Abstract
An important effector of Ca2+ signaling in animals and yeast is the Ca2+/calmodulin-dependent protein phosphatase calcineurin. However, the biochemical identity of plant calcineurin remained elusive. Here we report the molecular characterization of AtCBL (Arabidopsis thaliana calcineurin B-like protein) from Arabidopsis. The protein is most similar to mammalian calcineurin B, the regulatory subunit of the phosphatase. AtCBL also shows significant similarity with another Ca2+-binding protein, the neuronal calcium sensor in animals. It contains typical EF-hand motifs with Ca2+-binding capability, as confirmed by in vitro Ca2+-binding assays, and it interacts in vivo with rat calcineurin A in the yeast two-hybrid system. Interaction of AtCBL1 and rat calcineurin A complemented the salt-sensitive phenotype in a yeast calcineurin B mutant. Cloning of cDNAs revealed that AtCBL proteins are encoded by a family of at least six genes in Arabidopsis. Genes for three isoforms were identified in this study. AtCBL1 mRNA was preferentially expressed in stems and roots and its mRNA levels strongly increased in response to specific stress signals such as drought, cold, and wounding. In contrast, AtCBL2 and AtCBL3 are constitutively expressed under all conditions investigated. Our data suggest that AtCBL1 may act as a regulatory subunit of a plant calcineurin-like activity mediating calcium signaling under certain stress conditions.
Calcium signaling mechanisms are widely employed by all eukaryotic organisms to regulate gene expression and a variety of other cellular processes (1, 2). Ca2+ mediates intracellular signaling and regulation of differentiation predominantly through the activity of Ca2+-dependent protein kinases and phosphatases (3). One of the well characterized effector molecules in both animals and fungi is the serine/threonine-specific protein phosphatase calcineurin (4). Calcineurin is a heterodimer composed of a regulatory B subunit (CNB) and a catalytic A subunit (CNA). The enzyme is activated through interaction with Ca2+-stimulated calmodulin (CaM) and acts as an effector of Ca2+ signaling (5, 6). Identification of calcineurin as the molecular target for the immunosupressants cyclosporin A and FK506 has established these drugs as useful tools to dissect the function of this phosphatase in various eukaryotic organisms (7, 8). In animals, calcineurin is involved in a variety of signaling pathways, including those for T cell activation (7–9) and neuronal functions (10, 11). In yeast, calcineurin mediates recovery from pheromone arrest and adaptation to high-salt stress (12–15).
By applying immunosuppressive drugs as probes, we have shown that a calcineurin-like activity is critical for the modulation of a plasma membrane ion channel activity in guard cells (16). In addition, a calcineurin enzyme from animal sources regulates ion channels in the plasma membrane and tonoplast of guard cells and aleurone cells (16–18). A recent study (19) also showed that overexpression of yeast calcineurin enhances salt tolerance in transgenic tobacco. Despite increasing evidence for calcineurin function in higher plants, the molecular nature of the plant calcineurin-like activity has remained elusive.
We report here the molecular and biochemical characterization of a CNB-like protein (AtCBL) from Arabidopsis. This Ca2+-binding protein is most similar to CNB from animals and is able to interact with the CNA subunit from rat as revealed by protein–protein interaction studies in the yeast two-hybrid system, and this interaction restores the salt tolerance to yeast CNB mutant. AtCBL1 gene expression is strongly regulated by stress signals such as drought, cold, and wounding, consistent with a possible function of this protein in plant stress responses.
MATERIALS AND METHODS
Cloning, cDNA Sequence Analysis, and DNA Blot.
PCRs were performed for 35 cycles (1 min 94°C, 1 min 40°C, 1 min 72°C) using 2 μl of phage stock of an Arabidopsis cDNA library (20) as a template. One of the primer pairs (PCD1, 5′-ATCTA(C/T)GA(C/T)ATGGAN-3′; PCD2, 5′-(A/C)(A/C)CIAATIGTCTT(C/T)TCN-3′ yielded a PCR product of the expected size (shown in Fig. 1A). The PCR products were gel purified and cloned into PCRII TA vector as described by the manufacturer (Invitrogen). A 1- to 2-kb size-selected Arabidopsis cDNA library (20) was screened to isolate full-length cDNA clones. Clones were sequenced with an AutoRead Sequencing kit (Pharmacia) and analyzed on ALF (Pharmacia). Sequences were analyzed by using dnastar software, and comparison to databases was performed by using the blast program. DNA isolation and gel blot analyses were carried out as described (21).
Figure 1.
