Abstract
Recent outbreaks of avian influenza in humans have stressed the need for an improved nonhuman primate model of influenza pathogenesis. In order to further develop a macaque model, we expanded our previous in vivo genomics experiments with influenza virus-infected macaques by focusing on the innate immune response at day 2 postinoculation and on gene expression in affected lung tissue with viral genetic material present. Finally, we sought to identify signature genes for early infection in whole blood. For these purposes, we infected six pigtailed macaques (Macaca nemestrina) with reconstructed influenza A/Texas/36/91 virus and three control animals with a sham inoculate. We sacrificed one control and two experimental animals at days 2, 4, and 7 postinfection. Lung tissue was harvested for pathology, gene expression profiling, and proteomics. Blood was collected for genomics every other day from each animal until the experimental endpoint. Gross and microscopic pathology, immunohistochemistry, viral gene expression by arrays, and/or quantitative real-time reverse transcription-PCR confirmed successful yet mild infections in all experimental animals. Genomic experiments were performed using macaque-specific oligonucleotide arrays, and high-throughput proteomics revealed the host response to infection at the mRNA and protein levels. Our data showed dramatic differences in gene expression within regions in influenza virus-induced lesions based on the presence or absence of viral mRNA. We also identified genes tightly coregulated in peripheral white blood cells and in lung tissue at day 2 postinoculation. This latter finding opens the possibility of using gene expression arrays on whole blood to detect infection after exposure but prior to onset of symptoms or shedding.
Although influenza virus was one of the first human viruses isolated, it continues to pose a challenge by resurfacing each year with newly acquired genetic variation (83). This adaptive nature of the virus leads to new species specificity, tissue tropism, and replicative abilities through both genetic drift and shift that directly affect virulence and can potentially result in the emergence of pandemic strains (34, 55, 63, 81, 83). Although subtypes circulating in humans since 1918 have been limited to combinations of H1/H2/H3 and N1/N2 surface proteins, there are increasing concerns that (H5/H7) avian influenza viruses are adapting to other species, including humans, and causing morbidity in normally unaffected hosts (34, 82). International surveillance of influenza virus has increased, and research efforts have been adopted in an attempt to diagnose cases early, produce effective vaccines faster, and devise new antiviral drugs.
Using reverse genetics techniques (19, 32, 33, 53, 54), scientists can now make replicas of emerging influenza viruses and can generate novel recombinant viruses to study pathogenicity or to be used as vaccines. These techniques have already been used to investigate the virulence of reconstructed 1918 pandemic (4, 39, 41, 78, 79) and avian (30, 46) influenza viruses in mouse models. While the availability of murine reagents and genetically modified mouse models offers powerful tools for studying disease pathogenesis and for evaluating therapeutic and prophylactic strategies, influenza virus infection of mice does not precisely replicate the natural infection in the human host (75). Mice are not natural hosts of influenza virus, making their utility as transmission or immunoprotection models limited. Ferrets have been considered appropriate host models because they are outbred mammals that are naturally susceptible to infection with influenza viruses, resulting in disease that resembles that of human influenza (26, 85). Limitations include a shortage of influenza-virus-seronegative ferrets, a confounding higher body temperature, and the lack of available immunological reagents (48).
Although other animal models of influenza virus infection exist and are routinely used, none are as close to humans in physiology and DNA sequence as nonhuman primates. The close phylogenetic relationship between humans and nonhuman primates has driven their widespread use as models for human disease, and influenza is no exception. Macaques, in particular, have been used extensively to study viral respiratory diseases, including influenza (8, 42, 68, 69, 71, 72), severe acute respiratory syndrome (20, 44, 45, 50, 59, 70), and metapneumovirus (43), and to evaluate therapeutic and prophylactic strategies (18, 25, 28, 45, 60, 67). With the rapid advancement in sequencing of several nonhuman primate transcriptomes, primate resources have expanded and the ability to perform global gene expression and protein profiling is at hand (49). Recently, Baskin et al. examined the suitability of pigtailed macaques as models of influenza infection virus and disease in the context of transcriptional studies, by integrating clinical data and pathology with examination of global and immune-response-specific gene expression in affected tissues (3). In the present study, our goal was to more fully define the impact of host-virus interaction in lungs, with an emphasis on the early response. We address the question “What does influenza virus infection look like in a primate?” by using both classical infection study protocols augmented with the powerful technologies of functional genomics performed with macaque-specific oligonucleotide arrays and high-throughput proteomics. By processing multiple pulmonary samples from the same animal and from the same lesion, areas of localized infection (with viral mRNA present) as well as areas without direct evidence of infection (with viral mRNA absent) were detected. We observed that pulmonary transcriptional profiles, predominantly that of the innate immune response, were strongly affected by the amount of viral mRNA present. Very importantly, we have identified transcriptional markers of early infection in peripheral white blood cells.
MATERIALS AND METHODS
Animals.
Nine male pigtailed macaques, ranging in age from 6 to 9.5 years and in weight from 11.2 to 23.4 kg, were obtained from the Washington National Primate Research Center for the purpose of this study. None of these animals had received any major treatments in the past, and they were seronegative for type D simian retrovirus, simian T-cell leukemia virus, and simian foamy virus. A physical exam 2 weeks before scheduled inoculation, as well as a complete blood count and determination of serum chemistry (both processed at the Department of Laboratory Medicine, University of Washington Medical Center) and influenza A virus serum antibody titers (by measurement of hemagglutination inhibition titers against H1N1 A/New Cal/20/99, H3N2 A/Panama/2007/99, and H3N2 A/New Mexico/17/2003), ruled out any major health problems and prior exposure to influenza virus. All animals were moved from another part of the facility into the room assigned to the study 4 days prior to start of the experiment. All animals displayed behavior and appetite within normal limits, indicative of successful acclimation, by the start of the study. During the study, the room was under a biosafety level of 2+, and all procedures were performed according to guidelines approved by the University of Washington Environmental Health and Safety Committee, the Occupational Health Administration, the Primate Center Research Review Committee, and the Institutional Animal Care and Use Committee.
