes (31). In addition, we have mapped, in the same population, QTLs for 10 other life history traits related with fruit size, seed number, and plant resource availability to investigate which characters might be controlled by the same genes (pleiotropy), by linked genes, or by independently inherited genes.
MATERIALS AND METHODS
Plant Material.
The pure lines Ler and Cvi, a set of 162 RILs derived from reciprocal crosses between the two genotypes and the the isogenic line of the Ler strain Landsberg ERECTA (LER) (28) were used. These lines have been previously characterized for molecular markers (31).
To obtain seeds from reciprocal crosses, Ler and Cvi flowers were emasculated and hand-pollinated with pollen from the other genotype. Control parental seeds were derived by hand pollination with sister plants. To determine whether there is competition for seed growth between seeds within a fruit, flowers were hand-pollinated with different amounts of pollen from sister plants.
Growth Conditions and Measurement of Quantitative Traits.
Plants were grown under long-day light conditions in an air conditioned greenhouse. For the QTL mapping, the complete set of RILs, parental lines, and reciprocal F1 hybrids were evaluated in a single experiment with a blocked design. Two plants in each of the two blocks were measured per RIL, and 4–8 plants for the other genotypes. In each plant, a fruit (in crucifers called silique) from the main inflorescence at a position between 6 and 12 (counted from the lowest silique on the main stem) was dissected under a stereomicroscope, and the following traits were recorded: Fruit length (FL), seed number per fruit (SNF), and number of unfertilized ovules (Unf). The total number of ovules per fruit (ONF) was obtained as SNF + Unf, and the percentage of unfertilized ovules (Unf%) as the (Unf × 100)/ONF. Ovary length (OL) was measured from a flower at stage B3 or 13 (32, 33) at a similar position in the plant. The following traits also were recorded: number of fruits in the main inflorescence (FN), total number of side shoots or inflorescences (SSN) (number of branches in the main inflorescence plus the number of side shoots from the rosette), total leaf number (TLN) (rosette plus cauline leaves), length of the largest leaf (LLL), and plant height (PH). The total number of seeds produced by a plant (TSN) was estimated as TSN = (SSN + 1) × FN × SNF. The seeds from the main inflorescence were harvested, and the mean seed length (SL) of 10 seeds per plant (measured under a stereomicroscope equipped with an ocular calibrated in 5–10 μm units) was estimated. The seed weight (SW) of 100 seeds was determined by using a microbalance (Mettler UM3).
QTL Analyses.
Each trait was analyzed separately by using the mean values of the four plants per RIL, except for SW, which was obtained as a single measurement from a seed bulk of 12 plants. The data of TLN and Unf% were transformed (log10 and arcsin√, respectively) to improve the normality of the distributions. Two RILs showed very high sterility, which affected the phenotype of most traits under study; therefore, they were eliminated from most the analyses. A set of 99 markers covering nearly all of the Arabidopsis genetic map at average intervals of 5 centimorgan was selected from the Ler/Cvi RIL map (31). The computer program mapqtl version 3.0 [J. W. van Ooijen and C. Maliepaard, 1995; Centre for Plant Breeding and Reproduction Research–Dienst Landbouwkunding Onderzoek (CPRO-DLO), Wageningen, The Netherlands] was used to identify and locate QTLs linked to these molecular markers by using interval mapping and multiple-QTL model mapping methods as described in its reference manual (http://www.cpro.dlo.nl/cbw/mapping/). A logarithm-of-odds (LOD) score threshold of 2.8 was applied to declare the presence of a QTL, which corresponds to a genomewide significance α = 0.05 (J. W. van Ooijen, personal communication); two-LOD support intervals were established as ≈95% confidence intervals (34). The estimated additive effect and the percentage of variance explained by each QTL, and the total variance explained by all of the QTLs affecting a trait, were obtained with mapqtl in the final multiple-QTL model (MQM) in which one marker cofactor was fixed per QTL.
Two-way interactions were searched for among all pairwise combinations of the 99 nuclear markers as well as the cytoplasmic genotype, by using the computer program epistat (35) at a significance threshold of P < 0.0005.
Cytoplasmic effects in the RIL population were analyzed by using one-way ANOVA and by using multiple-factor linear models in combination with the QTL markers affecting each trait. The statistical package spss 7.5 was used for statistical comparisons.
Microscopic Analyses.
For the measurement of fruit and seed growth along seed development, single flowers from the main inflorescence at stage B3 or 13 (32, 33) were tagged daily in 14 Ler plants and 10 Cvi plants. A single plant was analyzed every 2–3 days, so at the end most of the developmental stages were collected from different fruit positions in both genotypes. The FL was measured, and the seeds were cleared for microscopy as described (22). The average developmental stage of the embryos from each fruit was determined, and the length of 10 seeds per fruit at that stage was measured under a Nikon microscope equipped with Nomarski optics. For the determination of cell number and cell size in the outer layer of the seed coat, ripe seeds sitting on their radicle side were directly photographed under a scanning electron microscope (Jeol JSM 5200). From these views, the number of cells in a single row along the largest seed length was counted, and the mean cell area of 10 cells was determined by using the UTHSCSA imagetool program (Health Science Center at San Antonio, University of Texas; ftp://maxrad6.uthscsa.edu). For the determination of cell number and size in the hypocotyls, embryos excised from ripe seeds were stained in a solution of 0.1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) and immediately photographed under a Nikon Fluorophot microscope. The number of epidermal cells in a single row along the hypocotyl and the mean cell area of 10 cells was determined.