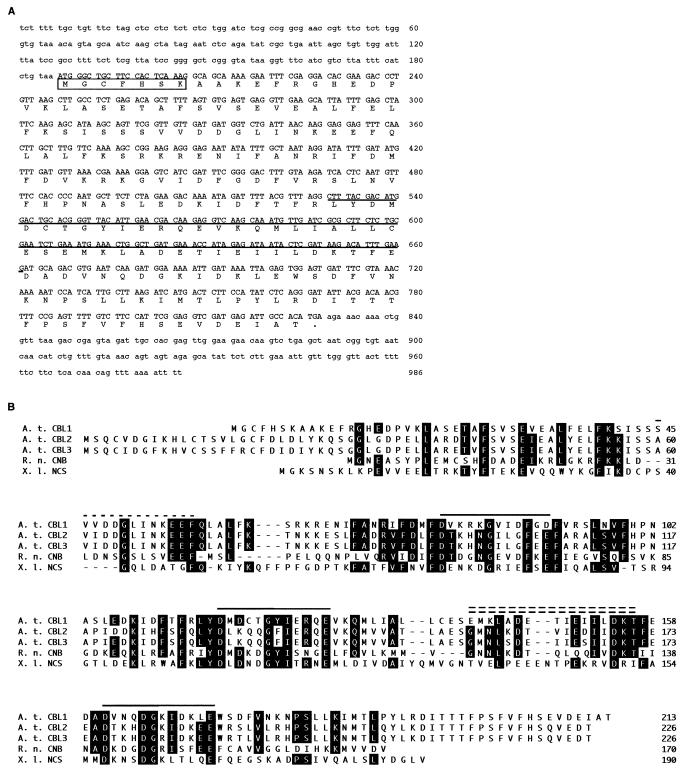
Sequence analyses of AtCBL1 cDNA and AtCBL proteins. (A) cDNA and deduced amino acid sequences of AtCBL1. The coding region of the cDNA is presented in capital letters and the noncoding regions in lowercase letters. The boxed sequence contains a typical myristoylation site (MGXXXSK). The underlined sequence represents the cDNA fragment identified by PCR (see Materials and Methods). An in-frame stop codon (taa) is immediately upstream from the ATG starting codon. (B) Alignment of the amino acid sequences of three AtCBL proteins from Arabidopsis (A. t. CBL1, A. t. CBL2, A. t. CBL3) with rat CNB (R. n. CNB), and Xenopus neuronal calcium sensor (X. l. NCS). Alignment was generated by using the clustal method with dnastar software. Amino acids identical to the consensus sequence of the alignment are shown on black background. The numbers on the right indicate the amino acid position. Dashes indicate gaps introduced to improve the alignment. Solid lines above the sequence indicate position of EF-hand regions. The single-dashed line represents a variation of the Ca2+-binding domain. The double-dashed line denotes the CNA interaction domain.
Stress Treatments and Northern Blots.
Five-week-old Arabidopsis seedlings grown under short-day conditions were used for the stress treatments. Wounding was performed by puncturing leaves with a hemostat. Typically, at least 80% of the leaves in the treated pot were wounded. Drought and cold treatments were performed as described (22). Samples were collected at time points indicated in Fig. 6. Total RNA was isolated, blotted, and hybridized with 32P-labeled cDNA probes according to a procedure described previously (21).
Figure 6.
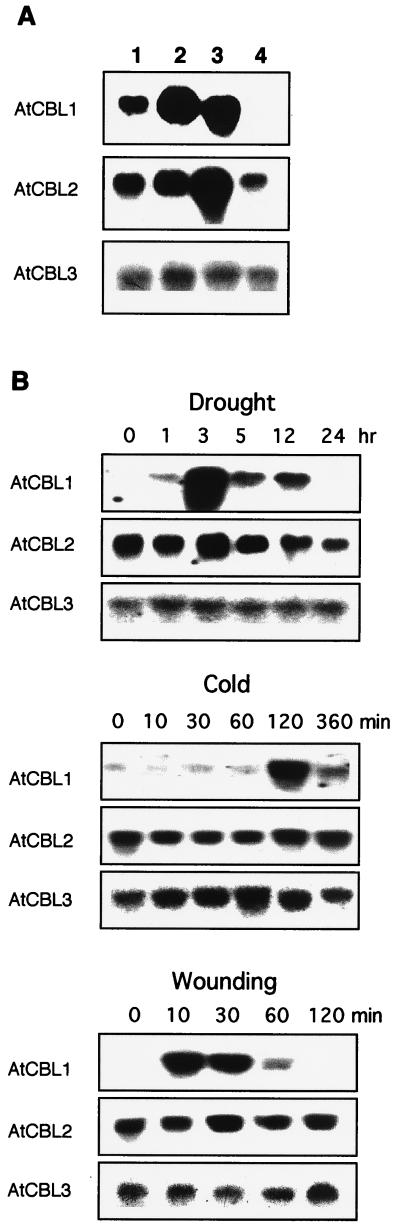
Expression pattern of three members of AtCBL gene family. (A) mRNA levels in leaves (lane 1), stems (lane 2), roots (lane 3), and flowers (lane 4). (B) mRNA accumulation under the stress conditions drought, cold, and wounding. A 10-μg sample of total RNA was loaded into each lane for RNA gel analysis using AtCBL cDNAs as hybridization probes.
Plasmid Construction.
Full-length cDNA of AtCBL1 was amplified by PCR using Pfu DNA polymerase and primer PCS1 (5′-CCATGGATCCAGGCTGCTTCCACTCAAAG-3′) and M13 reverse primer. The cDNA was cloned as a BamHI/PstI fragment into yeast vector pGBT9.BS (23), giving rise to pBDAtCBL1. To generate plasmid pBDrCNB, rat CNB cDNA (24) was cloned as a BamHI/XhoI fragment into the same vector after PCR amplification with primers PRCNBF (5′-CTAGTGGATCCCGGAAATGAGGCAAGT-3′) and PRCNBR (5′-TATTCTAGACTCGAGTCACACATCTACCAC-3′). Rat CNA cDNA (25) was amplified with primers PRCNAF (5′-CTAGTGGATCCCTCCGAGCCCAAGGCG-3′) and PRCNAR (5′-ATATCTAGAGAATTCTCACTGAATATTGCT-3′), cloned as BamHI/EcoRI fragment into plasmid pGAD.GH (23), and named pADrCNA. Primer PRCNAF and PRCNAR were used to generate plasmid pADdrCNA harboring nucleotide positions 1–1185 of the coding region of rat CNA in vector pGAD.GH. The nucleotide sequences of all constructs were verified by sequencing.