Protocol.
The protocol was adapted from that described by Rimmelzwaan et al. (67, 68, 69) and based on our previous work (3). Briefly, the nine animals were designated either control (sham-inoculated) animals (n = 3) or experimental animals (n = 6) and were matched for age and weight. Three sets of one control and two experimental animals were assigned endpoints of 2, 4, or 7 days postinoculation (p.i.). The control phase of the study was performed first, to avoid cross-infection by infected animals. Experimental animals were inoculated with 107 50% tissue culture infectious doses of reconstructed influenza A/Texas/36/91 virus. The sequence of the rescued Tx/91 virus was confirmed by reverse transcription (RT)-PCR and sequence analysis. Future studies will use this reconstructed Texas virus as a background to genetically engineer chimeras that will investigate host response to viral gene constellations in a systematic manner. Inoculation was done intratracheally and on tonsils and conjunctivae, which approximate the natural routes of infection and had resulted in the successful infection and reproduction of the human disease in past experiments (3). Animals were monitored for clinical signs by daily clinical observation and conventional blood work ancillary diagnostics. At necropsy, tissue samples were either immediately homogenized in solution D (4 M guanidinium thiocyanate, 25 mM sodium citrate, 0.5% sarcosyl, 0.1 M β-mercaptoethanol) as previously described (14, 39) for macaque oligonucleotide arrays, snap frozen for proteomics work and viral isolation (see below), or fixed in 10% formalin for histology and immunohistochemistry. Methods for viral isolation, serology, histopathology, and immunohistochemistry were provided in a previous article (3).
Tissue processing for macaque oligonucleotide arrays.
Total RNA was isolated from areas of the lungs that had gross lesions, i.e., discoloration due to the infection and in most cases consolidation as well. One animal (designated “Day 2 A”), sacrificed at day 2 p.i., had a more discrete and hyperemic lesion in the accessory lobe, suggesting acute inflammation. Tissues were collected at necropsy and immersed immediately in solution D. Tissue was homogenized immediately for 30 seconds with a Kinematica Polytron PT1200 instrument and a model PT-DA1212/2 generator (Kinematica, Lucerne, Switzerland) in 10-ml round-bottom polypropylene test tubes with 5 ml of solution D. In order to reduce the generation of aerosols during this process, the Polytron generator was passed through a hole in the test tube lid, which had been drilled in a manner that ensured a tight fit with the instrument. To further minimize possible contact with aerosols, a barrier shield was used in addition to positive-air-pressure respirators and full-protective personal protection equipment. The homogenized samples were then snap frozen on dry ice and stored at −70°C. Total RNA was subsequently extracted according to the current lab protocol (39). Whole blood was collected for arrays and processed with the PAXgene blood RNA system (PreAnalytix; QIAGEN, Valencia, CA) according to the manufacturer's instructions.
Oligonucleotide microarray analysis.
The experimental design for microarray analyses involved infected macaque samples being cohybridized with a reference mock sample to a macaque oligonucleotide array containing 131 viral probes, corresponding to 26 viruses, and 22,559 rhesus probes, corresponding to ∼18,000 rhesus genes. A complete description of this novel array is available upon request. The reference mock sample for pulmonary tissues was created by pooling equal mass quantities of total RNA extracted from lung samples of three mock animals. The common-reference mock sample for blood samples was created by pooling equal mass quantities of total RNA extracted from whole-blood samples of all nine animals at day 0 (prior to infection or mock infection). An Agilent 2100 bioanalyzer was used to check the purity of the total RNA prior to cRNA probe production with an Agilent low-RNA-input fluorescent linear amplification kit (Agilent Technologies Inc., Palo Alto, CA). Slides were scanned with an Agilent DNA microarray scanner, and image analysis was performed using Agilent feature extractor software (Agilent Technologies). Each microarray experiment was done with two technical replicates by reversing the dye hybridization for the experimental and reference samples (40). All data were entered into a custom-designed database, Expression Array Manager, and then uploaded into Resolver 4.0 (Rosetta Biosoftware), DecisionSite for Functional Genomics (Spotfire, Inc.), and Ingenuity Pathway Analysis (Ingenuity Systems) for analysis and mining. Initially, genes were selected to be included for transcriptional profile based on two criteria: a >99% probability of being differentially expressed (P ≤ 0.01) and an expression level change of twofold or greater. For study of gene expression in peripheral white blood cells, each experimental animal (at day 0, day 2, day 4, and day 7) was compared to a pool of six animals at day 0. Using the “re-ratio” feature of Resolver, we then compared each sample against that of the same animal at day 0 virtually and in this manner obtained a 7-day-p.i. time course experiment where each animal was its own control. Finally, biological gene sets (referred to as biosets) were compiled for key cellular processes by selecting genes of interest that both were represented on the microarray and had gene ontology annotation (29). In accordance with proposed standards (7), all data described in this report, including sample information, intensity measurements, gene lists, error analysis, microarray content, and slide hybridization conditions, are available in the public domain through Expression Array Manager at http://expression.microslu.washington.edu/expression/index.html.
qRT-PCR.