RESULTS
Variation for Life History Traits: Phenotypes of Ler, Cvi, and Their Reciprocal Hybrids.
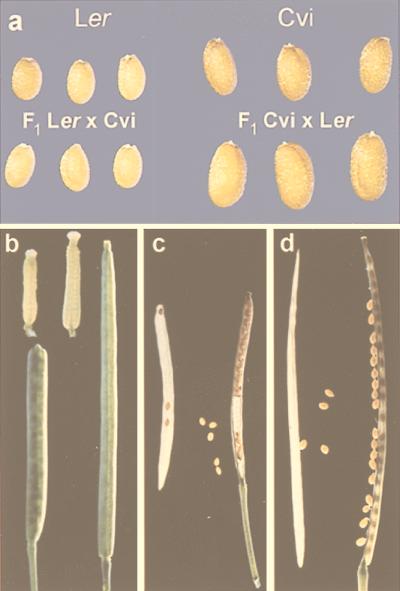
Seeds from the parental line Cvi were, on average, 21% longer and 81% heavier than Ler seeds (Table 1; Fig. 1). Hybrid seeds (Ler × Cvi) obtained by using Ler as mother plant were not significantly different in size from Ler seeds in several independent experiments, although they were consistently slightly smaller. In contrast, seeds from the reciprocal cross using Cvi as mother were on average 18% longer and 40% heavier than Cvi seeds. The reciprocal differences disappeared in the next generation (Table 1), the mean size of the seeds obtained from both reciprocal hybrid plants being similar and lying between the parental values.
Table 1.
Arabidopsis life history traits
| Genotype | Seed length, μm | Seed weight, mg per 100 seed | Fruit length, mm | Ovary length, mm | Ovule number per fruit | Seed number per fruit | Unfertilized percentage | Total seed number × 1,000 | Fruit number | Side shoot number | Plant height, cm | Largest leaf length, mm | Total leaf number |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cvi | 577 ± 6 | 3.51 ± 0.08 | 17.5 ± 1.6 | 2.43 ± 0.06 | 55.5 ± 5.2 | 42.9 ± 6.9 | 21.7 ± 7.8 | 9.9 ± 1.5 | 41.4 ± 2.6 | 4.6 ± 0.5 | 36.7 ± 1.6 | 50 ± 1.3 | 13.5 ± 2.3 |
| *606 ± 20 | — | 14.5 ± 1.9 | |||||||||||
| Ler | 478 ± 17 | 1.93 ± 0.10 | 13.0 ± 0.9 | 1.95 ± 0.15 | 66.4 ± 3.9 | 59.6 ± 8.4 | 9.2 ± 9.9 | 14.0 ± 2.5 | 46.5 ± 4.2 | 4.1 ± 0.8 | 29.5 ± 1.8 | 38.3 ± 2.1 | 10.0 ± 1.6 |
| *482 ± 4 | — | 12.7 ± 0.9 | |||||||||||
| LER | 495 ± 12 | 2.03 ± 0.06 | 16.6 ± 0.4 | 2.23 ± 0.17 | 79.4 ± 3.2 | 77.9 ± 3.4 | 1.7 ± 1.5 | 26.2 ± 3.9 | — | 3.7 ± 0.7 | 62.7 ± 2.8 | 48.8 ± 2.9 | 11.8 ± 1.7 |
| F1 Ler × Cvi | 18.9 ± 0.6 | 2.33 ± 0.17 | 72.3 ± 5.2 | 63.6 ± 4.5 | 1.9 ± 2.7 | 12.7 ± 2.1 | 45.5 ± 4.5 | 3.4 ± 0.5 | 45.6 ± 3.7 | 45.5 ± 3.8 | 11.5 ± 1.0 | ||
| *467 ± 12 | 1.85 ± 0.07 | 13.0 ± 0.8 | |||||||||||
| F1 Cvi × Ler | 18.6 ± 1.3 | 2.16 ± 0.07 | 71.7 ± 4.4 | 62.3 ± 5.6 | 6.2 ± 5.6 | 13.0 ± 2.2 | 40.6 ± 3.6 | 4.1 ± 0.4 | 42.7 ± 3.6 | 43.0 ± 2.9 | 9.7 ± 0.9 | ||
| *681 ± 20 | 4.91 ± 0.06 | 17.5 ± 3.5 | |||||||||||
| F2 Ler × Cvi | 540 ± 6 | 2.80 ± 0.07 | |||||||||||
| F2 Cvi × Ler | 540 ± 11 | 2.89 ± 0.04 | |||||||||||
| RIL mean | 514 ± 30 | 2.6 ± 0.47 | 12.4 ± 2.8 | 1.97 ± 0.23 | 53.3 ± 9.0 | 42.5 ± 12.3 | 17.4 ± 12.5 | 10.3 ± 5.7 | 40.0 ± 10.2 | 4.8 ± 1.8 | 29.1 ± 9.3 | 34.6 ± 9.8 | 11.8 ± 7.3 |
| RIL mean** | 438–611 | 1.45–3.73 | 7.4–20.1 | 1.48–2.74 | 33.5–79.5 | 19.0–74.3 | 0.8–57.5 | 2.5–28.3 | 19.8–69.8 | 2.5–12.8 | 12.6–55.9 | 12.6–55.9 | 5.7–36.4 |
| RILhb2 | 0.92 | — | 0.97 | 0.94 | 0.94 | 0.93 | 0.85 | 0.95 | 0.97 | 0.95 | 0.98 | 0.97 | 0.99 |
Mean phenotypic values ± SD for life history traits of Ler, Cvi, and LER lines, F1 hybrids, and the RIL population (four plants per RIL and eight for the rest of the genotypes were used). Seeds developed on parental lines and on F1 hybrids are referred to as F1 hybrid seeds or F2 seeds, respectively, the mother plant being indicated first in the cross.