Expression and Purification of Recombinant AtCBL1 Protein.
The coding region of AtCBL1 cDNA was amplified with Pfu DNA polymerase (Stratagene) and subcloned into pGEX-4T-3 vector (Pharmacia). The resulting clones were verified by DNA sequencing. Overexpression of glutathione S-transferase (GST)-AtCBL1 fusion protein was performed as described previously (21, 26) with the following modifications: upon induction with 0.25 mM isopropyl β-d-thiogalactopyranoside (IPTG), bacterial cultures were pelleted and resuspended in a buffer containing 100 mM NaCl, 50 mM Tris⋅HCl at pH 8.0, 2 μM PMSF, 1 mM benzamidine, and 2 mM EDTA before lysis by sonication. The cell lysate was pelleted at 15,000 × g to collect the supernatant containing the fusion protein, which was subsequently affinity purified by glutathione-Sepharose 4B (Pharmacia).
Gel-Shift and 45Ca2+-Overlay Assays.
GST-AtCBL1 fusion protein was prepared as described above and analyzed by SDS/PAGE in the presence of 5 mM EGTA or 5 mM CaCl2 buffer. Proteins were stained with Coomassie blue to monitor the mobility pattern of the proteins. For the 45Ca2+-overlay assay, recombinant proteins were analyzed by SDS/PAGE, transferred to nitrocellulose membrane, and blotted by 45Ca2+ as previously described (27).
Yeast Two-Hybrid Assays.
Yeast strain SMY3 (28) was transformed by the polyethylene glycol/lithium acetate method as described (29). Transformants were plated on synthetic complete (SC) medium lacking Trp and Leu. Interaction assays were performed on plates containing 25 mM 3-aminotriazole in SC without Trp, Leu, and His. β-Galactosidase activity was monitored in a filter-lift assay (30) and was also assayed with chlorophenyl-red β-d-galactopyranoside (Boehringer Mannheim) as substrate (31, 32). β-Galactosidase activity (units) was calculated as follows: A574 of the supernatant ×1000/[reaction time (min) × culture volume used for the assay (ml) × OD600 of the culture].
RESULTS
Cloning and Sequence Analysis of Arabidopsis CLB1.
Extensive peptide and DNA alignments of known CNA and CNB sequences were performed to develop a homology-based cloning strategy. Highly conserved domains identified in CNA proteins from different species are also conserved in several plant phosphatases that belong to the PP1/PP2A subtypes. In contrast, the CNB subunit is highly conserved and contains unique sequences that are absent from other Ca2+-binding proteins. Therefore, we focused our efforts on the identification of the regulatory subunit of plant calcineurin by a systematic PCR approach.
One of the primer pairs (PCD1, PCD2) produced a product of the expected size, and its sequence showed significant similarity to known CNB proteins in a blastx search (PAM 120 score of 61). When this cDNA was used to screen an Arabidopsis cDNA library, 12 clones were identified, one of which was fully sequenced. The 986-bp-long cDNA contains an ORF of 213 aa, corresponding to a predicted polypeptide of 22.5 kDa (Fig. 1A). Database analysis with the amino acid sequence revealed a significant similarity to CNB proteins from different organisms (PAM 120 scores of 136 to CNB from rat, mouse, and human a in blastp search), and a much lower degree of similarity to CaM and CaM-related sequences (PAM 120 score of 77 to CaM from rat and Arabidopsis). The identified clone was therefore termed AtCBL1 (Arabidopsis thaliana calcineurin B-like protein 1). Like CNB, AtCBL1 contains a conserved site for myristoylation at the N terminus (MGXXXSK). As shown in Fig. 1B, the amino acid sequence of AtCBL1 shows 32% identity (56% similarity) compared with CNB from rat. In addition, the AtCBL1 protein also shares significant similarity to another Ca2+-binding protein referred to as neuronal calcium sensor from Xenopus, although the homology is limited to the Ca2+-binding domains (33). In contrast, the similarity to CNB is not restricted to the Ca2+-binding regions of the proteins. In particular, the CNA-binding domain in CNB is highly conserved in the AtCBL1 protein (24), indicating that AtCBL1 may represent a plant homolog of CNB from other organisms.
AtCBL Is Encoded by a Small Gene Family in Arabidopsis.