Quantitative real-time RT-PCR (qRT-PCR) was used to validate the presence of influenza virus mRNA found by microarrays in lung tissue. Total RNA samples were treated with DNase by using a DNA-free DNase kit (Ambion, Inc., Austin, TX). cDNA was generated using reverse transcription reagents and random hexamers (Applied Biosystems, Foster City, CA). Primer and probe sets for each of the target influenza virus HA and M sequences are available at http://expression.microslu.washington.edu/expression/index.html. qRT-PCR was performed with the ABI 7500 real-time PCR system, using TaqMan chemistry (Applied Biosystems, Foster City, CA). Each target was run in quadruplicate, in 20-μl reaction volumes with TaqMan 2× PCR Universal Master Mix (Applied Biosystems, Foster City, CA). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and 18S were chosen as endogenous controls to normalize quantification of the target. Quantification of each gene, relative to the calibrator, was calculated by the instrument, using the 2−ΔΔCT equation within the Applied Biosystems Sequence Detections version 1.3 software.
Sample preparation for proteomics.
During necropsy, lung tissue samples set aside for proteomic analysis were thoroughly rinsed in saline and then snap frozen on dry ice. Subsequently, frozen samples were rinsed in cold (4°C) phosphate-buffered saline and homogenized in high-salt buffer (500 mM KCl, 20 mM MgCl2, 50 mM Tris, pH 8.0) by using repeating rounds in a Kinematica Polytron PT1200 instrument with a model PT-DA1212/2 generator (Kinematica, Lucerne, Switzerland) and Dounce homogenization. The resulting suspensions were centrifuged, and the supernatant was retained and dialyzed extensively in 25 mM NH4HCO3, with five changes of fresh buffer every 3 h. A bicinchoninic acid assay (Pierce Biotechnology, Inc., Rockford, IL) was performed to determine protein concentration. Equal amounts of total protein from multiple animals were independently pooled to create a sample representing an infected macaque and a sample representing an uninfected macaque to maximize protein coverage when creating a database for macaque pulmonary tissue. Each sample was reduced in volume with a SpeedVac to ∼0.5 ml, reduced (5 mM 2-methyl-2-tert-butylperoxy-propane [TBP]), denatured (50% 2,2,2-trifluoroethanol [TFE]), and incubated for 1 h at 60°C (∼1 ml total volume per sample). Samples were then diluted with 50 mM NH4HC03 to a final TFE concentration of 10% for trypsin digestion.
Trypsin digestion.
Sequencing grade-modified porcine trypsin was prepared as instructed by the manufacturer (Promega, Madison, WI) and added to all protein samples at a 1:50 (wt/wt) trypsin-to-protein ratio for overnight digestion at 37°C. Samples were reduced by ∼50% in volume with a SpeedVac to remove any remaining TFE, snap frozen in liquid nitrogen to interrupt trypsin activity, and stored at −80°C until time for analysis.
Peptide enrichment and separation.
Additional solid-phase extraction C18 (Discovery DSC-18; SUPELCO, Bellefonte, PA) cleanup was performed to ensure the purity of the peptide samples for the cysteinyl-peptide enrichment step. Peptides were eluted from the C18 column by using 80% acetonitrile with 0.1% trifluoroacetic acid. Peptide samples were concentrated in a SpeedVac, and a bicinchoninic acid protein assay was performed to determine the final peptide concentration. Cysteinyl-peptide enrichment was performed on two pooled samples, 800 μg each from influenza virus-infected and mock-treated animals, where both the captured cysteinyl-peptide fraction and the flowthrough non-cysteinyl-peptide fraction were retained for further analysis. Cysteinyl-peptide enrichment has been previously described in detail (36, 37), but briefly, the tryptic digests were reduced with 5 mM dithiothreitol for 30 min at 37°C, after which the samples were diluted 1:5 in coupling buffer (50 mM Tris buffer, pH 7.5, 1 mM EDTA) and incubated for 1 h at room temperature with thiopropyl Sepharose 6B thiol-affinity resin (Amersham Biosciences, Uppsala, Sweden) prepared per the manufacturer's instructions. The unbound non-cysteinyl-peptide fraction was collected from the resin by spinning the column at low speed, removing the supernatant, and then washing the resin in wash buffer (50 mM Tris buffer, pH 8.0, 1 mM EDTA), all of which was retained as the non-cysteinyl-peptide fraction. For cysteinyl-peptide release, a 20 mM dithiothreitol solution in washing buffer was added to the resin and incubated for 30 min at room temperature. The resin was further washed with 100 μl of 80% acetonitrile. The eluted sample was pH adjusted to 8.0, alkylated with 80 mM iodoacetamide for 30 min at room temperature, desalted with a solid-phase extraction C18 column as described above, and lyophilized to reduce volume.
The non-cysteinyl-peptide fractions were subjected to strong cation-exchange chromatography (SCX) by using a 200-mm by 2.1-mm, 5 μM, 300-Å PolySulfoethyl A column with a 10-mm by 2.1-mm guard column (PolyLC, Inc., Columbia, MD), and the details of this SCX peptide fractionation step have been previously described in detail (36, 37). The peptides were resuspended in 900 μl of mobile phase A and separated on an Agilent 1100 system (Agilent, Palo Alto, CA) equipped with a quaternary pump, degasser, diode array detector, peltier-cooledautosampler, and fraction collector (set at 4°C for both samples). A total of 25 fractions was collected for each sample, resulting in two sets of SCX fractions for the non-cysteinyl-peptide enrichments of both the influenza virus-infected and mock-infected lung tissue samples. No SCX fractionation was performed with the cysteinyl-peptide-enriched fractions due to the available amount of total recovered sample.
RPLC separation and MS-MS analysis.
