Abstract
Association of tristetraprolin (TTP) with mRNAs containing selected AU-rich mRNA-destabilizing elements (AREs) initiates rapid cytoplasmic degradation of these transcripts. The RNA-binding activity of TTP is mediated by an internal tandem zinc finger domain that preferentially recognizes U-rich RNA ligands containing adjacent UUAU half-sites, and is accompanied by conformational changes within the peptide. Here, we have used analogues of the TTP RNA-binding domain containing specific tryptophan substitutions to probe the Zn2+- and RNA substrate-dependence of conformational events within individual zinc fingers. Fluorescence methods demonstrate that the N-terminal, but not C-terminal zinc finger domain adopts a stably folded conformation in the presence of Zn2+. Denaturant titrations suggest that both the N- and C-terminal zinc fingers exhibit limited structural heterogeneity in the absence of RNA substrates, although this is more pronounced for the C-terminal finger. Binding to a cognate ARE substrate induced significant conformational changes within each zinc finger, which also included increased resistance to chemical denaturation. Studies with mutant ARE ligands revealed that a single UUAU half-site was sufficient to induce structural modulation of the N-terminal finger. However, RNA-dependent folding of the C-terminal zinc finger was only observed in the presence of tandem UUAU half-sites, suggesting that the conformation of this domain is linked not only to RNA substrate recognition, but also to the ligand occupancy and/or conformational status of the N-terminal finger. Coupled with previous structural and thermodynamic analyses, these data provide a mechanistic framework for discrimination of RNA substrates involving ligand-dependent conformational adaptation of both zinc fingers within the TTP RNA-binding domain.
In mammals, many mRNAs that encode proteins contributing to the control of cell proliferation, differentiation, apoptosis, and inter-cell communication are inherently unstable. This property limits the cytoplasmic concentration of these mRNAs, and hence their potential for translation, but also permits new steady-state mRNA levels to be attained rapidly following changes in the synthetic rate (1, 2). The degradation of many labile mRNAs is directed by the presence of AU-rich elements (AREs)1 within their 3′-untranslated regions (3′UTRs). AREs constitute a broad family of mRNA-destabilizing sequences ranging from approximately 40 to 150 bases in length and frequently containing one or more AUUUA motifs within a U-rich tract (3–5). While the sequences of these cis-acting elements show remarkable diversity across the mRNA population, AREs from specific mRNAs are often highly conserved across species (6–8), implying that variations in ARE character may facilitate gene-specific control of mRNA decay kinetics. mRNA turnover directed by AREs is initiated by rapid 3′→5′ exonuclease digestion of the polyA tail (9, 10), followed by 5′-decapping and decay of the mRNA body, which may involve both 3′→5′ and 5′→3′ exoribonucleolytic components (11–16).
Control of mRNA decay through AREs is mediated by the association of cellular transacting factors. To date, upwards of 20 distinct proteins (17, 18) and one microRNA (19) targeting AU-rich RNA sequences have been identified, although functional significance has been ascribed to only a small subset of these interactions. The ARE-binding factor tristetraprolin (TTP; also known as TIS11 and Nup475) is the prototype of a family of tandem CCCH zinc finger proteins (20) and promotes accelerated mRNA turnover in cells (10, 21), possibly via direct recruitment of RNA catabolic activities. Recent data indicate that TTP can interact with numerous mRNA-degrading factors, including some responsible for 3′→5′ deadenylation and decay, 5′-decapping, and 5′→3′ exonuclease activities (11, 13, 22). TTP may also target mRNAs for degradation via an RNA interference-based mechanism, by enhancing the association of microRNA miR16 with ARE-containing mRNA substrates (19). However, unlike some other ARE-binding proteins that readily bind diverse populations of U-rich RNA sequences (8, 23–25), the RNA-binding domain of TTP is highly selective, optimally binding U-rich RNA sequences containing AUUUA or AUUUUA motifs (25–27).
Previously, we demonstrated that a 73-amino acid peptide containing the TTP tandem zinc finger domain, termed TTP73, formed high affinity complexes with ARE substrates in a zinc-dependent manner (26, 28). Several observations based on these and other studies also suggested that ARE recognition by this peptide may proceed via an “induced-fit” or “selected-fit” mechanism (29, 30). First, heteronuclear single quantum coherence (HSQC) analyses permitted resonance assignments for most residues within the N-terminal zinc finger in the absence of an RNA substrate, indicative of structural ordering within this motif (28). By contrast, virtually no residues within the C-terminal domain could be assigned, consistent with conformational heterogeneity in this region. Second, TTP73 association with cognate RNA targets induced large peak shifts in N-terminal zinc finger residues by HSQC, and also permitted assignment of residues within the C-terminal finger (28). These data indicated that RNA binding was accompanied by dramatic changes in peptide conformation involving both zinc fingers. Third, independent structural analyses of the RNA-binding domain of a TTP family member, TIS11d, showed that both the N-terminal and C-terminal zinc fingers adopted similar folds when complexed with a 9-base recognition sequence (31). Finally, TTP73 binding to high affinity RNA substrates is associated with a large negative change in heat capacity (26). This thermodynamic signature of hydrophobic surface area burial (32) without substantial changes in RNA structure (26) is consistent with NMR predictions of protein folding linked to RNA:peptide complex formation.
In this work, we have used tryptophan (Trp)-substituted TTP73 analogues to evaluate localized conformational changes within each zinc finger motif in response to Zn2+ coordination and RNA substrate binding, exploiting the sensitivity of Trp fluorescence to changes in its local environment (33). The design of Trp-substituted variants of the wild type TTP73 peptide (TTP73 wt) was based on two principal criteria: [i] that the substituted Trp residues localize to regions likely to experience similar conformational changes in each zinc finger, and [ii] that the Trp substitutions not interfere with the RNA-binding activity of the peptide. Trp substitutions at Phe11 and Phe49 were selected as the most promising conformational sensors, based on the following rationale. First, Phe11 and Phe49 occupy homologous positions in the N- and C-terminal zinc fingers of TTP, respectively, between the first two Cys residues comprising each Cx8Cx5Cx3H motif (Figure 1A). Second, Phe11 and Phe49 are conserved in the TTP family member TIS11d, and are located in similar α-helical regions within each TIS11d zinc finger when bound to an ARE substrate (Figure 1B). Localization of these Phe residues within structured domains in the TIS11d-ARE complex indicated that modulation of peptide folding near these sites would likely influence the fluorescence properties of each Trp mutant, based on alterations in proximity to nonpolar surfaces and/or changes in solvent accessibility. In particular, Phe11 and Phe49 exhibit stacking potential with Zn2+-coordinated His residues His27 and His65, respectively, in the TIS11d-ARE complex (Figure 1C and 1D). Furthermore, these Phe residues are maintained in close proximity (< 3.5 ) to the zinc ions and conserved Zn2+-coordinated Cys residues (Cys17 and Cys55) of their respective zinc finger domains. Additional features indicated that Trp substitutions at Phe11 and Phe49 were unlikely to severely compromise the RNA-binding activity of the TTP tandem zinc finger peptides. Phe11 and Phe49 are not involved in coordination of Zn2+, which is absolutely essential for RNA binding by TTP (26, 34). Furthermore, these residues are distal to the RNA-binding interface in the TIS11d-ARE complex (Figure 1B).
FIGURE 1.