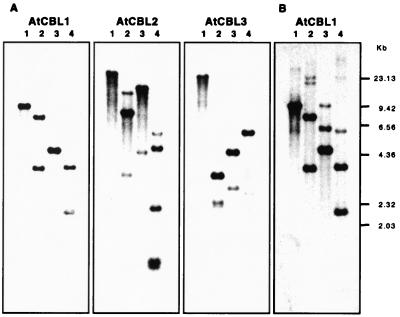
Comparative Southern blot analyses at high and low stringency using AtCBL1 cDNA as a probe (Fig. 2) indicated that AtCBL1 is a member of a small gene family. Partial sequencing of the 11 remaining cDNA clones from the primary library screening revealed a second closely related cDNA, which was designated AtCBL2. Database searches using AtCBL1 and AtCBL2 sequences identified several expressed sequence tags (ESTs) from Arabidopsis (GenBank accession nos. N65303, H77209, T20940, T21746). Further analysis revealed that N65303 is identical to AtCBL1 and H77209/T20940 are identical to AtCBL2, respectively. EST T21746, however, represents a previously unknown member of the AtCBL gene family and was, therefore, designated AtCBL3. The AtCBL2 and AtCBL3 cDNAs are both 1,043 bp long and contain ORFs of 216 aa each, corresponding to potential polypeptides of 25.8 kDa. As shown in Fig. 1B, the amino acid sequences of AtCBL proteins are highly conserved (63% identity and 79% similarity comparing AtCBL1 with AtCBL2 or AtCBL3; 91% identity and 96% similarity between AtCBL2 and AtCBL3). Comparative Southern blot analyses using all three AtCBL cDNAs as probes (Fig. 2) indicate that this gene family consists of five or six members with sufficient similarities to be detected under the stringency conditions used in our experiments (see Materials and Methods).
Figure 2.
Genomic organization of AtCBL genes. Aliquots (5 μg per digestion) of genomic DNA from Arabidopsis plants (Columbia ecotype) were digested with BamHI (lanes 1), BglII (lanes 2), EcoRI (lanes 3), and HindIII (lanes 4). (A) After agarose gel electrophoresis, DNA was blotted onto nylon membranes and hybridized under high-stringency conditions with 32P-labeled probes for AtCBL1, AtCBL2, and AtCBL3, respectively. (B) Low-stringency hybridization was performed using AtCBL1 cDNA as a probe. HindIII-digested phage λ DNA was used to provide molecular size markers.
AtCBL1 Encodes a Functional Ca2+-Binding Protein.
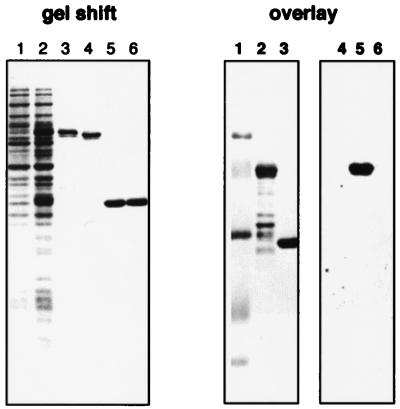
The presence of conserved EF-hand motifs in the predicted protein sequence of AtCBL suggests that it may function as a Ca2+-binding protein. To examine this possibility, we expressed the recombinant AtCBL1 protein as a GST fusion in Escherichia coli. Two approaches were used to confirm that the recombinant protein is a functional Ca2+-binding protein. One was a specific gel-shift assay in which all EF-hand-containing proteins migrate faster during gel electrophoresis when the loading buffer contains high levels of Ca2+ instead of EGTA (27). As shown in Fig. 3, the GST-AtCBL1 fusion protein migrates more slowly in the presence of EGTA compared with its migration in Ca2+-containing buffer. As a negative control, the mobility of GST was not affected by the presence of EGTA or Ca2+ in the buffer. As a second functional test, we performed a Ca2+-binding assay using the 45Ca2+-overlay technique (27). The GST-AtCBL1 fusion protein clearly binds Ca2+, whereas the GST protein and the molecular weight markers did not (Fig. 3).
Figure 3.
AtCBL1 is a functional Ca2+-binding protein. In the gel-shift panel, lane 1 and lane 2 were loaded with protein extract prepared before and after isopropyl β-d-thiogalactopyranoside (IPTG) induction, respectively. GST-AtCBL1 fusion protein was purified and incubated in EGTA- (lane 3) or calcium-containing (lane 4) buffer before being analyzed by SDS/PAGE. As a negative control, GST protein was analyzed in the same way (lanes 5 and 6). In the overlay assay, lanes 1–3 show Coomassie blue-stained molecular weight marker, GST-AtCBL1 (0.4 μg), and GST (0.2 μg), respectively. Lanes 4–6 indicate the same samples transferred to the nitrocellulose membrane and assayed by 45Ca2+ binding. Only the fusion protein in lane 2 shows Ca2+-binding capability (lane 5).
AtCBL1 Interacts in Vivo with Rat CNA and Complements a Yeast CNB Mutant.
Calcineurin forms a tightly associated heterodimer composed of regulatory B and catalytic A subunits (5, 6). Interaction between the two subunits is necessary for the function of the holoenzyme and the stabilization of its subunits. Since the identity of the AtCBL-interacting protein(s) from plants is unknown, we tested whether AtCBL can interact with the heterologous CNA subunit from rat in vivo in the yeast two-hybrid system. To avoid potential interference in our assay by the endogenous yeast CNB, we used a yeast strain (SMY3) harboring a disrupted CNB gene. AtCBL1 was fused to the GAL4 DNA-binding domain (pBDAtCBL1). Rat CNA was fused to the GAL4 transcriptional activation domain (pADrCNA). As a positive control, rat CNB was also cloned into the binding domain vector (pBDrCNB).