The method used in this study, with the coupling of a constant-pressure (5,000-lb/in2) reversed-phase capillary liquid chromatography (RPLC) system (150-μm inside diameter, 360-μm outside diameter, 65-cm capillary; Polymicro Technologies Inc., Phoenix, AZ) and a Finnigan LTQ ion trap mass spectrometer (MS; ThermoFinnigan, San Jose, CA) using an electrospray ionization source manufactured in-house, has been previously reported (74). Each SCX fraction (from the influenza virus-infected and mock-infected lung tissue samples) was analyzed via capillary RPLC-tandem mass spectrometry (MS-MS), for a total of 56 analyses, i.e., 25 SCX-based analyses for each non-cysteinyl-peptide preparation combined with 3 analyses each for the cysteinyl-enriched peptide fractions.
LC-MS-MS data analysis.
SEQUEST analysis software was used to match the MS-MS fragmentation spectra with sequences from the April 2005 IPI human database, containing 49,161 entries. The criteria selected for filtering followed methods based upon a human reverse-database false-positive model which has been shown to give ∼95% confidence for the entire protein data set (58). Briefly, protein identifications were retained if their identified peptide met the following criteria: (i) SEQUEST DelCN value of ≥0.10 and (ii) a SEQUEST correlation score (Xcorr) of ≥1.5 for charge state 1+ and full tryptic peptides, an Xcorr of ≥3.1 for charge state 1+ and partial tryptic peptides, an Xcorr of ≥1.9 for charge state 2+ and full tryptic peptides, an Xcorr of ≥3.8 for charge state 2+ and partial tryptic peptides, an Xcorr of ≥2.9 for charge state 3+ and full tryptic peptides, and an Xcorr of ≥4.5 for charge state 3+ and partial tryptic peptides. To remove redundantly identified proteins, the program ProteinProphet was utilized (52). All peptides which passed our filter criteria were given identical scores of 1 and entered into ProteinProphet for redundancy analysis only. This condensed the number of proteins detected from an initial 3,938 to a combined total of 3,548 proteins reported as identified, with >99% of these proteins identified by a full tryptic peptide. Proteins had to pass minimum criteria, which are based on the analysis performed in previous reports by our laboratory (36, 57), to be used in a quantitative nature. Briefly, a protein needed to have a minimum of five total peptide identifications in one of the samples, so as to eliminate the inclusion of proteins which are not detected with enough frequency to warrant quantitation, as well as at least a 3.0-fold increase/decrease in the relative abundance measurements between the two samples.
RESULTS
Progression and patterns of global gene expression from days 2 to 7 p.i. in influenza virus-infected macaques.
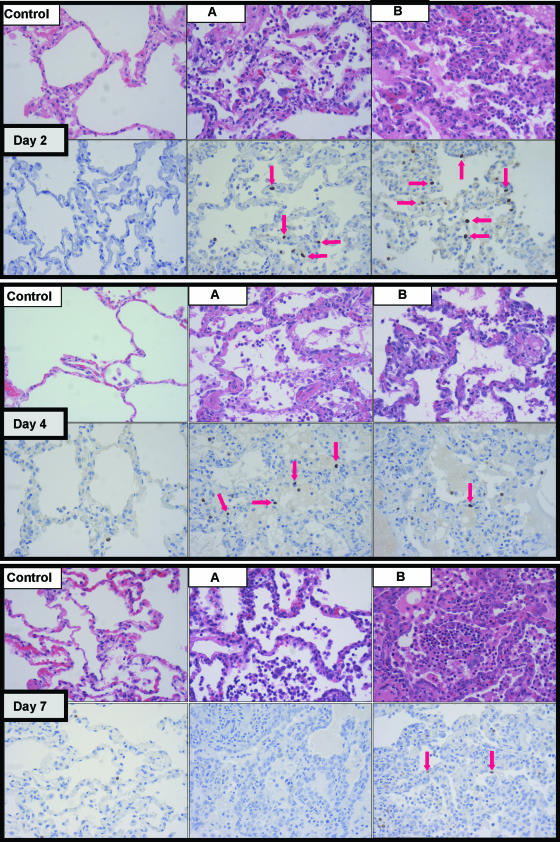
Six animals were inoculated with reconstructed A/Texas/36/91 influenza virus, a mildly pathogenic H1N1 strain in humans, at day 0. Clinical samples were collected during the study, and tissues were harvested at the assigned endpoints. The same was done for three mock-infected animals in a control phase of the study performed temporally apart to avoid cross-infection by infected animals. All animals in the experimental group showed signs of infection based on three criteria: clinical signs, gross and/or microscopic lung pathology, and tracheobronchial lymph node pathology. Foci of pneumonia, hyperemia, or consolidation were used for antigen staining and extraction of total RNA. We were successful in detecting influenza virus mRNA, by array and qRT-PCR (HA and M), from two infected animals (Day 2 A and Day 7 B) (data not shown) and intranuclear viral antigen in five (Day 2 A, Day 2 B, Day 4 A, Day 4 B, and Day 7 B, as shown in Fig. 1). Figure 1 illustrates representative histopathology and immunohistochemistry (using an antibody against the viral nucleoprotein [NP]) for all experimental animals. Clinical signs, pathology, antigen detection, and influenza virus mRNA positivity results are summarized in Table 1 and were reported in detail (http://expression.microslu.washington.edu/expression/index.html).
FIG. 1.
For each day postinfection (day 2, day 4, and day 7), progressive microscopic histopathological changes (top panels) and influenza virus antigen staining (bottom panels) are shown. Red arrows point to discrete intranuclear staining in respiratory epithelium and denote the presence of viral NP antigen, indicative of recent active infection. Samples from a mock-infected animal are included for each day to show lack of pathology (top panels) and nonspecific cytoplasmic staining of neutrophils (bottom panels). All photographs were taken at 40× optical magnification.