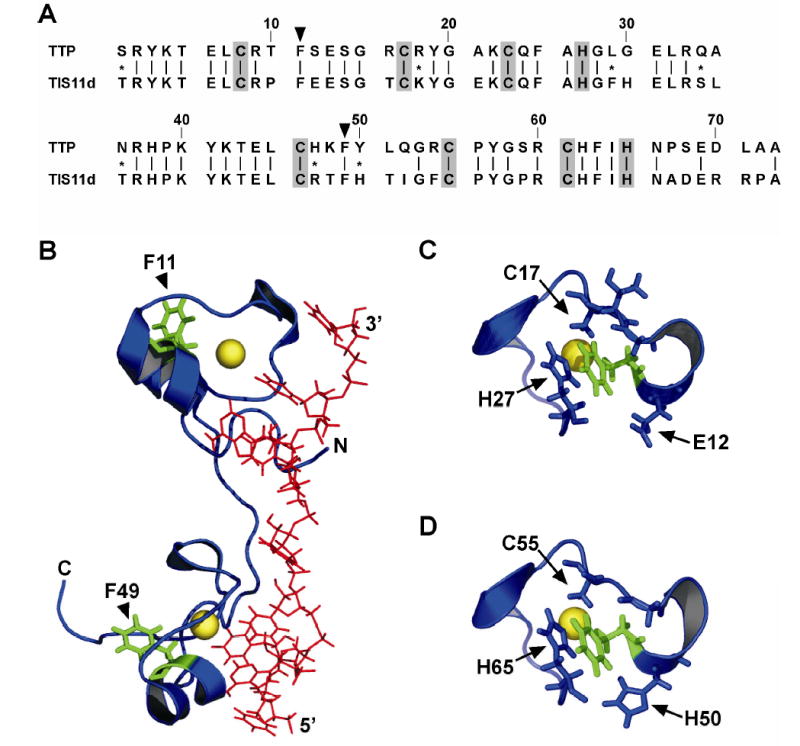
Selection of tryptophan substitution sites within the TTP tandem zinc finger domain. (A) Sequence alignment between the zinc finger domains of human TTP (GenBank accession number NP_003398; residues 102–174) and human TIS11d (GenBank accession number AAA91778; residues 152–224) was solved using the ALIGN program at the San Diego Supercomputer Center Biology Workbench (www.workbench.sdsc.edu). Lines indicate identical residues, while conservative substitutions are denoted by asterisks. Zn2+-coordinating residues comprising each CCCH motif are shaded, and the positions of Phe residues mutated to Trp (F11 and F49) are indicated by arrowheads. (B) Structure of the TIS11d tandem zinc finger domain complexed with an ARE substrate (PDB number 1RGO from Ref. 31) showing the location of residues corresponding to F11 and F49 of TTP73 in the folded peptide:RNA complex (green). The RNA substrate is in red and zinc ions are indicated by yellow spheres. Enlarged views show the local environments of F11 (C) and F49 (D) (green) of TIS11d in the peptide:RNA complex.
Based on these considerations, TTP73 mutant peptides F11W and F49W were used to evaluate Zn2+- and RNA-dependent conformational changes in the N- and C-terminal zinc finger domains of TTP, respectively. Alterations in local peptide structure were monitored by changes in the fluorescence quantum yields, emission peak wavelengths, and solvent accessibilities of Trp residues in each zinc finger. The relative stabilities of peptide structural transitions were compared using denaturant titration analyses. These studies indicate that the N- and C-terminal zinc finger domains exhibit very different structural responses to Zn2+ binding in the absence of RNA. However, local peptide structures within both zinc finger domains exhibit extreme sensitivity to denaturant prior to RNA binding, consistent with conformational sampling between weakly folded states. Association with a cognate ARE substrate stabilized local peptide folding in both zinc fingers, based on resistance to chemical denaturation and dramatic decreases in both the quantum yields and solvent accessibilities of substituted tryptophan residues. By contrast, only the N-terminal zinc finger exhibited a similar structural response in the presence of ARE mutant substrates lacking either UUAU half-site. Considering these data together with previous NMR and thermodynamic analyses of interactions between the tandem zinc finger domain of TTP and RNA substrates, a model for ARE recognition involving conformational sampling and remodeling within the TTP zinc finger domains is discussed.
EXPERIMENTAL PROCEDURES
Materials
The RNA substrates used in this study were denoted ARE9 (5′-UUAUUUAUU-3′), ARE13 (5′-AUUUAUUUAUUUA-3′), ARE13-AU3U (5′-AUUUAUUUUUUUA-3′), ARE13-UU3A (5′-AUUUUUUUAUUUA-3′), and UC13 (5′-UCUCUCUCUCUCU-3′). Variants of RNA oligonucleotides containing 5′-linked fluorescein moieties are indicated by the prefix “Fl-“. Each RNA substrate was synthesized, 2′-hydroxyl deprotected, and purified by Dharmacon Research (Lafayette, CO) or Integrated DNA Technologies (Coralville, IA). Lyophilized pellets were resuspended in 10 mM TrisHCl [pH 8.0] and quantified by A260/A495 as previously described (35, 36). Where required, 5′-hydroxyl RNA substrates were radiolabeled to specific activities of 3–5 × 103 cpm/fmol using T4 polynucleotide kinase and [γ-32P]ATP as described (24).
All synthetic peptides were synthesized, purified, and refolded by Albachem Ltd. (Edinburgh, Scotland) as described previously (28). TTP73 is a 73-amino acid peptide corresponding to the tandem zinc finger domain (residues 102–174) of human TTP (GenBank accession number NP_003398). Two additional peptides, termed TTP73 F11W and F49W, are TTP73 mutants containing individual Phe→Trp mutations at positions 11 and 49, respectively. Lyophilized peptides were resuspended in 10 mM TrisHCl [pH 8.0] containing 2 mM dithiolthreitol and frozen in aliquots at −80 ºC, and quantified by Coomassie Blue-stained SDS-PAGE against a titration of BSA.
RNA-Peptide Binding Studies
Interactions between TTP73 peptides and RNA substrates were qualitatively evaluated using gel mobility shift assays (GMSAs). Binding reactions (10 μl total volume) containing varying concentrations of peptide and 0.2 nM 32P-labeled RNA substrate were assembled in RNA binding buffer (10 mM TrisHCl [pH 8.0] with 100 mM KCl, 2 mM dithiolthreitol, 0.1 μg/μl bovine serum albumin and 10% glycerol). Heparin (0.2 μg/μl) was included to inhibit non-specific interactions between peptides and RNA ligands. ZnCl2 (5 μM) or EDTA (0.5 mM) were also added to binding reactions where indicated. Prior to addition of RNA substrates, wild type or mutant TTP73 peptides were incubated for 20 minutes at room temperature in binding mixtures to ensure coordination of Zn2+. Previous experiments indicated that the RNA-binding activity of TTP73 was significantly impaired without this Zn2+ pre-incubation step (data not shown). Following the addition of RNA, binding reactions were incubated on ice for 15 minutes and fractionated by electrophoresis through 6% (40:1 acrylamide:bis-acrylamide) native gels at 4ºC. Gels were then dried and reaction products visualized by Phosphorimager scan (Amersham Biosciences, Piscataway, NJ).
Binding constants describing interactions between TTP73 peptides and fluorescein-conjugated RNA substrates were quantitatively measured using a fluorescence anisotropy-based assay, essentially as described (26). Fluorescence anisotropy was measured using a Beacon 2000 Fluorescence Polarization System (Panvera, Madison, WI) equipped with fluorescein excitation (λex = 490 nm) and emission (λem = 535 nm) filters. In all experiments reported in this work, protein binding did not induce a change in the fluorescence quantum yield of Fl-RNA substrates. As such, the measured fluorescence anisotropy (At) of each binding reaction was interpreted as a function of the intrinsic anisotropy (Ai) and fractional concentration (fi) of each fluorescent species using Equation 1 (37, 38):
| (1) |
Since the TTP73 peptide binds RNA substrates ≤ 13 nucleotides in length via 1:1 stoichiometry (26), measurement of At across samples containing varying concentrations of peptide where RNA concentration is limiting (ie: [TTP73]free · [TTP73]total) thus permits resolution of binding constants by Equation 2:
| (2) |
Here, AR and APR represent the intrinsic anisotropy values of the free and protein-associated Fl-RNA substrates, respectively, while K represents the equilibrium association constant. AR was measured directly from binding reactions lacking peptide (n ≥ 3), while all other constants were solved by non-linear least squares regression of At versus [TTP73] plots using PRISM v3.0 (GraphPad, San Diego, CA).
Tryptophan Fluorescence and Quenching Analyses
Tryptophan fluorescence was measured using a Cary Eclipse spectrofluorometer (Varian Instruments, Walnut Creek, CA) fitted with a sub-micro cell. Samples (60 μl total volume) containing peptide (2 – 2.5 μM) were prepared with ZnCl2 (25 μM) or EDTA (0.5 mM) as described for fluorescence anisotropy analyses (above). Heparin was omitted from reactions employed for tryptophan fluorescence assays, since protein contaminants in commercially available heparin preparations were found to contribute significant background fluorescence (data not shown). RNA substrates were added at equimolar concentrations where indicated, and tryptophan emission spectra were measured using 295 nm excitation with 10 nm bandwidth for both excitation and emission monochromators. For measurements of solvent accessibility, varying concentrations of potassium iodide or acrylamide were added after 30 minutes of RNA:peptide complex formation. Stern-Volmer quench constants (KSV) were extracted from measured emission at 350 nm (F) as a function of quencher concentration ([Q]) using Equation 3, where F0 represents the fluorescence intensity in the absence of the quencher (33):
| (3) |
For denaturant titration experiments, varying concentrations of GnHCl were added after 30 minutes of RNA:peptide complex formation, then incubated for a further 30 minutes at room temperature prior to measurement of tryptophan emission spectra.