, RIL mean given as range of minimum to maximum values. RIL hb2, Broad-sense heritability in the RIL population. ∗, Fruits obtained by hand pollination.
Figure 1.
(a) Seeds of Ler, Cvi, and the reciprocal crosses; (b) Carpels and closed fruits of Ler (Left) and Cvi (Right); Dissected fruits of Ler (c) and Cvi (d).
Ler and Cvi also differed in the estimated TSN (Table 1). Ler plants produced on average more ovules per fruit (20%) and had more fruits on the main inflorescence (12%), but they had fewer lateral inflorescences (12%) than Cvi plants. Furthermore, Cvi plants showed consistently a higher percentage of unfertilized ovules than Ler plants, whereas the F1 hybrids were more fertile. It is estimated that Ler produced in total about 41% more seeds than Cvi plants. FL and OL also were significantly different, Cvi showing on average 34% longer fruits and comparably longer ovaries. The length of fruits obtained in reciprocal crosses was similar to that of the mother plant fruits, indicating a maternal control of fruit size. Consequently, the ovule/seed density within the ovaries/fruits of Ler plants is on average higher than in Cvi (Fig. 1). In addition, Ler fruits are characterized by their blunt tip, which is due to the erecta mutation (28). This characteristic was also present in the ovaries, the overall shape and size of the fruit correlating with the ovary shape and size (Fig. 1). Furthermore, Cvi plants showed larger mean values for the vegetative traits TLN, LLL, and PH (Table 1).
The reciprocal hybrid plants showed small but significant differences for FN, TLN, and SSN (the same number of branches in the main inflorescence but more side shoots from the rosette). In addition, heterotic effects were observed for FL, ONF, SNF, Unf%, and PH.
Developmental Variation for Seed and Fruit Size in Ler and Cvi.
Competition between seeds within a fruit. To analyze whether seeds within a fruit compete for factors affecting final seed size, we investigated the relationship between SNF and the average SL per fruit. As has been previously shown for Arabidopsis (36), the final fruit length was highly correlated with the number of seeds developed within it in both Ler and Cvi (r = 0.95 and 0.94 respectively; n = 90). In Ler plants, there was no significant correlation between mean SL and the SNF, whereas in Cvi plants, the SNF was correlated with mean SL (r = 0.3; P < 0.01). Therefore, the number of seeds developed within a fruit affected little the final mean SL in the fruit.
Seed and fruit growth.
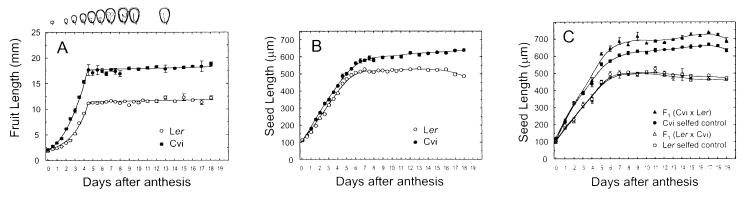
FL and mean SL per fruit were measured in both parental lines throughout the 19 days of their development (Fig. 2). No difference was observed between Ler and Cvi in the timing of fruit growth, the fruits of both genotypes reaching their final length sharply between 4 and 5 days after anthesis, which corresponded approximately to an early heart stage of their embryos. Seed coat and endosperm growth preceded embryo growth, determining the overall final length of the embryo and the seed. The mean ovule size of Ler and Cvi did not differ significantly at time of anthesis, but because of the considerable developmental variation observed within a fruit, it is hard to estimate this at the time of fertilization. For that, Ler and Cvi flowers were emasculated and the ovule size was measured 3 days later. At that time, nonfertilized Ler ovules showed the amphilotropous shape (37), and were slightly longer and narrower than Cvi ovules (3.8%). Therefore, ovule size differences could not account for the final Ler/Cvi seed size variation. After fertilization, the ovule integuments and the endosperm grew rapidly, and 48 hours after anthesis, the ovule had acquired the shape of the premature seed (Fig. 2; ref. 37). No major morphological difference was observed in the seed coat, both lines showing the five typical layers with approximately comparable thickness (22, 37). The timing of embryo development was very similar in Ler and Cvi, but they differed in the rate and timing of the integument and endosperm growth. The developing seeds of Ler reached their final length 6–7 days after anthesis, when the embryo is at an early walking-stick stage. The integuments of Cvi grew faster than the Ler ones and showed a fast growth phase during slightly longer time (12–24 hours), until embryos had reached a late walking stick stage. In addition, Cvi seeds continued growing very slowly but steadily during the cell expansion and storage phase of embryo development (38) and they did not reduce their length, as Ler seeds did, during the desiccation period late in seed development. Measurements of fresh and dry weight did not show any significant difference in relative water content of ripe seeds between both genotypes (Ler 6.2%; Cvi 6.3%) indicating that the continuous seed size increase of Cvi is not caused by reduced water loss but probably by accumulation of storage products. Therefore, the larger size of Cvi seeds compared with Ler is mainly because of the faster and prolonged growth of the integuments and the endosperm during the growth phase of seed development, and this size difference is slowly enlarged during the later phases.
Figure 2.