TABLE 1.
Clinical signs, pathologies, antigen detection, and influenza mRNA positivity results for this studya
| Animal group and designation | Clinical feature(s) | Lung pathologyb | TB node pathology | Presence of IHC (NP) antigen | Result for indicated viral mRNA analysis
|
|
|---|---|---|---|---|---|---|
| Array | qRT-PCR | |||||
| Mock infected | ||||||
| Day 2 | 3.9% Weight loss | Negative | Negative | Negative | Negative | Negative |
| Day 4 | 1.5% Weight loss | Negative | Negative | Negative | Negative | Negative |
| Day 7 | 1.35% Weight loss | Negative | Negative | Negative | Negative | Negative |
| Influenza infected | ||||||
| Day 2 A | Rhinorrhea, reduced feces production, 1.14% weight loss | Acute-subacute bronchopneumonia (accessory lobe) | Reactive | Positive +++ | Positive +++ | Positive +++ |
| Day 2 B | Reduced feces production | Acute-subacute diffuse bronchopneumonia | Reactive | Positive + | Negative | Negative |
| Day 4 A | Increased respiratory sounds, rhinorrhea, 8% weight loss | Acute-subacute bronchopneumonia (L caudal lobe) | Reactive | Positive + | Negative | Negative |
| Day 4 B | 3.3% Weight loss | Bronchopneumonia (L caudal lung lobe) | Reactive | Positive + | Negative | Positive |
| Day 7 A | Increased respiratory sounds, rhinorrhea, conjunctivitis, rising WBC count, 6.6% weight loss | Subacute bronchopneumonia (accessory lobe) | Reactive | Negative | Negative | Negative |
| Day 7 B | Rhinorrhea, conjunctivitis, rising WBC count, 3.22% weight loss | Subacute bronchopneumonia (R caudal and accessory lobes) | Grossly normal but reactive on histology | Positive ++ | Positive + | Positive + |
IHC, immunohistochemistry; WBC, white blood cell; TB, tracheobronchial; +, weakly detected; ++, detected; +++, strongly detected. The table summarizes clinical features of control and experimental animals, pathology, and positivity of lung samples for influenza virus mRNA. Live influenza virus was found on a pharyngeal swab from animal Day 7 B at day 2 p.i., and a complete blood count showed a rise in absolute and relative lymphocyte numbers at day 4 p.i. in that animal and the same in animal Day 7 A. All infected animals showed increased bronchial lavage fluid cellularity, particularly at day 2 and day 4 p.i. We were unable to isolate replicating influenza virus above plaque assay detection levels from the lung lesions, but this was not surprising since the disease was considered mild and the tissue sampled for viral isolation was on the periphery of the lesions which were first and foremost set aside for assays, proteomics, and histology. Detection of viral antigen staining was performed using one tissue sample, while detection of viral mRNA was performed using another tissue sample. One sample could not be used for both, due to unique sample preparation protocols and this explains why detection of viral antigen is not always concordant with detection of viral mRNA. Of note, one of the mock-infected animals had a transient nasal discharge (slight and clear), and all control animals had mild diffuse pulmonary inflammation, most likely due to the bronchial lavages. Finally, mock-infected animals experienced a mild weight loss attributed to the stress of experimental manipulation.
L, left; R, right.
In order to test the hypothesis that the presence or absence of influenza viral mRNA within a sample had a significant effect on global gene expression, we performed array analysis on samples from areas that were close to the main lesion yet looked distinct from one another by visual examination. Because animal Day 2 A had a very discrete and hyperemic lesion in the accessory lobe, multiple samples were taken from within this acute lesion and the immediate surrounding area. Three samples from this animal were selected for inclusion in transcriptional profiling, including two which were positive for viral mRNA. These profiles were obtained by comparing total RNA isolated from affected lung areas to that isolated from the lungs of mock-infected animals. With a ≥2-fold change and a P value of ≤0.01 selected as cutoff parameters, samples from animals with detectable viral mRNA showed greater gene expression changes (∼1,300 genes) than samples from animals with nondetectable viral mRNA (∼200 genes), indicating a definite response of lung tissue to the virus.
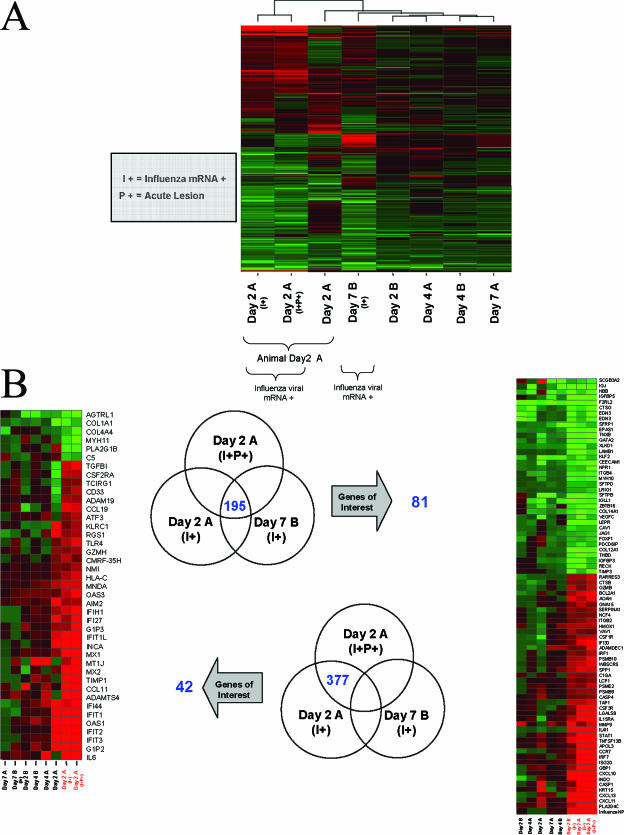
Unsupervised hierarchical clustering methods were used to arrange rows (genes) and columns (samples) for identifying groups of genes or samples with similar expression patterns (13, 73). Sample Day 2 A (I+P+) was positive for viral mRNA (I+) and within the main acute lesion (P+), Day 2 A (I+) was positive for viral mRNA and adjacent to the lesion, and Day 2 A was negative for viral mRNA and adjacent to the lesion as well. Sample Day 7 B (I+) was also positive for viral mRNA and was harvested from an area of consolidation in that animal. These data were plotted as a heat map where each matrix entry represents a gene expression value. Red corresponds to a higher gene expression than that of the controls; green corresponds to a lower gene expression (Fig. 2A). Since it was anticipated that factors such as the presence or absence of viral mRNA, timing after inoculation, and genetic similarity between samples from the same animal could all impact clustering, cutoff parameters were further restricted so that they held true for any one gene in at least two of the eight samples. This analysis yielded 1,373 genes, with the two samples (I+ and I+P+) from animal Day 2 A clustered next to one another and with the third sample from animal Day 2 A in close proximity, corroborating the assumed influence of genetic similarities among samples from the same animal. The expression profile for sample Day 7 B (I+) was within close proximity of those for the three samples mentioned above, indicating that the presence of influenza virus mRNA was indeed a key factor in determining the transcription of cellular genes, independently of timing after infection.