RESULTS
The Zinc Fingers of TTP Exhibit Different Structural Responses upon Coordination of Zn2+
Zn2+-dependent changes in the conformation of the TTP zinc finger domains were initially assessed by measuring Trp emission spectra of the TTP73 F11W and F49W peptides in the presence and absence of zinc (Figure 2). Fluorescence emission from the wild type TTP73 peptide was barely detectable above background. Since TTP73 wt lacks Trp residues, this confirmed that other aromatic residues (Phe and Tyr) do not significantly contribute to total fluorescence emission from the TTP73 mutant peptides measured under these conditions. The intensity of fluorescence emission from TTP73 F11W was decreased by 50% in the presence of Zn2+, indicating that the conformation of the N-terminal zinc finger was sensitive to the presence of the cation. The F11W fluorescence emission peak was also blue-shifted from 356 nm to 346 nm (Table 1), consistent with transition of the Trp residue side chain to a less polar environment in a Zn2+-dependent manner (33). By contrast, neither the fluorescence quantum yield nor emission peak wavelength of the F49W mutant was significantly modified by Zn2+, indicating that the C-terminal zinc finger does not experience the same conformational transition as the N-terminal finger in the presence of the cation.
FIGURE 2.
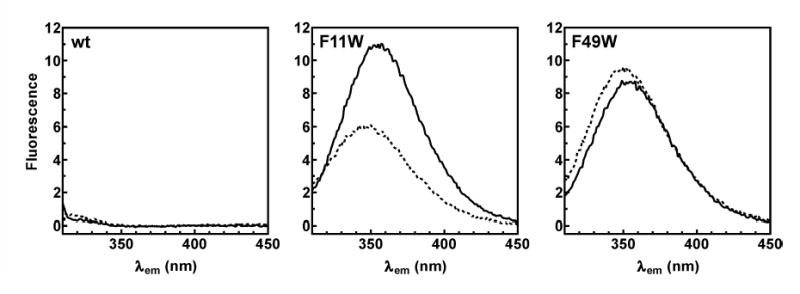
Fluorescence emission spectra of TTP73 peptides. Blank-corrected emission spectra (λex = 295 nm) of wild type TTP73 (wt; left panel) and TTP73 mutant peptides F11W (center panel) and F49W (right panel) were measured following incubation for 20 minutes at room temperature in RNA binding buffer containing EDTA (0.5 mM; solid line) or ZnCl2 (25 μM; dotted line). Peptide concentrations were 2 μM for each sample. Spectral normalization was omitted to permit comparison of relative fluorescence quantum yields between peptides.
Table 1.
Emission Properties of TTP73 Phe→Trp Mutant Peptides
| Peptide | Zn2+ | RNA | Emission peak (nm)a | Relative intensity at 350 nma,b |
|---|---|---|---|---|
| F11W | − | - | 356 ± 1 | 1.0 |
| + | - | 346 ± 1 | 0.51 ± 0.06 | |
| + | UC13 | 347 ± 2 | 0.54 ± 0.02 | |
| + | ARE13 | 347 ± 1 | 0.30 ± 0.04 | |
| + | ARE13-AU3U | 348 ± 1 | 0.30 ± 0.03 | |
| + | ARE13-UU3A | 347 ± 2 | 0.33 ± 0.05 | |
| F49W | − | - | 354 ± 1 | 1.0 |
| + | - | 352 ± 1 | 1.11 ± 0.06 | |
| + | UC13 | 352 ± 1 | 0.98 ± 0.04 | |
| + | ARE13 | 354 ± 1 | 0.68 ± 0.06 | |
| + | ARE13-AU3U | 352 ± 2 | 1.01 ± 0.05 | |
| + | ARE13-UU3A | 352 ± 1 | 0.99 ± 0.05 |
mean ± σn−1 for n ≥ 3 independent experiments
intensity relative to -Zn2+ for each peptide
As an independent measure of Zn2+-dependent conformational changes in the TTP zinc finger domains, the solvent accessibilities of Trp residues at positions 11 and 49 were analyzed by collisional quenching in the presence or absence of Zn2+. For the TTP73 F11W peptide, Zn2+ decreased Stern-Volmer constants (KSV) derived from titrations of iodide or acrylamide quenchers by 56% and 40%, respectively (Figure 3 and Table 2), indicating significant loss of solvent accessibility concomitant with Zn2+-dependent conformational changes in the N-terminal finger. Conversely, Zn2+ induced no significant changes in KSV values for the TTP73 F49W peptide, supporting the model of little or no net Zn2+-dependent alterations of peptide conformation in this region of the C-terminal zinc finger domain. Control experiments with free tryptophan confirmed that the Zn2+ concentrations employed in this study did not systematically influence the fluorescence quantum yield or solvent accessibility of this amino acid (data not shown). For both TTP73 mutant peptides, fluorescence emission from Trp residues was more efficiently quenched by acrylamide versus iodide (Table 2). This observation is consistent with previous studies showing that acrylamide quenches the fluorescence of a tryptophan analogue by electron transfer whereas iodide quenching relies on a collisional heavy atom effect (39). As a result, acrylamide can quench fluorescent compounds from greater distances in solution, rendering it a more effective quenching agent.
FIGURE 3.
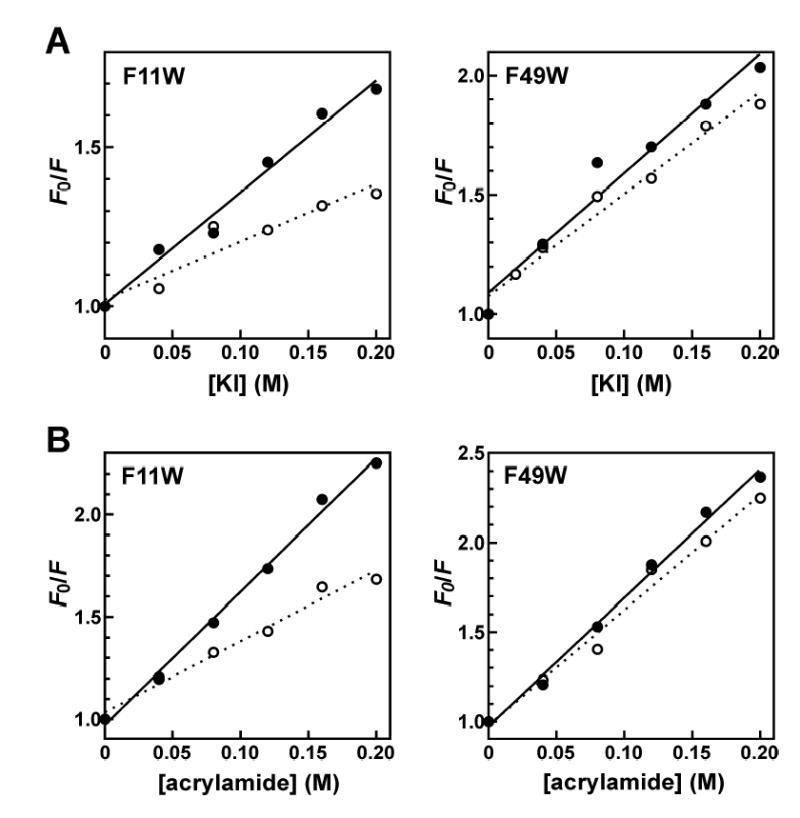
Effects of Zn2+ coordination on solvent accessibility of Trp residues at TTP73 positions 11 and 49. Stern-Volmer plots were prepared from fluorescence emission (λex = 295 nm; λem = 350 nm) of TTP73 F11W or F49W peptide samples (2.5 μM each) containing varying concentrations of potassium iodide (A) or acrylamide (B) as described under “Experimental Procedures”. Prior to assembly of quenching reactions, TTP73 mutant peptides were incubated with EDTA (solid circles, solid line) or ZnCl2 (open circles, dotted line) as in Figure 2.