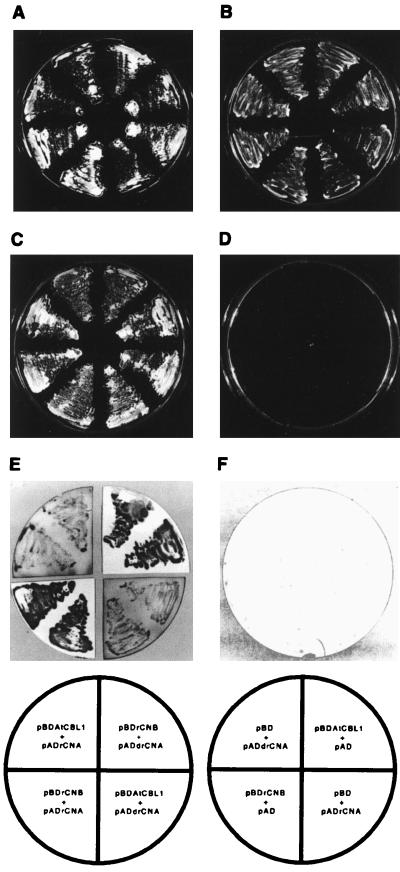
AtCLB1 interacted with the full-length rat CNA in vivo, as indicated by growth on selective medium and activation of β-galactosidase activity (Fig. 4 C and E). As expected, the interaction between the rat CNA and CNB subunits appeared to be much stronger (Fig. 4 C and E). Control experiments with pBDAtCBL1 and pBDrCNB constructs showed no activation of the yeast reporter gene system (Fig. 4 and Table 1).
Figure 4.
Interaction between AtCBL1 and rat CNA protein in the yeast two-hybrid system. Growth of yeast transformants on SC −Trp −Leu medium (A and B), SC −Trp −Leu −His containing 25 mM 3-aminotriazole (C and D), and the colony color of the transformants determined by the filter lift assay (E and F) are shown. Plates containing histidine, but lacking tryptophan and leucine, select for the presence of the two plasmids containing the Gal4 DNA-binding and the Gal4 activation domain. Plates lacking tryptophan, leucine, and histidine and supplemented with 25 mM 3-aminotriazole select for a positive interaction between the fusion proteins. The array of the yeasts containing the different constructs is indicated in the scheme at the bottom. The symbols for the constructs were described in the text.
Table 1.
Quantitative analyses of AtCBL1–rat CNA interactions
| Gal4 DNA-binding domain (BD) | Gal4 activating domain (AD) | Colony color | β-Galactosidase activity, units* |
|---|---|---|---|
| BD | AD | White | 0.012 |
| BDrCNB | AD | White | 0.013 |
| BD | AD | White | 0.018 |
| BD | ADrCNA | White | 0.255 |
| BDAtCBL1 | AD | White | 0.133 |
| BDrCNB | ADdrCNA | Blue | 133.31 |
| BDrCNB | ADdrCNA | Pale blue | 7.43 |
| BDAtCBL1 | ADdrCNA | Pale blue | 6.58 |
| BDAtCBL1 | ADrCNA | Pale blue | 1.00 |
The β-galactosidase activity was mesured as described in Materials and Methods. The numbers shown are the means of three independent measurements.
CNA interacts not only with the CNB subunit but also with CaM. To exclude potential binding of AtCBL1 to the CaM-binding domain of rat CNA, we generated an additional construct (pADdrCNA). This plasmid expresses a truncated version of the rat CNA catalytic subunit lacking the C-terminal CaM-binding domain. Interestingly, the truncated version of rat CNA (pADdrCNA) lacking the CaM-binding domain interacted more strongly with both rat CNB and AtCLB1. This might be because of a lack of interference from the yeast CaM or better steric accessibility of the CNB-binding domain in the truncated rat CNA protein.
To exclude the possibility that the GAL4 DNA-binding or activation domain contributed to the observed interactions, domain swapping experiments were performed by replacing the DNA-binding domain of each construct with the transcriptional activating domain and vice versa. Identical results were obtained when these constructs were used (data not shown).
Quantitative analysis of the in vivo interactions revealed a reproducible increase in β-galactosidase expression when the truncated version of rat CNA was used in the interaction assay (Table 1). For the interaction with AtCBL1, β-galactosidase activity increased from 8-fold (pBDAtCBL1 + pADrCNA) to 50-fold (pBDAtCBL1 + pADdrCNA) compared with the background level of expression (pBDAtCBL1 + pAD). The interactions between the rat calcineurin subunits were even more robust. The estimated activities of β-galactosidase were 7-fold (pBDrCNB + pADrCNA) or 20-fold (pBDrCNB + pADdrCNA) higher than the corresponding activities observed with AtCBL1. The observed activity resulting from the interaction between AtCBL1 and the truncated rat CNA further suggests that the AtCBL1 protein most likely binds to the CNB-binding domain of rat CNA. Because none of the other Ca2+-binding proteins has been shown to interact with the CNB-binding region of CNA, the results shown here indicate that AtCBL1 may function as a regulatory subunit of a plant calcineurin-like protein.