Fruit and seed growth. (A) Fruit length growth; (B) Seed length growth in Ler and Cvi; (C) Seed length growth in reciprocal crosses. Crosses (fruits hand-pollinated) are compared with self-pollinated fruits from the same plants at one position below, which had approximately synchronous development. The developmental stage of the seed is depicted schematically above A. Means ± SE of a minimum of four (A and B) or three (C) fruits are shown, and distance-weighted least squares curves were fitted.
Compared with Cvi, hybrid seeds (Cvi × Ler) (Fig. 2C) showed similar timing of testa/endosperm growth, but they grew faster during days 4–8 after anthesis, i.e., after the fruit had reached its final length and up to the time that the embryo growth phase ended.
Relationship Between Seed Size and Cell Size and Cell Number.
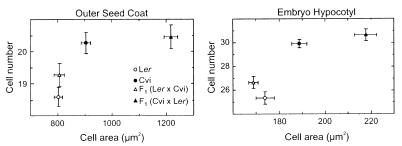
Cell size and cell number of seeds of Ler, Cvi and their reciprocal crosses were estimated in the outer layer of the seed coat and the epidermis of the embryonic hypocotyl (Fig. 3). Ler and Cvi seeds differed significantly in cell size and cell number, both characters correlating with SL. Nevertheless, cell number was the major factor contributing to the Ler/Cvi SL difference, because Cvi had on average 10% and 18% more cells than Ler in the seed coat and hypocotyl, respectively, whereas these cells were on average only 6% and 4% longer. Seeds developed on a Ler mother plant with a Ler or a (Ler × Cvi) hybrid embryo were similar in cell size and number. However, the very large hybrid seeds developed on a Cvi mother plant had a similar cell number than Cvi seeds, but larger cells. Therefore, the heterotic maternal effect observed for seed size is mostly due to an increase in cell size.
Figure 3.
Relationship between cell size and cell number in seeds of Ler, Cvi, and the reciprocal crosses, in two tissues: the outer layer of the seed coat and the epidermis of the embryo hypocotyl. Mean ± SE of 10 seeds are shown.
Variation for Life History Traits: Phenotypes of the RIL Population.
The analysis of the various traits in the Ler/Cvi RIL population showed a large genetic component for all traits, with broad-sense heritabilities varying between 0.85 and 0.99 (Table 1). Transgression in both directions was found for all characters, with the largest transgressions appearing for seed number traits and the smallest ones for seed size traits.
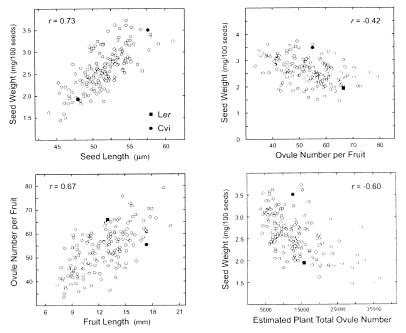
The two seed size parameters, SL and SW, were highly correlated (Fig. 4), and the difference between both reflected variation in seed shape. Seed size traits were negatively correlated with the three ovule/seed number-related traits (r coefficients for SW and ONF/SNF, FN, and SSN between −0.42 and −0.48; P < 0.001) as well as with the vegetative traits TLN and LLL (r values with SW of −0.39 and −0.47. respectively). FN, SSN, TLN, and LLL were all positively correlated (r between 0.5 and 0.65) but ONF/SNF was only correlated with LLL and PH (r between 0.48 and 0.55). In contrast, no significant correlation was observed between SL/SW and FL, although FL correlated strongly with ONF/SNF (Fig. 4). In addition, ONF was highly correlated with SNF (r = 0.81), and ONF also showed a negative correlation with Unf% (r = −0.3; P < 0.001).
Figure 4.
Correlation of RIL means among some life history traits in the Ler/Cvi population. ■ and ● correspond to Ler and Cvi mean values, respectively.
Mapping QTLs for Life History Traits.
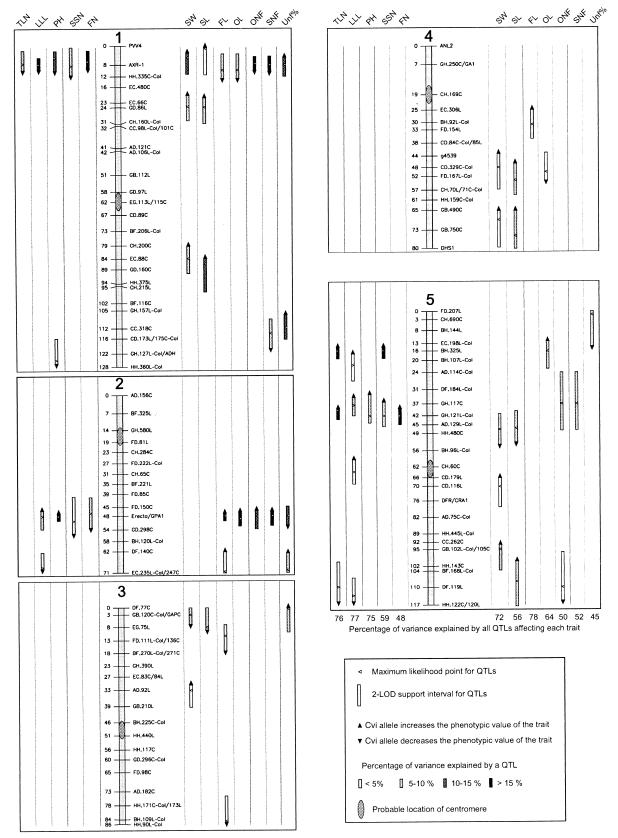
Fig. 5 shows the Ler/Cvi RIL core genetic map with the QTLs identified. Eleven putative loci were detected that affect SW and/or SL, each one explaining between 2.5% and 12% of the phenotypic variance. These loci accounted in total for 56.2% and 71.5% of the phenotypic variance for SL and SW, respectively. The lower arms of chromosomes 4 and 5 showed (with interval mapping or single marker methods) small effects on seed size practically throughout their length, and therefore the number of QTLs detected in these regions might be underestimated. In addition, most of the seed size QTLs showed genetic linkage to other seed size QTLs and, although the use of the MQM facilitated their detection, linkage might have given rise to slight underestimations of the additive effects (van Ooijen and Maliepaard, 1995; http://www.cpro.dlo.nl/cbw/mapping/). At nine of the detected QTLs, Cvi allele increased seed size, in agreement with the small transgression observed in the RIL population.