Table 2.
Stern-Volmer Quench Constants for TTP73 Phe→Trp Mutant Peptides
| Peptide | Zn2+ | RNA | Quencher | KSV (M−1)a | n |
|---|---|---|---|---|---|
| F11W | − | - | KI | 3.4 ± 0.2 | 3 |
| + | - | KI | 1.5 ± 0.2 | 3 | |
| − | - | acrylamide | 6.0 ± 0.5 | 3 | |
| + | - | acrylamide | 3.6 ± 0.3 | 3 | |
| + | UC13 | acrylamide | 4.4 ± 0.2 | 3 | |
| + | ARE13 | acrylamide | 1.4 ± 0.3 | 3 | |
| + | ARE13-AU3U | acrylamide | 1.5 ± 0.4 | 2 | |
| + | ARE13-UU3A | acrylamide | 1.5 ± 0.3 | 2 | |
| F49W | − | - | KI | 4.7 ± 0.3 | 3 |
| + | - | KI | 4.2 ± 0.1 | 4 | |
| − | - | acrylamide | 6.4 ± 0.4 | 4 | |
| + | - | acrylamide | 6.4 ± 0.6 | 4 | |
| + | UC13 | acrylamide | 7.3 ± 0.6 | 3 | |
| + | ARE13 | acrylamide | 1.3 ± 0.2 | 3 | |
| + | ARE13-AU3U | acrylamide | 5.9 ± 0.2 | 2 | |
| + | ARE13-UU3A | acrylamide | 5.6 ± 0.4 | 3 |
mean ± σn−1 for n independent experiments where n > 2, or mean ± spread for n = 2.
TTP73 F11W and F49W Form High Affinity Complexes with ARE Substrates
Previously, we demonstrated that the TTP73 wt peptide forms high affinity (Kd < 20 nM) interactions with RNA substrates of the form UUAUnAUU, where n ranges from 2 to 5 (26). To ensure that the Trp substitutions at positions 11 and 49 of TTP73 did not dramatically modify its RNA-binding activity, we employed gel mobility shift assays (GMSAs) and fluorescence anisotropy-based methods to characterize interactions between RNA substrates and the TTP73 F11W or F49W mutants. By GMSA, both the TTP73 F11W and F49W peptides formed single, co-migrating RNA:peptide complexes with either 9-base (ARE9; Figure 4A) or 13-base (ARE13; data not shown) RNA substrates containing canonical TTP binding motifs. Sequence specificity for ARE ligands did not appear to be compromised, since no peptide binding was detected to an irrelevant 13-base RNA substrate (UC13; Figure 4A), even at peptide concentrations up to 2.5 μM (data not shown). Quantitative analyses using fluorescence anisotropy (Figure 4B) indicated that association of TTP73 F11W and F49W peptides with fluorescein-labeled ARE substrates was well resolved by a binary equilibrium binding model (Equation 2). Derived bimolecular association constants showed a small but not statistically significant decrease in the affinity of TTP73 F11W for each of the 9- and 13-base ARE substrates relative to the wild type peptide (Table 3). Binding of the F49W mutant to each ARE ligand returned slightly weaker dissociation constants than for the F11W peptide (Kd = 6.8 – 8.3 nM); however, binding affinities yielding Kd values of up to 20 nM have been observed between TTP73 wt and other ARE substrates exhibiting potent mRNA-destabilizing activity (10, 28). This demonstration of specific, high affinity interactions between each TTP73 Phe→Trp mutant and ARE substrates validated the use of these peptides as probes of local protein structure in peptide:ARE complexes.
FIGURE 4.
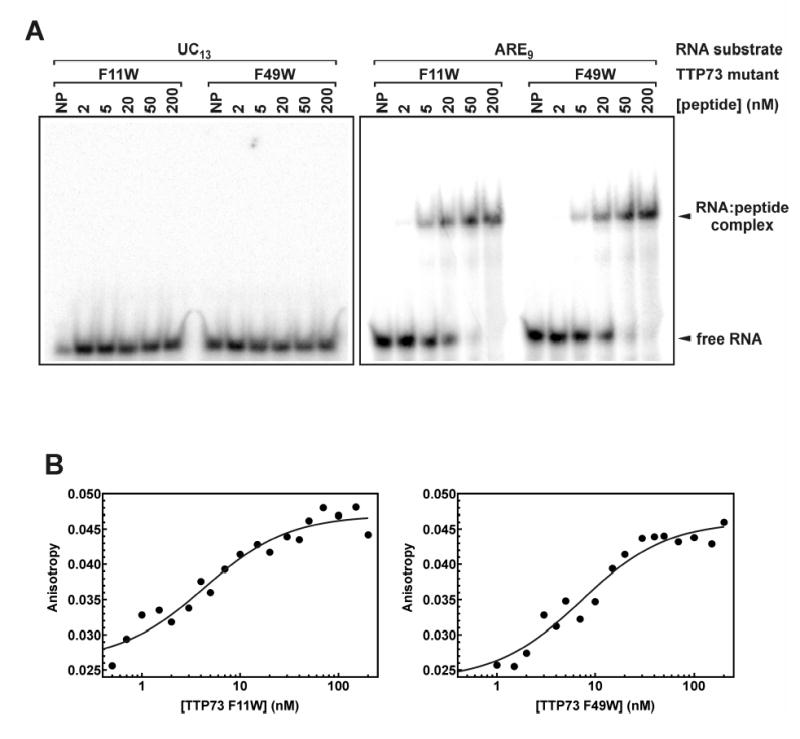
Association of TTP73 mutant peptides with RNA substrates. (A) Binding reactions containing 32P-labeled RNA substrates (0.2 nM) and varying concentrations of TTP73 F11W or F49W peptides were fractionated by native gel electrophoresis. The position of the complex formed by peptide binding to the ARE9 substrate is indicated. (B) Fluorescence anisotropy (λex = 490 nm; λem = 535 nm) was measured for binding reactions containing the Fl-ARE9 RNA substrate (0.2 nM) and titrations of TTP73 F11W (left panel) or F49W (right panel) peptides assembled as described under “Experimental Procedures”. Association constants describing the interaction of each peptide with the Fl-ARE9 ligand were resolved by nonlinear regression using Equation 2 (solid lines).
Table 3.
Equilibrium Binding of TTP73 Phe→Trp Mutant Peptides to RNA Substrates
| RNA | Peptide | K (× 108 M−1)a | Kd (nM)b | n |
|---|---|---|---|---|
| Fl-ARE9 | wt | 3.3 ± 0.4c | 3.0 | 3 |
| F11W | 2.23 ± 0.05 | 4.5 | 2 | |
| F49W | 1.46 ± 0.07 | 6.8 | 2 | |
| Fl-ARE13 | wt | 2.8 ± 0.5c | 3.6 | 4 |
| F11W | 1.7 ± 0.2 | 5.9 | 2 | |
| F49W | 1.2 ± 0.1 | 8.3 | 2 |
mean ± sn−1 for n independent experiments where n = 2, or mean ± spread for n = 2.
Kd estimated as 1/K.
wild type TTP73 binding data are from Ref. 26.
Cognate ARE Binding Induces Conformational Changes within Both Zinc Fingers of TTP
Following the demonstration that the TTP73 F11W and F49W peptides were competent for high affinity ARE binding, fluorescence emission and collisional quenching studies were performed to identify RNA substrate-dependent changes in the conformations of each TTP zinc finger domain. RNA and peptide components were assembled at concentrations at least 300-fold above Kd (2.5 μM of each reagent) to promote quantitative formation of RNA: peptide complexes. Fluorescence emission from both the F11W and F49W mutants was decreased by approximately 40% upon binding the ARE13 substrate relative to reactions supplemented with Zn2+ alone (Figure 5A and Table 1). RNA-dependent changes in the fluorescence of each peptide were specific for the high affinity ARE ligand, since substitution of a non-specific RNA substrate (UC13) did not significantly influence the fluorescence properties of either mutant relative to reactions lacking RNA. This demonstrates that the diminished fluorescence emission from the F11W and F49W mutants is a consequence of ARE substrate binding, and not a trivial result of collisional quenching by RNA in solution. Trp residues in both the 11 and 49 positions of TTP73 also displayed dramatic decreases in solvent accessibility when bound to an ARE ligand (Figure 5B). ARE13 substrate binding decreased Stern-Volmer constants derived from acrylamide titrations by 60% for the F11W peptide and 80% for the F49W mutant (Table 2). Like the RNA-dependent changes in fluorescence emission, restriction of solvent accessibility within each zinc finger domain was specific for ARE binding, since acrylamide quenching efficiency was not significantly modulated by the presence of the UC13 substrate for either TTP73 mutant peptide. Taken together, the decreased fluorescence emission and restricted solvent accessibility of Trp residues in both the TTP73 F11W and F49W peptides in response to ARE binding indicate that both zinc finger domains are structurally modified by association with cognate ARE substrates.