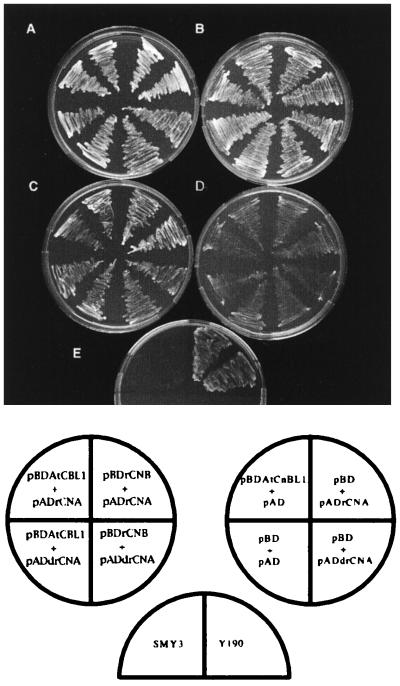
To further support this idea, we performed yeast complementation experiments. Yeast calcineurin mutant strains are highly sensitive to high-salt conditions (13–15). To test whether interaction of AtCBL1 with rat CNA is functionally significant, the yeast strains with various plasmid combinations were grown on either normal or high-salt medium. As shown in Fig. 5, CNB mutant (SMY3) transformed with different plasmids grew well in the absence of LiCl (Fig. 5 A and B). In the presence of 200 mM LiCl, mutant or mutant transformed with control plasmids either did not grew or grew very poorly (Fig. 5 D and E). In contrast, yeast strains that contained rat CNA/CNB or rat CNA/AtCBL1 plasmids grew at a rate similar to that of wild-type strain on high-salt medium (Fig. 5 A and C). This result suggests that AtCBL1 together with rat CNA can complement the defect of yeast CNB mutant in salt tolerance. Because AtCBL1 alone did not complement yeast CNB mutant, we speculate that AtCBL1 does not efficiently interact with yeast CNA.
Figure 5.
Complementation of yeast CNB mutant by interaction of rat CNA and AtCBL1. Yeast CNB mutant strain SMY3 (28) was transformed with various combinations of plasmids indicated in the circles. Annotations of the plasmids are shown in Materials and Methods and are the same as used in Fig. 4 and Table 1. The same combinations were used in A and C (left circle in the diagram at the bottom), and in B and D (right circle). Mutant (SMY3) and a wild-type (Y190) controls in E are shown in the half-circle. Yeast strains were grown on the synthetic medium containing 0 mM (A and B) and 200 mM (C, D, and E) LiCl.
Differential Expression Pattern of AtCBL Genes in Response to Stress Conditions.
Tissue specificity or signal responsiveness of gene expression often reflect the function of the corresponding gene products in plant development and signaling. Members of the AtCBL gene family may be expressed differently and therefore have distinct functions. We investigated the expression pattern of all three AtCBL genes by Northern blot analyses using total RNA from various tissues of Arabidopsis plants. As shown in Fig. 6A, the AtCBL1 gene is highly expressed in roots and stems. The expression level is lower in leaves and undetectable in flowers. In contrast, AtCBL3 is expressed at similar levels in all tissues analyzed, whereas AtCBL2 is preferentially expressed in roots. Together, these results suggest that AtCBL genes are differentially expressed in different tissues under normal growth conditions.
To test whether plant calcineurin-like proteins could play a role in plant stress responses, we examined the expression patterns of these genes under various stress conditions, including drought, wounding, cold, heat shock, and mechanical touch. As shown in Fig. 6B, wounding, drought, and cold treatment strongly increased mRNA levels of AtCBL1, whereas heat shock or mechanical touch had no effect (data not shown). In contrast, expression of AtCBL2 and AtCBL3 did not respond to these stimuli. These experiments may indicate that different isoforms of the AtCBL protein may have distinct distribution patterns in Arabidopsis plants and may function in different pathways. Note that induction of AtCBL1 in response to wounding, drought, and cold stress corresponds to the well established Ca2+ involvement in these signaling pathways.
DISCUSSION
Increasing evidence indicates that many fundamental steps and components of the cellular signaling machinery are conserved during the evolution of eukaryotic organisms. The detailed mechanisms, however, may become adapted to function in divergent processes in animals, yeast, and plants. Recent studies have suggested that this could also be true for the Ca2+-dependent phosphatase calcineurin, an important regulator of various cellular processes in animals and yeast.
In this study, we characterized AtCBL, a potential regulatory subunit of a calcineurin-like activity from Arabidopsis. Different isoforms of this protein are encoded by a small gene family consisting of at least six genes. We identified and characterized three members of this family. The cDNAs encode proteins with significant similarity to CNB from diverse organisms. Although the amino acid sequences of the Ca2+-binding EF-hand motifs are highly conserved, the sequence conservation also extends to the entire protein. Most strikingly, the region required for interaction with the CNA subunit is most highly conserved. This domain is located upstream of the fourth EF-hand domain and forms a helical contact surface for CNA interaction in animal homologues (24, 34, 35). The observed similarity to CaM is significantly lower than the observed degree of conservation with CNB. Another Ca2+-binding protein, neuronal calcium sensor, shares significant sequence homology with AtCBL, but the homology is limited to the EF-hand regions (Fig. 1B). These sequence features suggest that AtCBL is structurally most similar to CNB.