FIG. 2.
(A) Unsupervised clustering of gene expression profiles. The profiles for animal Day 2 A show similarities based on the fact that they were sampled from the same animal, while viral-mRNA-positive samples showed similarities as well. Day 2 A (I+P+) was positive for viral mRNA and within the main lesion, Day 2 A (I+) was positive for viral mRNA and adjacent to the lesion, and Day 2 A was negative for viral mRNA and adjacent to the lesion as well. All gene expression profiles are the results of comparing gene expression in the lungs of individual experimental animals versus gene expression in the lungs of mock-infected animals (pooled), and genes were included if they met the criterion of a twofold or greater change (P ≤ 0.01). A two-of-eight strategy allowed samples to cluster together if profile similarities existed based on timing of inoculation (n = 2 samples for each day). A three-of-eight strategy allowed samples to cluster together if profile similarities existed based on the presence or absence of viral mRNA (n = 3 samples with viral mRNA) or based on the samples being from the same animal (n = 3 samples from animal Day 2 A). Very similar clusters were obtained in both cases, so only one is represented here. (B) Venn diagrams showing genes with changes of ≥2-fold (P ≤ 0.01) in samples Day 2 A (I+P+), Day 2 A (I+), and Day 7 B (I+) (upper Venn diagram) and in samples Day 2 A (I+P+) and Day 2 A (I+) but not in sample Day 7 B (I+) (lower Venn diagram). Although there were fewer genes within the intersection of the three samples (195 genes), a greater proportion of these genes was annotated and had functions relevant to the immune response, infectious disease, or pulmonary stress. Heat maps show the same genes regulated in day 2, 4, and 7 lung samples. All gene expression profiles are the results of comparing gene expression in the lungs of individual experimental animals versus gene expression in the lungs of mock-infected animals (pooled).
Impact of influenza viral mRNA on the expression of genes relevant to the innate and adaptive immune responses.
Not surprisingly, the presence of influenza viral mRNA in lung tissue, suggesting recent viral replication, had a strong influence on the induction of the innate immune response and subsequent T-cell and B-cell activations. For an extension of global profiling, we focused on the effect of the virus by identifying differentially expressed genes common to both the Day 2 A (I+P+) and the Day 2 A (I+) positive samples and expanded the analysis by looking for commonalities between these two samples and the positive sample Day 7 B (I+). Genes of interest preferentially induced by the virus, with a ≥2-fold change and a P value of ≤0.01 selected as cutoff parameters, are shown [only for Day 2 A (I+P+) and Day 2 A (I+)] on the left panel of Fig. 2B, whereas the ones induced in Day 2 A (I+P+), Day 2 A (I+), and Day 7 B (I+) are shown on the right panel. The intersection of Day 2 A (I+P+) and Day 2 A (I+) holds a total of 572 differentially expressed genes, including 195 genes coregulated in Day 2 A (I+P+), Day 2 A (I+), and Day 7 B (I+). Both heat map panels should represent what took place during a localized host response to infection (where viral mRNA is present). The left panel focuses on transcriptional events present only early in infection in one animal. The right panel, with virus-positive samples from animal Day 2 A and animal Day 7 B, represents those changes occurring at both early and later stages of infection and suggests what consistent strategies the host uses to combat viral replication for the duration of the infection and in different animals.