FIGURE 5.
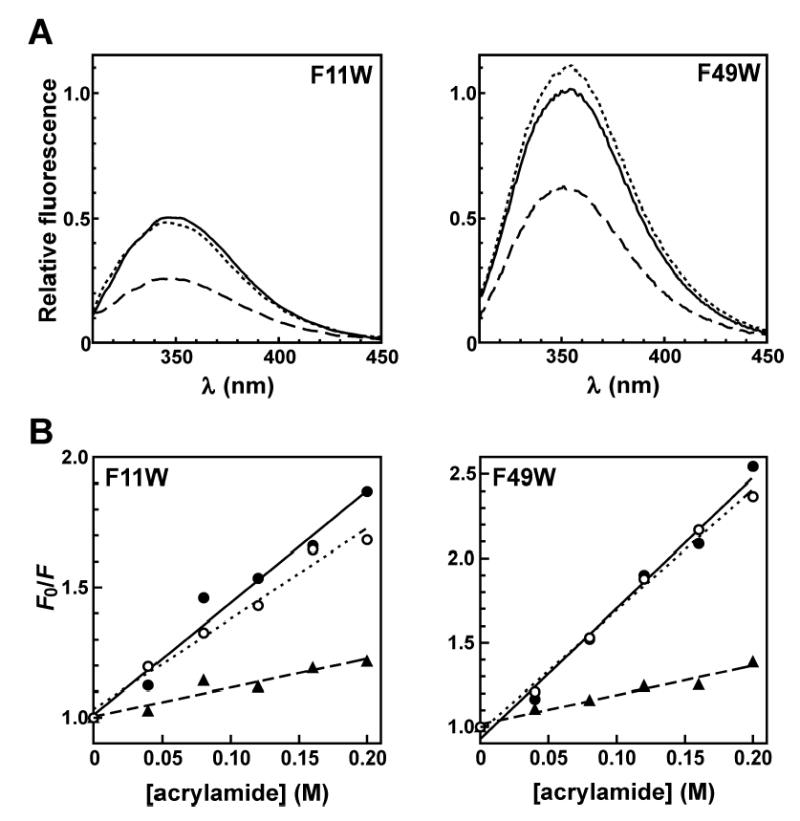
RNA-dependent changes in fluorescence emission and solvent accessibility of Trp residues at TTP73 positions 11 and 49. (A) Blank-corrected emission spectra (λex = 295 nm) of TTP73 F11W (2.5 μM; left panel) or F49W (2.5 μM; right panel) pre-incubated with ZnCl2 (25 μM) in the absence (dotted line) or presence of RNA substrates (2.5 μM) UC13 (solid line) or ARE13 (dashed line). Fluorescence spectra from each sample are shown relative to the emission of reactions containing EDTA (Figure 2). (B) Acrylamide quenching of Zn2+-conjugated TTP73 F11W (left panel) or F49W (right panel) in the absence (open circles, dotted line) or presence of RNA substrates UC13 (solid circles, solid line) or ARE13 (triangles, dashed line) resolved by Stern-Volmer plot. The sensitivity of TTP73:ARE equilibria to ionic strength precluded the use of potassium iodide as a collisional quencher in these experiments (26).
Additional experiments investigated the conformational events resulting from TTP73 peptide association with sub-optimal RNA substrates. For ARE ligands lacking adenosine residues defining either the 5′- (ARE13-UU3A) or 3′-(ARE13-AU3U) UUAU half-sites, wild-type TTP73 exhibits impaired but still significant binding activity (Kd ≈ 55 nM) (26). TTP73 F11W exhibited significant decreases in both Trp fluorescence emission and solvent accessibility upon binding to either of these ARE half-site mutant ligands, suggesting structural consequences similar to those observed with the cognate ARE substrate (Tables 1 and 2). However, the fluorescence characteristics of the TTP73 F49W peptide were not significantly influenced by association with either ARE half-site mutant. These data indicate that conformational adaptation of the N-terminal finger can occur independently of the C-terminal finger, and that RNA-dependent folding of the C-terminal finger may structurally discriminate TTP73:RNA complexes containing cognate versus near-cognate ARE substrates.
ARE Binding Stabilizes Folded States Involving Both Zinc Fingers of TTP
The preceding experiments indicated that at least one structural transition is experienced by Trp in position 49 of TTP73 in response to binding a cognate ARE ligand. By contrast, at least two distinct folding events were observed for Trp in position 11; one dependent on Zn2+ and another on binding to an ARE substrate. In order to evaluate the relative stabilities of each conformational transition, the fluorescence properties of TTP73 F11W and F49W peptides were monitored in the presence and absence of RNA substrates across titrations of the chemical denaturant, guanidine hydrochloride (GnHCl).
In the presence of Zn2+ but the absence of RNA, the TTP73 F49W peptide displayed enhanced fluorescence emission with increasing concentrations of GnHCl (Figure 6A). Emission from the F49W mutant increased by approximately 70% at 100 mM GnHCl (Figure 6B) but was not significantly elevated beyond this intensity, even at GnHCl concentrations as high as 3 M (data not shown). The increase in fluorescence of F49W as a function of denaturant concentration is consistent with structural relaxation near the Trp residue in the C-terminal zinc finger. Curiously, this transition was extremely sensitive to GnHCl, with the half-maximal effect observed at only 1–2 mM denaturant. However, the lack of approach to a lower asymptote at minimal denaturant concentrations precluded quantitative resolution of domain unfolding free energies by linear extrapolation (40, 41).
FIGURE 6.
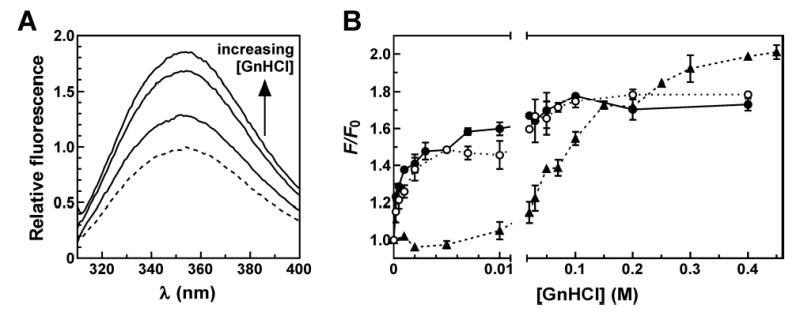
Denaturation of TTP73 F49W by GnHCl titration. (A) TTP73 F49W (2.5 μM) samples pre-incubated with ZnCl2 (25 μM) were treated with 0.5, 10, or 100 mM GnHCl for 30 minutes at room temperature before measurement of fluorescence emission spectra (λex = 295 nm). Blank-corrected spectra are shown normalized to the sample lacking GnHCl (dashed line). (B) Fluorescence emission (λex = 295 nm; λem = 350 nm) of TTP73 F49W across a titration of GnHCl in the absence (open circles, dotted line) or presence of equimolar concentrations of RNA substrates UC13 (solid circles, solid line) or ARE13 (triangles, dashed line). Fluorescence measurements were normalized to the emission of each F49W:RNA combination lacking GnHCl (F0). Points represent the mean ± spread of duplicate independent experiments.