Figure 5.
QTL mapping of life history traits. The ONF/SNF QTLs detected as interacting with the top of chromosome 1 are represented as blocks lacking the allele effect arrow and spanning the statistically significant region at P < 0.0005.
For the rest of traits, the number of putative QTLs identified varied between three and six, their additive effects explaining in total between 45% and 78% of the phenotypic variance. Searches for two-way QTL interactions were performed for all traits. The seed size QTLs located on top of chromosome 3 and bottom of chromosome 4 showed conditional epistasis (P = 0.0001). In addition, a new ONF and SNF QTL was identified on chromosome 5 around the marker GH.117C as interacting with PVV4, on top of chromosome 1 (P = 0.00001), where a QTL had already been located. This interaction accounted for 6.7% of the variance. In addition, a synergistic interaction was found between the two genomic regions on chromosome 5 affecting TLN, LLL, and SSN. Similar interaction has been previously described for flowering time QTLs in this population (39), suggesting pleiotropic effects of the same loci.
Because the RILs were obtained from reciprocal crosses (31), cytoplasmic contributions could be analyzed, but no significant effect was detected on any of the traits, either additive or as interacting with any of the chromosomal markers.
Colocation of QTLs for different traits (defined as overlapping of their two-LOD support intervals) was observed in multiple genomic regions, in agreement with the positive and negative correlations described above. The large number of QTLs colocating on chromosome 2 were caused by pleiotropy of the ERECTA locus (28, 40), which segregates in the RIL population, as seen from the analysis of the LER line (Table 1).
DISCUSSION
Genetic Control of Life History Traits.
Maternal effects and heterosis. Maternal and nonmaternal genetic factors are involved in the Ler/Cvi seed size allelic variation, as shown by the large difference between reciprocal crosses and the heterosis observed when Cvi is the female parent. No RIL was found with a mean seed size as large as that of the (Cvi × Ler) hybrid seeds. Moreover, the additive effects of the detected QTLs were in agreement with the observed transgression in the RIL population, not accounting for the large hybrid seeds. These results suggest the presence of interactions between the maternal and the nonmaternal genotypes. Significant cytoplasmic effects were detected neither when comparing F1 and F2 seed sizes nor in the RIL population, in contrast to previous work with other Arabidopsis accessions (27). Nevertheless, we could not discard that cytoplasm might contribute to maternal × nonmaternal interactions. In addition, imprinted genes could be implicated, as proposed by Scott et al. (41) for the seed size variation in Arabidopsis interploidy crosses. The large size observed in the (Cvi × Ler) hybrid seeds, and the slightly smaller than Ler mean size of the reciprocal (Ler × Cvi) hybrid seeds could result from the presence of paternally imprinted genes at which Ler alleles increase seed size. However, the seed size differences observed in single fruit crosses, formally, are not directly comparable to seed sizes obtained in RILs, in whole plants, (where all the fruits contain seeds with similar genotypes) because maternal effects might be different. Therefore, even the simplest genetic mechanism of dominance at nonmaternal loci and allele dispersion in the parental lines, in the absence of interactions, cannot be conclusively discarded as an explanation for the reciprocal differences.
Maternal effects, although small, also were detected for traits beyond seedling development (TLN, FN, and SSN). F1 hybrid plants developed from large seeds flowered earlier and with fewer leaves, had fewer fruits in the main inflorescence, and formed more lateral shoots than F1 plants derived from the reciprocal cross. This suggests that maternal differences may affect processes occurring at the shoot meristem that determine the number of organs. It is unknown whether these effects are physiological consequences related to seed size, but it is tempting to think so and to speculate that they may be determined by the amount of hypothetical cotyledon-specific factors such as those regulating node number and flowering time in pea (42).
Heterotic effects were observed for FL, ONF, SNF, Unf%, and PH. However, for all of the traits, transgressive RILs were obtained with higher phenotypic values than the hybrids in agreement with the detection of Ler and Cvi alleles increasing and decreasing each trait. Therefore, dominance at the loci found is the most likely genetic mechanism underlying these phenotypes.
QTL number and QTL effects.
Variation for the mean seed size in the main inflorescence showed a large genetic component and was probably determined by at least 11 loci with relatively small additive effects. The identification of seven SL QTLs at similar positions as SW QTLs indicates that most of these loci affect both parameters. Nevertheless, four of the seed size QTLs were detected only with one of the two parameters, and these loci probably are responsible for seed shape variation. A large genetic component also was found for all of the remaining traits but, in contrast, a smaller number of loci (between three and six) were identified controlling each character, and relatively larger effect QTLs were found for most of them. The relatively low variance explained by the additive effects of the identified QTLs in comparison with the large broad-sense heritabilities estimates for SL, ONF/SNF, Unf%, and FN suggests that (i) other smaller effect loci are involved and (ii) allelic interactions may play an important role in the genetic control of these traits. In agreement, despite the limited power to detect epistasis, several digenic interactions were detected for some of these traits.