The two samples from animal Day 2 A showed remarkable states of interferon (IFN) induction, with several genes upregulated >5-fold (the IFITL1, MX1, MX2, IFI44, IFIT1, and OAS1 genes) and even >10-fold (the IFIT2, IFIT3, and G1P2 genes), as shown in Fig. 2B. Of note, similar but much attenuated expression patterns could be observed in the other day 2 animal (Day 2 B) and day 4 animals (Day 4 A and Day 4 B), indicating consistent but progressively decreased induction of this response. On the left panel, of all the other samples, Day 2 A is the most similar to Day 2 A (I+) and Day 2 A (I+P+), suggesting that the IFN signature is also present adjacent to lesions but in areas where viral infection has not been detected, consistent with the paracrine effects of IFN. When focusing on localized host response for the duration of the infection by looking at genes differentially expressed exclusively in the two positive samples from animal Day 2 A and at the same time in the positive sample from animal Day 7B, fewer genes were found (195 versus 377 exclusive to the positive Day 2 A samples), but more of these were annotated as having functions in the immune response to infectious disease or in generic pulmonary stress. Many of these genes, upregulated at least 5- or 10-fold (the LGALS9, IRF7, ISG20, GBP1, CXCL10, KRT15, CXCL13, CXCL11, and PLA2G4C genes), showed similar expression levels in other animals and at other time points, perhaps representing common markers of infection. Interestingly, the SCGB3A2 gene (encoding secretoglobin, family 3A, member 2) was downregulated >10-fold in all influenza virus mRNA-positive samples. This gene has been implicated in controlling the transcription of lung surfactant proteins and is found to be expressed by Clara-like cells in the bronchial epithelium (6). In conclusion, an interferon signature response was induced early in infection while other immune and cytokine responses were sustained throughout the course of the infection.
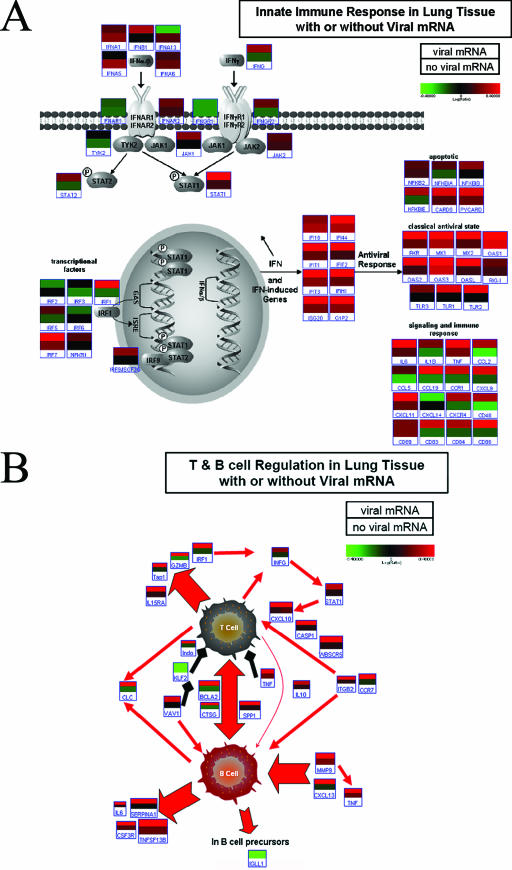
Because viral mRNA was shown to be a key factor in determining the transcription of cellular genes, two critical immune pathways were investigated by recombining expression data for samples Day 2 A (I+P+), Day 2 A (I+), and Day 7 (I+) in silico to generate a “+viral mRNA” average and by recombining expression data for samples Day 2 A, Day 2 B, Day 4 A, Day 4 B, and Day 7 A in silicoto generate a “no viral mRNA” average. We performed a pathway-based analysis by looking at genes specifically regulated as part of the innate immune response and involved in the regulation of T- and B-cell activation and proliferation. Figures 3A and B illustrate the differences between samples positive for influenza mRNA and samples negative for influenza mRNA. Many genes classically associated with interferon signaling were shown to be activated in animals with pulmonary viral mRNA, e.g., the genes encoding IFN-γ, IFN-β, STAT1, ISGF3G, IRF7, and NFKB1, among other genes induced either as part of the JAK-STAT pathway or through the interferon-stimulated response element. Among genes that were most affected were those coding for chemotactic factors, regulating cell adhesion (CCL-, CCR-, and CXCL-), and those associated with dendritic cell (CD83) or natural killer cell (CD48) function (Fig. 3A). Interestingly, MX1, OAS1, and OAS3 were shown to be upregulated in all animals, regardless of viral mRNA presence, again suggesting that the IFN signature is also present adjacent to lesions but in areas where viral infection has not been detected, consistent with the paracrine effects of IFN.
FIG. 3.
(A) Innate immune response in lung tissue with or without viral mRNA. The top bar of the individual gene heat maps was created by combining expression data for samples Day 2 A (I+P+), Day 2A (I+), and Day 7 B (I+), where viral mRNA was detected by array. The bottom bar was created by combining expression data for samples Day 2 A, Day 2 B, Day 4 A, Day 4 B, and Day 7 A, where no viral mRNA was detected. As in classical heat maps, brighter colors indicate stronger gene induction in combined samples, with red signifying upregulation and green downregulation. This illustrates the significant impact of the presence (top bar) or absence (bottom bar) of influenza viral mRNA on gene expression, even among essentially similarly affected portions of the same lung or among animals that were all infected. We selected probes with the strongest signals for any one gene for the purpose of these diagrams. All shown data are the results of comparing the combined gene expression in the lungs of experimental animals, as described above, versus gene expression in the lungs of mock-infected animals (pooled). This figure was produced using Resolver 4.0 (Rosetta Biosoftware) and Pathway Builder Tool (Protein Lounge, San Diego, CA). (B) T- and B-cell regulation in lung tissue with or without viral mRNA. All red arrows indicate activation and/or secretion. All black arrows indicate inhibitory interactions. The top bar of each heat map anchor was created by combining data from ratio experiments for Day 2 A (I+P+), Day 2 (I+), and Day 7 B (I+), in which viral mRNA was detected by array. The bottom bar of each heat map anchor was created by combining data from ratio experiments for Day 2 A, Day 2 B, Day 4 A, Day 4 B, and Day 7 A. This illustrates the significant impact of the presence or absence of influenza viral mRNA on gene expression, even among essentially similarly affected portions of the same lung or among animals that were all infected. We selected probes with the strongest signals for any one gene for the purpose of these diagrams. All heat map anchors are the results of comparing the combined gene expression in the lungs of individual experimental animals, as described above, versus gene expression in the lungs of mock-infected animals (pooled). This figure was produced using Resolver 4.0 (Rosetta Biosoftware) and Pathway Builder Tool (Protein Lounge, San Diego, CA).