Similar GnHCl denaturation experiments performed with TTP73 F49W in the presence of the UC13 RNA substrate closely mirrored the unfolding transition of TTP73 F49W in the absence of RNA. However, association of the F49W peptide with the ARE13 substrate significantly stabilized a peptide conformation in the C-terminal finger, with half-maximal unfolding observed at approximately 100 mM GnHCl (Figure 6B). The amplitude of the fluorescence change in the presence of the ARE13 ligand is enhanced relative to the other data sets because the fluorescence intensity of the TTP73 F49W:ARE13 complex in the absence of GnHCl (F0) is lower than those of the F49W peptide alone or in the presence of the UC13 substrate (Table 1). The extreme sensitivity of TTP73 F49W fluorescence to GnHCl suggests that the C-terminal zinc finger is very weakly folded or presents a dynamic population of conformations near position 49 in the absence of an ARE. The significant decrease in denaturant sensitivity exhibited in the presence of the ARE13 substrate may thus represent stabilization of one or a small subset of peptide conformations within the C-terminal zinc finger by association with a cognate RNA ligand.
Like the F49W mutant, TTP F11W displayed enhanced fluorescence emission with increasing concentrations of GnHCl in the presence of Zn2+ but absence of RNA (Figure 7A). Unlike F49W, however, monitoring fluorescence intensity at 350 nm as a function of denaturant concentration revealed two distinct conformational transitions for the F11W peptide. The weakest transition enhanced fluorescence intensity by approximately 30% (Figure 7B inset, open circles; cf. 0 M GnHCl versus 0.01 M), with half-maximal change observed at 1 mM GnHCl. The second transition was more gradual, with fluorescence emission increasing by approximately 100% across a broad range of GnHCl concentrations above 0.6 M. In addition to increases in fluorescence emission, high concentrations of GnHCl also reversed the blue-shifted position of the F11W peak induced by Zn2+ coordination (Figure 7A). Evaluation of F11W emission spectra across the GnHCl titration showed that loss of the blue-shifted emission peak accompanied the second (ie: more stable) transition, and was most pronounced between 0.6 – 2 M GnHCl (Figure 7C). The concurrent increase in fluorescence intensity and loss of the emission peak blue-shift of TTP73 F11W observed at GnHCl concentrations above 0.6 M indicates that this more stable conformational transition likely involves the release of Zn2+. By contrast, the weaker transition may represent conformational heterogeneity near position 11 of TTP73, similar to the model proposed for F49W (above). However, the amplitude of fluorescence change accompanying this weaker folding transition is smaller for F11W than for F49W, suggesting that the distribution or flexibility of local peptide conformations may be more constrained in the N-terminal finger relative to the C-terminal domain.
FIGURE 7.
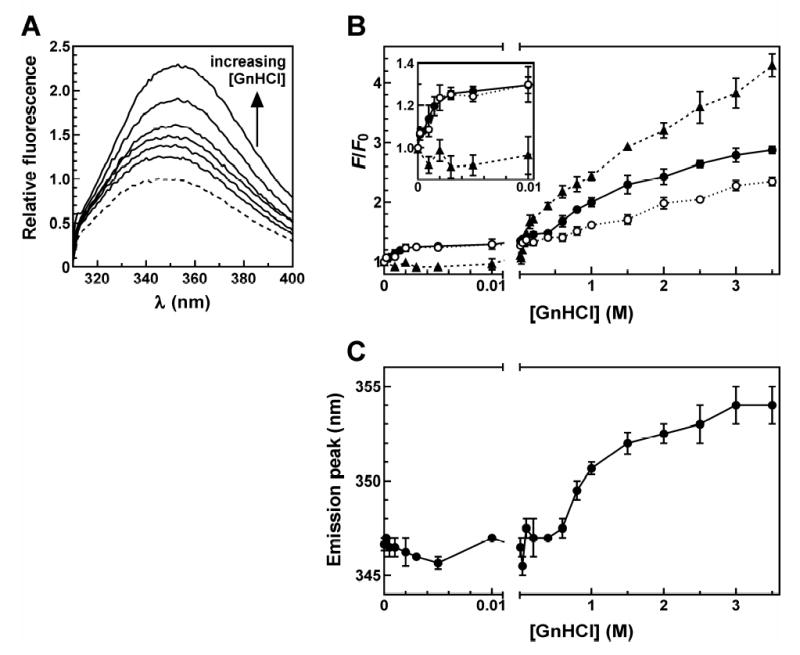
Denaturation of TTP73 F11W by GnHCl titration. (A) TTP73 F11W (2.5 μM) samples pre-incubated with ZnCl2 (25 μM) were treated with 0.005, 0.02, 0.1, 1, 2, or 3 M GnHCl for 30 minutes at room temperature before measurement of fluorescence emission spectra (λex = 295 nm). Blank-corrected spectra are shown normalized to the sample lacking GnHCl (dashed line). (B) Fluorescence emission (λex = 295 nm; λem = 350 nm) of TTP73 F11W across a titration of GnHCl in the absence (open circles, dotted line) or presence of equimolar concentrations of RNA substrates UC13 (solid circles, solid line) or ARE13 (triangles, dashed line), normalized to emission from each F11W:RNA combination lacking GnHCl (F0). Points represent the mean ± σn−1 of triplicate independent experiments. (C) Positions of emission intensity peaks were extracted from emission spectra and plotted for TTP73 F11W samples across a titration of GnHCl in the absence of RNA. Each point represents the mean ± σn−1 of triplicate independent experiments.
Similar to the F49W example, inclusion of the non-binding UC13 RNA substrate did not significantly influence the sensitivity of TTP73 F11W fluorescence to GnHCl (Figure 7B, closed circles). Addition of the ARE13 substrate, however, dramatically stabilized the weaker transition, delaying denaturant-induced increases in fluorescence intensity to GnHCl concentrations >100 mM (Figure 7B, triangles). Also similar to F49W, the change in fluorescence accompanying the initial F11W unfolding event in the presence of the ARE ligand exhibited a greater amplitude than reactions lacking this substrate, owing to the decreased fluorescence of the F11W:ARE13 complex in the absence of GnHCl (F0; Table 1). The addition of RNA substrates did not appear to significantly influence the more stable conformational transition of F11W, based on similarities in the amplitude of fluorescence change, the GnHCl concentration-dependence of this transition (Figure 7B), or loss of the emission peak blue-shift (data not shown). Together, these data support a model whereby the conformation of the N-terminal zinc finger of TTP is substantially more ordered by the coordination of Zn2+ than the C-terminal finger. Second, the GnHCl titration experiments indicate that even in the presence of Zn2+, local peptide structures within both the N- and C-terminal zinc finger domains of TTP retain some degree of flexibility or structural heterogeneity, but are further constrained and/or stabilized concomitant with high affinity RNA ligand binding.
DISCUSSION
To date, no high resolution molecular structure has been reported for the tandem zinc finger domain of TTP or any related CCCH zinc finger peptide in the absence of RNA. In this study, we provide evidence for distinct structural consequences within each zinc finger of TTP in response to association of zinc ions or RNA substrates. Furthermore, the RNA sequence-dependence of folding events within C-terminal zinc finger likely contributes to discrimination of cognate versus near-cognate RNA ligands.
Zn2+-dependent changes in conformation of the TTP zinc finger domains
Zn2+-dependent folding of the N-terminal zinc finger was revealed by decreased quantum yield of TTP73 F11W, significantly blue-shifted emission from this peptide, and decreased solvent accessibility of the Trp residue at position 11 in the presence of zinc (Tables 1 and 2). Denaturant titration experiments also demonstrated considerable folded stability for the Zn2+-associated N-terminal zinc finger. Half-maximal reversal of the Zn2+-induced F11W emission peak blue-shift ([GnHCl]1/2) was observed at approximately 0.8 M GnHCl, concomitant with increases in fluorescence intensity. By comparison, the structures of some well-characterized globular proteins show similar sensitivities to this denaturant, including α-lactalbumin ([GnHCl]1/2 = 1.1 M) (42) and maltose binding protein ([GnHCl]1/2 = 1.0 M) (43). Together, these data indicate that the N-terminal zinc finger of TTP73 adopts a stable conformation following Zn2+ coordination that includes partial burial of the residue at position 11. A previous study also suggested a defined Zn2+-dependent conformation for the N-terminal zinc finger of the TTP-related protein TIS11d, since HSQC spectra could not be resolved in the absence of Zn2+, but were well dispersed in the presence of the cation (31). Also, HSQC analyses permitted resolution of most residues in the N-terminal zinc finger of human (28) and murine (44) TTP in presence of Zn2+ but absence of RNA.