Several QTL mapping studies for SW have been performed in crosses between divergent varieties of domesticated plant species or in crosses between cultivated and related wild species (10–17). Relatively dense genetic maps have identified, for different crops, between three and eight QTLs, explaining between 35% and 78% of the phenotypic variance. Large-effect QTLs accounting for >20% of the variance have been found in several species (10, 12, 15, 16). Two of the loci that we identified in the present work had relatively large effects, explaining >10% of the variance. Given the linkage relationships among the various SW QTLs, because of the small genome of Arabidopsis, these effects will probably be underestimated. Therefore, comparable allelic variation appeared to be selected for in domesticated crops and naturally in Arabidopsis for this complex trait, i.e., a relatively large number of small-effect alleles and a small number of larger effect alleles. In addition, we have found large effect alleles (>15% of the variance explained) for several other life history traits such as ONF/SNF, SSN, LLL, and TLN (not taking into account the ERECTA locus because its allelic variation was artificially induced), and they have also been found for flowering time and TLN in other Arabidopsis crosses (39). Most of these traits are affected pleiotropically by allelic variation at the so-called flowering time loci (see below), which indicates that large effect mutations at them control much of the genetic variation for life history phenotypes in natural populations of Arabidopsis.
Comparative QTL Mapping of Life History Traits.
Relationship between seed size and other life history traits. Five of the seed size QTLs colocated with QTLs for some other traits (Fig. 5), their allele effects being in agreement with ecological, physiological, and developmental theory. Hence, these data suggest that the Ler/Cvi allelic variation at these genomic regions control seed size via maternal factors that affect ovule number and/or carpel development, ovule development, or reproductive resource availability in the mother plant.
Three genomic regions (top of chromosome 1, middle and bottom of chromosome 5) contained seed size QTLs as well as QTLs for several components of the ovule/seed number and for several vegetative traits, suggesting that these loci constrain seed size through the ovule/seed number produced by a plant. Thus, these genes will cause a trade-off between seed size and number in Arabidopsis. In addition, flowering time QTLs were previously mapped in these regions by using the same Ler/Cvi RIL population (39). Flowering time mutations are well characterized in Arabidopsis and affect pleiotropically TLN, LLL, SSN, total seed number (43, 44); several of them also affect FN and PH. Because all of the allelic effects on these traits were in agreement with the phenotypes observed in the Arabidopsis mutants, it is very likely that the colocation of most of the ovule/seed number and vegetative trait QTLs is caused by pleiotropy of life history mutations at these three flowering time loci. However, given the relatively low resolution of the QTL mapping, it is not possible to know whether the effects of these regions on seed size and ONF/SNF also reflect pleiotropy at a single flowering locus or represent several closely linked loci. A preliminary analysis of six late-flowering mutants (fca-1, co-3, fve-1, ft-1, gi-3, and fwa-1; data not shown) showed that they have a slightly larger mean SL than the wild type. This indicates that the greater number of seeds produced by induced late-flowering mutations (through SSN and FN) generally does not constrain seed size, probably because of the compensation of more resources in the larger mother plant. The negative correlations observed between seed size and flowering time, TLN, SSN, and FN in the Ler/Cvi population suggest that at least some of these genomic regions contain a second mutation affecting seed size and ONF/SNF closely linked to the flowering mutation. Alternatively, some of these three regions may carry particular alleles at a single flowering locus that also influences carpel development (ONF) and seed size, as suggested by the Arabidopsis late-flowering mutant sin1 (45), which also is affected in fruit and ovule development. In any case, ONF/SNF is probably the main seed number component constraining seed size in the Ler/Cvi variation. This constraint may be exerted physically by the density of ovules in the ovary, because these regions did not affect (or affected relatively less) the OL/FL. The ERECTA gene, the only other locus that affected significantly ONF, also influenced strongly the OL/FL, suggesting that a physical constraint within the fruit is not present in this case. Nevertheless, an energetic constraint on seed size cannot be discarded.
The region on top of chromosome 1 and the ERECTA locus affected not only the ONF but also Unf%. However, two arguments indicate that this partial sterility per se does not constrain significantly the seed size in the Ler/Cvi population. (i) SL within a fruit was weakly or not correlated with the SNF in Cvi and Ler, respectively. (ii) Most of the Unf % QTLs showed no significant effect on seed size. Furthermore, both regions contained the major QTLs affecting OL and FL and, in addition, the effect of the erecta mutation on fruit shape also was present in the ovary. Therefore, even though the actual size of the fruit was determined by the number of seeds developed in it, its potential size and shape is largely established by the initial prefertilization ovary size and shape. A similar relationship has been found in some flesh fruit species such as tomato (46). From ovary to final fruit, both parental lines increased their size to a comparable extent and at a similar rate, suggesting that the number and timing of cell divisions during fruit growth is similar in Ler and Cvi. Therefore, the cell size and number in the fruit are likely to be determined to some extent in the prefertilized ovary, although we do not know whether these size differences involve cell size and/or cell number. In conclusion, the allelic variation at the loci involved in carpel development will probably show pleiotropic effects on seed size and/or ONF and may be allelic to some of the Arabidopsis mutants affected in carpel and fruit development (47).
Another genomic region (top of chromosome 3) showed colocation of seed size and Unf% QTLs, suggesting that allelic variation at a locus involved in ovule development may contribute to the Ler/Cvi seed size differences, as it has also been observed for Arabidopsis ovule development mutants affected in seed shape and size (19, 22). Furthermore, a seed size QTL on chromosome 5 showed an effect on LLL, suggesting that this locus may affect seed size through the reproductive resource availability.