AtCBL contains putative EF-hand motifs for Ca2+ binding, an intrinsic feature to CNB function as a regulatory subunit of the protein phosphatase. Both gel-shift experiments and 45Ca2+-overlay assays revealed that recombinant AtCBL1 is capable of binding calcium. A stringent criterion to establish whether a protein can serve as a regulatory subunit of calcineurin is the demonstration of its ability to interact specifically with the catalytic CNA subunit and form a functional complex. We have investigated the interaction between AtCBL and rat CNA in the yeast two-hybrid system. Our data indicate that AtCBL is capable of interacting with rat CNA in vivo, although the affinity is reduced compared with the binding between the two calcineurin subunits from rat. This is not unexpected and may reflect a divergence resulting from the coevolution of both subunits within one organism. As we demonstrated by using a C-terminal truncation of rat CNA, the observed interaction is mediated most likely by the CNA domain required for CNB interaction (24, 35). This result excludes the possibility of an interaction between AtCBL and rat CNA by means of the CaM-binding domain of the catalytic subunit, and it is consistent with the higher structural similarity between AtCBL and CNB as compared with CaM. Indeed, there has not been any known example that shows interaction between a Ca2+-binding protein (other than CNB) and the CNB-binding domain of CNA. The yeast strain we used in the two-hybrid experiments lacks an endogenous active calcineurin because of a disruption of the yeast CNB gene. This mutation renders the strain hypersensitive to LiCl, a well established phenotype of calcineurin mutants in yeast (13–15). We found that the coexpression of AtCBL1 with full-length or truncated rat CNA restored the Li resistance of this yeast strain (Fig. 5). Taken together, the results support the idea that AtCBL1 may serve as a regulatory subunit of a calcineurin-like activity in plants.
The existence of multiple isoforms of AtCBL in Arabidopsis adds to the complexity of AtCBL function in plants. All identified isoforms of AtCBL interact with rat CNA in the yeast two-hybrid system in a similar manner (J.K., unpublished result), which could indicate functional redundancy of AtCBL isoforms. Alternatively, differences in protein structure and gene expression pattern of these isoforms may represent mechanisms for differential regulation of Ca2+ signaling in plant cells. For example, AtCBL1, but not AtCBL2 and AtCBL3, contains a conserved N-terminal site for myristoylation that may regulate the localization of the protein. In addition, only the AtCBL1 gene is induced in response to specific stress factors, such as drought, wounding, and cold, implicating this isoform in stress response. The fact that only a subset of stress factors induce AtCBL1 expression suggests that it has a specific function in the cellular response to these stimuli. This interpretation is further supported by the rapid and transient induction pattern of AtCBL1 mRNA levels during stress responses. Also consistent with a function of AtCBL1 in stress response, studies have revealed the occurrence of cellular calcium transients in response to drought and cold (36, 37).
After this work was submitted for publication, Liu and Zhu (38) reported that a salt-tolerance gene in Arabidopsis (SOS3) encodes a putative Ca2+-binding protein with a high amino acid similarity to the AtCBL proteins. This finding is consistent with calcineurin function in salt tolerance in yeast (13–15) and most likely in plants as well (19). Together, our study on AtCBL proteins, especially stress-responsive expression of AtCBL1, and study on the SOS3 gene product (38) emphasize a role for calcineurin-like proteins in stress signal transduction. Interestingly, the AtCBL proteins clearly play different roles as compared with SOS3 because they do not complement the defect caused by mutations in SOS3. In addition, unlike AtCBL1, SOS3 gene expression is not regulated by stress conditions (S.L., unpublished results). These studies suggest that AtCBLs and SOS3 are highly related but functionally distinct members of a family of Ca2+-binding proteins in higher plants. Although studies in this report provide strong evidence that this family of proteins potentially serve as a regulatory subunit of calcineurin-like protein phosphatase, confirmation of the functional identity of AtCBLs and SOS3 requires identification of their endogenous protein targets in higher plants. Irrespective of what their functional targets may be, characterization of AtCBL proteins provides an important stepping stone for further understanding the regulation of Ca2+ signal transduction in higher plants.
Acknowledgments
We thank Dr. Joseph Heitman for providing yeast strain SMY3, Juan Zhang for technical assistance, and the Arabidopsis stock center for providing expressed sequence tag clones and libraries. We also thank Drs. Robert Hayes and Ralph Bock for helpful discussions and critical reading of the manuscript. J.K. and K.H. were supported by fellowships from the Deutsche Forschungsgemeinschaft (Ku 931/1-3 and Ha 2146/2-1). This work was supported jointly by the Department of Energy (85ER13375 to W.G) and the National Institutes of Health (GM-52826 to S.L.).