While efficient and stable coordination of Zn2+ to the N-terminal zinc finger domain was anticipated from the literature and corroborated by this study, no data describing Zn2+-dependent conformational changes in the TTP C-terminal zinc finger have been reported. Although residues within the C-terminal zinc finger domain of TIS11d were resolved by HSQC in the presence of Zn2+ and absence of RNA (31), this was not the case for the wild type TTP73 peptide under these conditions (28). In the current study, the Trp residue inserted at position 49 of TTP73 revealed no significant changes in quantum yield, emission peak wavelength, or solvent accessibility in the presence of Zn2+ (Tables 1 and 2). Based on the TIS11d:ARE structure (31), Trp residues in positions 11 and 49 of TTP73 were anticipated to exist in similarly folded environments (Figure 1C and D), particularly with respect to potential stacking interactions between each Trp and Zn2+-coordinated His residues (His27 and His65). While this or a similar conformation is possible for the N-terminal zinc finger domain in the presence of Zn2+ alone (above), we conclude that such an arrangement for the TTP73 C-terminal finger is highly improbable in the absence of RNA given the insensitivity of TTP73 F49W fluorescence to Zn2+. High affinity binding between TTP73 F49W and ARE substrates (Table 3) refutes the possibility that the F49W mutation substantially impairs or precludes Zn2+-coordination to the C-terminal zinc finger, since: [i] association of TTP73 with ARE substrates is absolutely dependent on Zn2+ (26), [ii] mutation of Zn2+-coordinating residues in the C-terminal zinc finger domain abrogate RNA-binding activity (34), and [iii] RNA-binding by the N-terminal finger alone exhibits only micromolar affinity (45). An alternate possibility is that Zn2+ binds but might not tightly coordinate with the C-terminal zinc finger in the absence of an RNA substrate. This would account for the independence of TTP73 F49W fluorescence with respect to Zn2+, the extreme sensitivity of F49W fluorescence to GnHCl in the absence of RNA, and the lack of resolvable HSQC assignments in this domain (28). Furthermore, Co2+ titrations performed by Worthington et al. suggest sub-stoichiometric interaction of metal ions with the C-terminal zinc finger of TTP (46). While 1:1 binding was observed between Co2+ and the N-terminal zinc finger of TTP, comparable experiments with the tandem zinc finger domain yielded an average binding ratio of 1.7 moles metal ion per mole peptide. Taken together, these data suggest that the conformation of the C-terminal finger might sample from a population of structures, possibly involving dynamic association of Zn2+, which may be coupled to the specificity of RNA substrate recognition (discussed below).
RNA-dependent changes in conformation of the TTP zinc finger domains
Previous NMR and thermodynamic studies indicated that association with a cognate RNA substrate was accompanied by conformational changes within both zinc finger domains of TTP or TIS11d, and defined a cognate substrate as an RNA ligand containing tandem UUAU motifs (26, 28, 31). From the current study, this model is supported by significant decreases in fluorescence emission from both TTP73 F11W and F49W when bound to the cognate ARE13 substrate (Table 1). Furthermore, the F11W and F49W peptides exhibited virtually equivalent Stern-Volmer quench constants upon binding the ARE13 ligand (Table 2), consistent with placement of each Trp residue in a similar structural environment. Localization of the Trp substitutions distal to the RNA-binding interface of each zinc finger indicates that the ARE-dependent changes in Trp fluorescence likely result from conformational changes in the peptide rather than steric effects from the associated RNA ligand. However, two observations illustrate subtle differences in folding between the TTP zinc finger domains, even when bound to RNA. First, emission from the F49W peptide was not blue-shifted by ARE-induced peptide folding, suggesting a more polar environment for the substituted Trp in the C-terminal versus N-terminal zinc fingers (Table 1). Second, the F49W peptide retained an increased fluorescence quantum yield relative to F11W in the presence of the ARE (Table 1). Differences in local environments could be due to the residues flanking the proposed Trp:His stack in each zinc finger (Figure 1). Based on the TIS11d:ARE structure, the Trp in position 11 would be positioned within 3.4 Å of Glu12 (Ser in TTP) in the peptide:ARE complex, while the Trp in position 49 would be juxtaposed by His50 (3.2 Å; Tyr in TTP). The adjacent residues could alter the orientation or flexibility of Trp residue side chains in the folded zinc finger domains, or could differentially quench Trp emission (47).
While both zinc finger domains exhibited conformational changes upon binding to a cognate RNA ligand, only the N-terminal finger displayed similar folding behavior when associated with near-cognate ARE substrates, in which either the 5′- or 3′-UUAU motif was mutated (Tables 1 and 2). One possibility is that the “Zn2+-primed” N-terminal finger binds the RNA ligand first, and hence occupies the first accessible UUAU half-site. Recognition of a cognate site would then be achieved by comparable structural rearrangements within the C-terminal finger, requiring a second UUAU motif immediately upstream of the first. In this manner, the conformational status of the C-terminal finger may constitute the final step in discrimination of RNA ligands. While U-rich RNA sequences containing a single UAUU motif can bind the TTP tandem zinc finger domain, these ligands bind 10- to 20-fold more weakly than a cognate RNA binding site (26), and are incapable of directing rapid cytoplasmic mRNA decay through TTP (10).
Might RNA substrate recognition involve conformational sampling within TTP zinc finger domains?
The fluorescence of Trp residues in both positions 11 and 49 of TTP73 exhibited extreme sensitivity to GnHCl in the absence of an ARE ligand (Figures 6 and 7). These data could denote either thermodynamically weak local peptide conformations and/or dynamic populations of heterogeneously folded structures within each zinc finger. With respect to the N-terminal zinc finger, some support for the latter possibility comes from previous NMR studies where superimposing refined structures of the Zn2+-coordinated N-terminal zinc finger of murine TTP revealed significant structural heterogeneity in the absence of RNA (44). The distribution of local peptide conformations was particularly diverse among amino acid side chains not involved in coordination of Zn2+. For the C-terminal zinc finger domain, the lack of defined HSQC peaks in the absence of an RNA substrate (28) coupled with insensitivity of TTP73 F49W fluorescence to Zn2+ (this study) also support a model of heterogeneous peptide conformations prior to ARE binding. However, several observations suggest that association of the TTP RNA-binding domain with cognate RNA substrates is coupled to restriction of conformational heterogeneity within each zinc finger domain. First, emission from Trp residues in positions 11 and 49 of TTP73 became significantly less sensitive to GnHCl upon binding to an ARE ligand (Figures 6B and 7C), consistent with RNA-dependent contributions to the thermodynamic stability of peptide folding within each zinc finger. Second, resolution of HSQC peaks to residues in the C-terminal zinc finger following ARE binding (28) supports RNA substrate-dependent stabilization of a single or small population of related peptide structures. Finally, thermodynamic characterization of TTP73 binding to high affinity ARE substrates indicated a significant negative change in heat capacity accompanying peptide:RNA complex formation. This change in heat capacity was manifested as an enhanced entropic penalty for cognate ARE ligand binding relative to non-canonical RNA substrates, but was compensated by improved binding enthalpy when an upstream UAUU motif was available for association with the C-terminal zinc finger domain (26). Taken together, the structural resistance to denaturant, resolution of HSQC peaks, and unfavorable changes in entropy associated with TTP73 binding to cognate ARE substrates are consistent with restricted local peptide mobility in the TTP73: ARE complex relative to the unbound TTP73 peptide.
Data from this study supports a model for ARE substrate recognition by the TTP RNA-binding domain whereby the Zn2+-coordinated zinc fingers exhibit significant local flexibility in the absence of an ARE ligand, particularly within the C-terminal finger. It remains possible that such conformational sampling may be less pronounced in the full length protein, where structural restraints from flanking protein sequences may limit the conformational heterogeneity of the zinc finger domain. Binding to a cognate ARE substrate is associated with stabilization of a unique or small population of folded states within each zinc finger, consistent with “induced-fit” or “selected-fit” models of macromolecular complex formation (29, 30). Characteristics of these stabilized folded states include residue side chain burial (denoted by decreased fluorescence quantum yield and collisional quenching constants) at sites distal to the RNA-binding interface (this study) and improved binding potential for cognate RNA substrates, reflected in increased enthalpy of binding (26). Furthermore, these studies establish the potential of TTP73 site-directed Trp mutants as powerful tools for forthcoming structural and energetic investigations of RNA recognition by this tandem zinc finger domain. For example, can RNA-dependent folding of the C-terminal zinc finger occur independently of the folded N-terminal zinc finger? Can tandem N-terminal (or C-terminal) zinc finger domains convey RNA binding affinity and specificity comparable to the wild-type peptide? Finally, is adaptive RNA recognition by the wild-type TTP tandem zinc finger domain obligate for the mRNA-destabilizing activity of this protein? We anticipate that future studies including analyses of additional site-specific Trp mutants of the TTP73 peptide will permit resolution of these questions.