Relationship between seed size and seed development.
About half of the putative seed size QTLs did not show a significant effect on any other trait studied, indicating that a large portion of the Ler/Cvi seed size variation is independent of the maternal ovule/seed number and resource availability components discussed above. Therefore, at least part of this allelic variation is likely to be involved specifically in seed development processes, i.e., may be mutations at loci controlling the resource allocation between the mother plant and the seeds. The control of seed size can be envisioned as the control of growth (cell division and elongation) in the testa, the endosperm, and the embryo. The seed coat and the endosperm growth preceded embryo growth (37, 38), and the seed coat reached almost its final size at about 7 days after anthesis, establishing by then the overall embryo and seed volume. The Ler/Cvi seed size difference was, to a large extent, caused by changes in the rate and duration of seed coat/endosperm growth, although this difference increased throughout seed development from ovule maturation until seed desiccation. Therefore, it is likely that the Ler/Cvi allelic variation affects multiple processes of seed development, involving several seed tissues and interactions. In addition, the Ler/Cvi variation involved changes in the final cell number and cell size of both the testa and the embryo, although the seed coat differences may be determined during ovule development. The cell number variation is controlled mainly by maternal factors, whereas the nonmaternal allelic variation affects mostly the cell size, as indicated by the reciprocal crosses. This cell size variation may involve, hypothetically, endoreduplication (48, 49).
It is likely that some of the loci identified in the present work will correspond to genes that have been recently identified as involved in the metabolic control of cell division and differentiation during seed development or in the developmental control of the different ovule and/or seed components (see the Introduction). It can be expected that the use of Arabidopsis as a model plant for genome analysis (50) will enable the identification of genes corresponding to some of these QTLs, contributing to the further understanding of the natural genetic variation and the evolution of seed size.
Acknowledgments
We thank Dr. P. van Tienderen for critical comments on the manuscript; Dr. G. P. Rédei for providing the LER genotype; Dr. J. W. van Ooijen for supplying mapqtl and for his helpful assistance in the QTL mapping; Dr. K. M. Léon-Kloosterziel for help in the microscopy techniques; Leonie Bentsink for providing the RIL seed weight data. M.K. and H.B.-V. were supported by the Human Frontier Science Program (RG-303/95) and C.A.-B. was supported by the Biotechnology Program of the European Union (BIO4CT965008).
ABBREVIATIONS
- SW
seed weight
- SL
seed length
- ONF
ovule number per fruit
- SNF
seed number per fruit
- Unf%
percentage of unfertilized ovules
- FL
fruit length
- OL
ovary length
- FN
fruit number in the main inflorescence
- SSN
side shoot number
- TLN
total leaf number
- LLL
largest leaf length
- PH
plant height
- QTL
quantitative trait locus
- RIL
recombinant inbred line
References
- 1.Harper J L, Loveli P H, Moore K G. Annu Rev Ecol Syst. 1998;1:327–356. [Google Scholar]
- 2.Silvertown J. Trends Ecol Evol. 1989;4:24–26. doi: 10.1016/0169-5347(89)90013-X. [DOI] [PubMed] [Google Scholar]
- 3.Westoby M, Jurado E, Leishman M. Trends Ecol Evol. 1992;7:368–372. doi: 10.1016/0169-5347(92)90006-W. [DOI] [PubMed] [Google Scholar]
- 4.Krannitz P G, Aarssen L W, Dow J M. Am J Bot. 1991;78:446–450. [Google Scholar]
- 5.Stanton M. Ecology. 1984;65:1105–1112. [Google Scholar]
- 6.Venable D L. Am Nat. 1992;140:287–304. [Google Scholar]
- 7.Roach D, Wulff R. Annu Rev Ecol Syst. 1987;18:209–235. [Google Scholar]
- 8.Tanksley S D. Annu Rev Genet. 1993;27:205–233. doi: 10.1146/annurev.ge.27.120193.001225. [DOI] [PubMed] [Google Scholar]
- 9.Prioul J L, Quarrie S, Causse M, Devienne D. J Exp Bot. 1997;48:1151–1163. [Google Scholar]
- 10.Grandillo S, Tanksley S D. Theor Appl Genet. 1996;92:935–951. doi: 10.1007/BF00224033. [DOI] [PubMed] [Google Scholar]
- 11.Lu C, Shen L, Tan Z, Xu Y, He P, Chen Y, Zhu L. Theor Appl Genet. 1997;94:145–150. doi: 10.1007/s001220050393. [DOI] [PubMed] [Google Scholar]
- 12.Maughan P J, Saghai Maroof M A, Buss G R. Theor Appl Genet. 1996;93:574–579. doi: 10.1007/BF00417950. [DOI] [PubMed] [Google Scholar]
- 13.Mian M A, Bailey M A, Tamulonis J P, Shipe E R, Carter T E, Jr, Parrot W A, Ashley D A, Hussey R S, Boerma H R. Theor Appl Genet. 1996;93:1011–1016. doi: 10.1007/BF00230118. [DOI] [PubMed] [Google Scholar]