ABBREVIATIONS
- CNA
calcineurin A (catalytic) subunit
- CNB
calcineurin B (regulatory) subunit
- CaM
calmodulin
- GST
glutathione S-transferase
Footnotes
References
- 1.Clapham D E. Cell. 1995;80:259–268. doi: 10.1016/0092-8674(95)90408-5. [DOI] [PubMed] [Google Scholar]
- 2.Trewavas A, Read N, Campbell A K, Knight M. Biochem Soc Trans. 1996;24:971–974. doi: 10.1042/bst0240971. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 4.Guerini D. Biochem Biophys Res Commun. 1997;235:271–275. doi: 10.1006/bbrc.1997.6802. [DOI] [PubMed] [Google Scholar]
- 5.Klee C B, Draetta G F, Hubbard M J. Adv Enzymol. 1988;61:149–200. doi: 10.1002/9780470123072.ch4. [DOI] [PubMed] [Google Scholar]
- 6.Perrino B A, Ng L Y, Soderling T R. J Biol Chem. 1995;270:340–346. doi: 10.1074/jbc.270.1.340. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Farmer J J D, Lane W L, Friedman J, Weissman I, Schreiber S L. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 8.Schreiber S L, Crabtree G R. Immunol Today. 1992;13:136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- 9.Rao A, Luo C, Hogan P G. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 10.Chang H Y, Takei K, Sydor A M, Born T, Rusnak F, Jay D G. Nature. 1995;376:686–690. doi: 10.1038/376686a0. [DOI] [PubMed] [Google Scholar]
- 11.Tong G, Shepherd D, Jahr C E. Science. 1995;267:1510–1512. doi: 10.1126/science.7878472. [DOI] [PubMed] [Google Scholar]
- 12.Foor F, Parent S A, Morin N, Dahl A M, Ramadan N, Chrebet G, Bostian K A, Nielsen J B. Nature. 1992;360:682–684. doi: 10.1038/360682a0. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura T, Liu Y, Hirata D, Namba H, Harada S, Hirokawa T, Miyakawa T. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardenas M E, Muir R, Scott L, Breuder T, Heitman J. EMBO J. 1995;14:2772–2783. doi: 10.1002/j.1460-2075.1995.tb07277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Withee J L, Mulholland J, Jeng R, Cyert M S. Mol Biol Cell. 1997;8:263–277. doi: 10.1091/mbc.8.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luan S, Li W, Rusnak F, Assmann S M, Schreiber S L. Proc Natl Acad Sci USA. 1993;90:2202–2206. doi: 10.1073/pnas.90.6.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen G J, Sanders D. Plant Cell. 1995;7:1473–1483. doi: 10.1105/tpc.7.9.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bethke P, Jones R L. Plant J. 1997;11:1227–1235. [Google Scholar]
- 19.Pardo J M, Reddy M P, Yang S, Maggio A, Huh G-H, Matsumoto T, Coca M A, Paino-D’Urzo M, Koiwa H, Yun D-J, Watad A A, Bressan R A, Hasegawa P M. Proc Natl Acad Sci USA. 1998;95:9681–9686. doi: 10.1073/pnas.95.16.9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieber J J, Rothenberg M, Roman G, Feldmann K A, Ecker J R. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 21.Luan S, Kudla J, Gruissem W, Schreiber S L. Proc Natl Acad Sci USA. 1996;93:6964–6969. doi: 10.1073/pnas.93.14.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elledge S J, Mulligan J T, Ramer S W, Spottswood M S, Davis R W. Proc Natl Acad Sci USA. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe Y, Perrino B A, Soderling T R. Biochemistry. 1996;35:562–566. doi: 10.1021/bi951703+. [DOI] [PubMed] [Google Scholar]
- 25.Perrino B A, Fong Y L, Brickey D A, Saitoh Y, Ushio Y, Fukunaga K, Miyamoto E, Soderling T R. J Biol Chem. 1992;267:15965–15969. [PubMed] [Google Scholar]
- 26.Luan S, Lane W S, Schreiber S L. Plant Cell. 1994;6:885–892. doi: 10.1105/tpc.6.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krinks M H, Klee C B, Pant H C, Gainer H. J Neurosci. 1988;8:2172–2182. doi: 10.1523/JNEUROSCI.08-06-02172.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cardenas M E, Hemenway C, Muir R S, Ye R, Fiorentino D, Heitman J. EMBO J. 1994;13:5944–5957. doi: 10.1002/j.1460-2075.1994.tb06940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gietz D, Jean A, Woods R A, Schiesh R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breeden L, Nasmyth K. Cold Spring Harbor Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- 31.Guarente L. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 32.Iwabuchi K, Li B, Bartel P L, Fields S. Oncogene. 1993;8:1693–1696. [PubMed] [Google Scholar]
- 33.Olafsson P, Wang T, Lu B. Proc Natl Acad Sci USA. 1995;92:8001–8005. doi: 10.1073/pnas.92.17.8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kissinger C R, Parge H E, Knighton D R, Lewis C T, Pelletier L A, Tempczyk A, Kalish V J, Tucker K D, Showalter R E, Moomaw E W, Gastinel L N, Habuka N, Chen X, Maldonado F, Barker J E, Bacquet R, Villafranca J E. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 35.Griffith J P, Kim J L, Kim E E, Sintchak M D, Thomson J A, Fitzgibbon M J, Fleming M A, Caron P R, Hsiao K, Navia M A. Cell. 1995;82:507–522. doi: 10.1016/0092-8674(95)90439-5. [DOI] [PubMed] [Google Scholar]
- 36.Knight H, Trewavas A J, Knight M R. Plant J. 1997;12:1067–1078. doi: 10.1046/j.1365-313x.1997.12051067.x. [DOI] [PubMed] [Google Scholar]
- 37.Knight H, Trewavas A J, Knight M R. Plant Cell. 1996;8:489–503. doi: 10.1105/tpc.8.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Zhu J-K. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]