Acknowledgments
We thank Dr. Kristin Varney for assistance with the molecular modeling of the TIS11d: RNA complex.
Footnotes
This work was supported by NIH/NCI grant CA102428 and a Scientist Development Grant from the American Heart Association (to G.M.W.).
Author e-mail: gwils001@umaryland.edu
Abbreviations: ARE, AU-rich element; GMSA, gel mobility shift assay; GnHCl, guanidine hydrochloride; HSQC, heteronuclear single quantum coherence; TTP, tristetraprolin; UTR, untranslated region
References
- 1.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 2.Hargrove JL, Schmidt FH. The role of mRNA and protein stability in gene expression. FASEB J. 1989;3:2360–2370. doi: 10.1096/fasebj.3.12.2676679. [DOI] [PubMed] [Google Scholar]
- 3.Chen CYA, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 4.Bakheet T, Williams BRG, Khabar KSA. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? . Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shabalina SA, Ogurtsov AY, Lipman DJ, Kondrashov AS. Patterns in interspecies similarity correlate with nucleotide composition in mammalian 3′UTRs. Nucleic Acids Res. 2003;31:5433–5439. doi: 10.1093/nar/gkg751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tholanikunnel BG, Raymond JR, Malbon CC. Analysis of the AU-rich elements in the 3′-untranslated region of β2-adrenergic receptor mRNA by mutagenesis and identification of the homologous AU-rich region from different species. Biochemistry. 1999;38:15564–15572. doi: 10.1021/bi9913348. [DOI] [PubMed] [Google Scholar]
- 8.Fialcowitz EJ, Brewer BY, Keenan BP, Wilson GM. A hairpin-like structure within an AU-rich mRNA-destabilizing element regulates trans-factor binding selectivity and mRNA decay kinetics. J Biol Chem. 2005;280:22406–22417. doi: 10.1074/jbc.M500618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu N, Chen CYA, Shyu AB. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai WS, Carrick DM, Blackshear PJ. Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. J Biol Chem. 2005;280:34365–34377. doi: 10.1074/jbc.M506757200. [DOI] [PubMed] [Google Scholar]
- 11.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brewer G. Evidence for a 3′-5′ decay pathway for c-myc mRNA in mammalian cells. J Biol Chem. 1999;274:16174–16179. doi: 10.1074/jbc.274.23.16174. [DOI] [PubMed] [Google Scholar]
- 13.Chen C-Y, Gherzi R, Ong S-E, Chan EL, Raijmakers R, Pruijn GJM, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 14.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 15.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO reports. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao M, Wilusz CJ, Peltz SW, Wilusz J. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 2001;20:1134–1143. doi: 10.1093/emboj/20.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson GM, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acids Res Mol Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T, Kruys V, Huez G, Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-acting factors. Biochem Soc Trans. 2002;30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]
- 19.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin S-C, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 20.Blackshear PJ. mRNA degradation: an important process in controlling gene expression. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 21.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson GM, Sun Y, Lu H, Brewer G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J Biol Chem. 1999;274:33374–33381. doi: 10.1074/jbc.274.47.33374. [DOI] [PubMed] [Google Scholar]
- 25.Raghavan A, Robison RL, McNabb J, Miller CR, Williams DA, Bohjanen PR. HuA and tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. J Biol Chem. 2001;276:47958–47965. doi: 10.1074/jbc.M109511200. [DOI] [PubMed] [Google Scholar]
- 26.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: Conformational changes coupled to the bipartite nature of AU-rich mRNA-destabilizing motifs. J Biol Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 27.Worthington MT, Pelo JW, Sachedina MA, Applegate JL, Arseneau KO, Pizarro TT. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/Tristetraprolin protein. J Biol Chem. 2002;277:48558–48564. doi: 10.1074/jbc.M206505200. [DOI] [PubMed] [Google Scholar]
- 28.Blackshear PJ, Lai WS, Kennington EA, Brewer G, Wilson GM, Guan X, Zhou P. Characteristics of the interaction of a synthetic human tristetraprolin tandem zinc finger peptide with AU-rich element-containing RNA substrates. J Biol Chem. 2003;278:19947–19955. doi: 10.1074/jbc.M301290200. [DOI] [PubMed] [Google Scholar]
- 29.Williamson JR. Induced fit in RNA-protein recognition. Nature Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 30.Wang C, Karpowich N, Hunt JF, Rance M, Palmer AG. Dynamics of ATP-binding cassette contribute to allosteric control, nucleotide binding and energy transduction in ABC transporters. J Mol Biol. 2004;342:525–537. doi: 10.1016/j.jmb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Hudson BD, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nature Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 32.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 33.Lakowicz JR. Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum; New York, NY: 1999. [Google Scholar]
- 34.Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA: Non-binding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J Biol Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- 35.Wilson GM, Sutphen K, Moutafis M, Sinha S, Brewer G. Structural remodeling of an A+U-rich RNA element by cation or AUF1 binding. J Biol Chem. 2001;276:38400–38409. doi: 10.1074/jbc.M106509200. [DOI] [PubMed] [Google Scholar]
- 36.Heyduk T, Ma Y, Tang H, Ebright RH. Fluorescence anisotropy: rapid, quantitative assay for protein-DNA and protein-protein interaction. Methods Enzymol. 1996;274:492–503. doi: 10.1016/s0076-6879(96)74039-9. [DOI] [PubMed] [Google Scholar]
- 37.Otto MR, Lillo MP, Beechem J. Resolution of multiphasic reactions by the combination of fluorescence total-intensity and anisotropy stopped-flow kinetic experiments. Biophys J. 1994;67:2511–2521. doi: 10.1016/S0006-3495(94)80741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jameson DM, Sawyer WH. Fluorescence anisotropy applied to biomolecular interactions. Methods Enzymol. 1995;246:283–300. doi: 10.1016/0076-6879(95)46014-4. [DOI] [PubMed] [Google Scholar]
- 39.Zelent B, Kusba J, Gryczynski I, Johnson ML, Lakowicz JR. Time-resolved and steady-state fluorescence quenching of N-acetyl-L-tryptophanamide by acrylamide and iodide. Biophysical Chemistry. 1998;73:53–75. doi: 10.1016/s0301-4622(98)00137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 41.Manyusa S, Whitford D. Defining folding and unfolding reactions of apocytochrome b5 using equilibrium and kinetic fluorescence measurements. Biochemistry. 1999;38:9533–9540. doi: 10.1021/bi990550d. [DOI] [PubMed] [Google Scholar]
- 42.Salamanca S, Chang J-Y. Unfolding and refolding pathways of a major kinetic trap in the oxidative folding of α-lactalbumin. Biochemistry. 2005;44:744–750. doi: 10.1021/bi048182n. [DOI] [PubMed] [Google Scholar]
- 43.Sheshadri S, Lingaraju GM, Varadarajan R. Denaturant mediated unfolding of both native and molten globule states of maltose binding protein are accompanied by large ΔCp's. Protein Science. 1999;8:1689–1695. doi: 10.1110/ps.8.8.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amann BT, Worthington MT, Berg JM. A Cys3His zinc-binding domain from Nup475/tristetraprolin: A novel fold with a disklike structure. Biochemistry. 2003;42:217–221. doi: 10.1021/bi026988m. [DOI] [PubMed] [Google Scholar]
- 45.Michel SLJ, Guerrerio AL, Berg JM. Selective RNA binding by a single CCCH zinc-binding domain from Nup475 (tristetraprolin) Biochemistry. 2003;42:4626–4630. doi: 10.1021/bi034073h. [DOI] [PubMed] [Google Scholar]
- 46.Worthington MT, Amann BT, Nathans D, Berg JM. Metal binding properties and secondary structure of the zinc-binding domain of Nup475. Proc Natl Acad Sci USA. 1996;93:13754–13759. doi: 10.1073/pnas.93.24.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y, Barkley MD. Toward understanding tryptophan fluorescence in proteins. Biochemistry. 1998;37:9976–9982. doi: 10.1021/bi980274n. [DOI] [PubMed] [Google Scholar]