- 14.Paterson A H, Lin Y-R, Li Z, Schertz K F, Doebley J F, Pinson S R M, Liu S-C, Stansel J W, Irvine J E. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 15.Rami J-F, Dufour P, Trouche G, Fliedel G, Mestres C, Davrieux F, Blanchard P, Hamon P. Theor Appl Genet. 1998;97:605–616. [Google Scholar]
- 16.Timmerman-Vaughan G M, McCallum J A, Frew T J, Weeden N F, Russel A C. Theor Appl Genet. 1996;93:431–439. doi: 10.1007/BF00223187. [DOI] [PubMed] [Google Scholar]
- 17.Xiao J, Li J, Yuan L, Tanksley S D. Theor Appl Genet. 1996;92:230–244. doi: 10.1007/BF00223380. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell-Olds T. Trends Ecol Evol. 1995;10:324–328. doi: 10.1016/s0169-5347(00)89119-3. [DOI] [PubMed] [Google Scholar]
- 19.Gasser C S, Broadhvest J, Hauser B A. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:1–24. doi: 10.1146/annurev.arplant.49.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Grossniklaus U, Vielle-Calzada J-P, Hoeppner M A, Gagliano W B. Science. 1998;280:446–450. doi: 10.1126/science.280.5362.446. [DOI] [PubMed] [Google Scholar]
- 21.Hong S K, Kitano H, Satoh H, Nagato Y. Development (Cambridge, UK) 1996;122:2051–2058. doi: 10.1242/dev.122.7.2051. [DOI] [PubMed] [Google Scholar]
- 22.Léon-Kloosterziel K M, Keijzer C J, Koornneef M. Plant Cell. 1994;6:385–392. doi: 10.1105/tpc.6.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller E M, Chourey P S. Plant Cell. 1992;4:297–305. doi: 10.1105/tpc.4.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohad N, Margossian L, Hsu Y, Williams C, Repetti P, Fischer R L. Proc Natl Acad Sci USA. 1996;93:5319–5324. doi: 10.1073/pnas.93.11.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swain S M, Reid J B, Kamiya Y. Plant J. 1997;12:1329–1338. doi: 10.1046/j.1365-313x.1997.11030443.x. [DOI] [PubMed] [Google Scholar]
- 26.Weber H, Borisjuk L, Wobus U. Trends Plant Sci. 1997;2:169–174. [Google Scholar]
- 27.Corey L A, Matzinger D E, Cockerham C C. Genetics. 1976;82:677–683. doi: 10.1093/genetics/82.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rédei G P. Z Vererbungsl. 1962;93:164–170. [Google Scholar]
- 29.Rédei G P. In: Methods in Arabidopsis Research. Koncz C, Chua N, Schell J, editors. Teaneck, NJ: World Scientific; 1992. pp. 1–15. [Google Scholar]
- 30.Lobin W. Arabidopsis Inf Serv. 1983;20:119–123. [Google Scholar]
- 31.Alonso-Blanco C, Peeters A J M, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper M T R. Plant J. 1998;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- 32.Müller A. Kulturpflanze. 1961;9:364–393. [Google Scholar]
- 33.Smyth D R, Bowman J L, Meyerowitz E M. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Ooijen J W. Theor Appl Genet. 1992;84:803–811. doi: 10.1007/BF00227388. [DOI] [PubMed] [Google Scholar]
- 35.Chase K, Adler F R, Lark K G. Theor Appl Genet. 1997;94:724–730. [Google Scholar]
- 36.Barendse G W, Kepczynski J, Karssen C M, Koornneef M. Physiol Plant. 1986;67:315–319. [Google Scholar]
- 37.Schneitz K, Hulskamp M, Kopczak S D, Pruitt R E. Development (Cambridge, UK) 1997;124:1367–1376. doi: 10.1242/dev.124.7.1367. [DOI] [PubMed] [Google Scholar]
- 38.Mansfield S G. In: Arabidopsis: An Atlas of Morphology and Development. Bowman J, editor. New York: Springer; 1994. pp. 349–398. [Google Scholar]
- 39.Alonso-Blanco C, El-Assal S E D, Coupland G, Koornneef M. Genetics. 1998;149:749–764. doi: 10.1093/genetics/149.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komeda Y, Takahashi T, Hanzawa Y. J Plant Res. 1998;111:283–288. [Google Scholar]
- 41.Scott R J, Spielman M, Bailey J, Dickinson H G. Development. 1998;125:3329–3341. doi: 10.1242/dev.125.17.3329. [DOI] [PubMed] [Google Scholar]
- 42.Weller J L, Reid J B, Taylor S A, Murfet I C. Trends Plant Sci. 1997;2:412–418. [Google Scholar]
- 43.Koornneef M, Alonso-Blanco C, Peeters A J M, Soppe W. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:345–370. doi: 10.1146/annurev.arplant.49.1.345. [DOI] [PubMed] [Google Scholar]
- 44.van Tienderen P H, Hammad I, Zwaal F. Am J Bot. 1996;83:169–174. [Google Scholar]
- 45.Lang J D, Ray S, Ray A. Genetics. 1994;137:1101–1110. doi: 10.1093/genetics/137.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho L C. In: Fruit and Seed Production. Marshall C, Grace J, editors. Cambridge, U.K.: Cambridge Univ. Press; 1992. pp. 101–124. [Google Scholar]
- 47.Alvarez J, Smyth D R. J Plant Res. 1998;111:295–298. [Google Scholar]
- 48.Davies D R. Heredity. 1977;39:153–163. [Google Scholar]
- 49.Gendreau E, Höfte H, Brown S, Traas J. Plant J. 1998;13:221–230. doi: 10.1046/j.1365-313x.1998.00030.x. [DOI] [PubMed] [Google Scholar]
- 50.Meinke D W, Cherry J M, Dean C, Rounsley S D, Koornneef M. Science. 1998;282:662–682. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]