An important effect of autocrine and paracrine exposure to the virus, as defined by the response of infected cells versus the response of neighboring cells to type I interferon, was the strong induction of T-cell proliferation and activation, as shown in Fig. 3B. This activation encompassed both cytotoxic T cells (Tap1, GZMB, and IFN-γ) and T helper cells, as evidenced by the concurrent stimulation of B cells. This robust T-cell response in infected lung tissue would be expected to be moderated by the strong expression of tumor necrosis factor alpha (TNF-α), known to decrease chemotaxis and increase apoptosis of T cells (1, 21), and INDO (indoleamine 2,3-dioxygenase), which decreases T-cell proliferation (35). In fact, it would appear from the induction of genes coding for B-cell products, including interleukin-6 (IL-6), and from the downregulation of IGLL1, produced by B-cell precursors, that the B-cell response was well under way even at day 2 p.i.
Coregulation of genes in peripheral blood and infected lung tissue.
Currently, there are adequate methods for diagnosing influenza soon after the onset of viral shedding, yet these diagnostic tests demonstrate limited specificities or sensitivities (56, 66, 76) and do not hold prognostic value. While gene expression profiling for lung tissue may provide valuable insights into influenza pathogenesis and successful host response, gene profiling for blood could complement or even replace in some instances lung tissue collection, help in the identification of the pathogen, and perhaps predict pathology to come. So far, there have been only a few studies investigating the effects of pulmonary infections on gene expression in peripheral white blood cells in vivo (62, 84). Most often, transcriptional studies are performed on white blood cells after in vitro culture and a number of other manipulations. We were interested in using blood as a snapshot of the ongoing lung infection and potentially as an indicator of disease progression in the animal as a whole. Our study included the use of blood collection tubes (PAXgene blood RNA system) that stabilize RNA in whole blood, so the effects of handling and storage on active transcripts were minimized (11, 16, 61). In this manner, we were able to study gene expression in all white blood cells as opposed to only peripheral mononuclear cells.
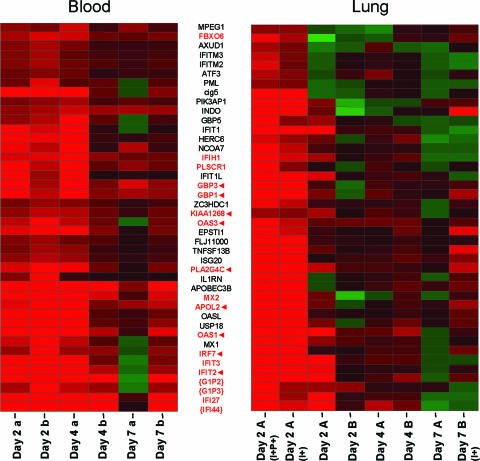
In order to begin investigating the effects of influenza virus infection in pulmonary tissue on peripheral blood, expression profiles were obtained by comparing total RNA isolated from whole-blood samples of animals Day 7 A and Day 7 B at day 2, day 4, and day 7 postinfection to a pool of total RNA isolated from whole-blood samples of all nine animals at day 0. Using a “re-ratio” feature in silico, we then compared each sample against the day 0 sample from that same animal, and in this manner, we obtained a 7-day-p.i. time course experiment where each animal was its own control. Figure 4 illustrates the close regulation between genes in blood samples taken at day 2 p.i. (Fig. 4, left panel, first two columns) and genes regulated in viral-mRNA-positive lung samples, also harvested at day 2 p.i. (Fig. 4, right panel, first two columns) but from a different animal. This gene expression similarity in the blood samples (left panel) was sustained throughout the course of infection, albeit to a lesser extent. This expression profile included a considerable proportion of interferon-induced genes (encoding IFI-, G1P-, GBP-, IRF7, INDO, and cig5) and antiviral genes encoding MX1, MX2, OASL, OAS1, and OAS3 (also induced by interferon). A number of these genes were similarly expressed in blood at days 4 and 7 (Fig. 4, red font), which, if these findings were confirmed, could expand the useful window for a prognostic test based on transcriptional profiling of peripheral white blood cells. Several of the genes shown to be regulated in blood were also regulated at multiple time points in the lungs (Fig. 4, red arrows), as shown in the right panel. APOL2 and PLA2G4C are known to be involved in the acute-phase response (23), with PLA2G4C being regulated by both collagen and interferon. IRF7, INDO, and GBP1 play roles in the immune response, especially inflammatory stress. Both GBP1 and IRF7 are well-known components of induction of host response to viruses (2, 51). While FBXO6 is not as closely related to the host immune response, there is some precedence for the assumption that F-box proteins play a critical role in the controlled degradation of cellular regulatory proteins (10).
FIG. 4.
Coregulated genes in blood and infected tissue. The heat map on the left indicates differential expression of blood samples taken from two animals (Day 7 A and Day 7 B) at day 2, day 4, and day 7. To indicate that these are blood samples, a and b are lowercase. The heat map on the right indicates differential expression of genes in pulmonary tissue collected from six animals. To indicate that these are pulmonary samples, A and B are uppercase. This figure shows strong and early coregulation of a set of genes in peripheral white blood cells and in lung tissue where influenza viral mRNA was detected by arrays, even in different animals. This set was obtained by unsupervised clustering of genes in blood and in lung tissue positive for viral mRNA and selecting genes that were regulated by a factor of twofold or more (P ≤ 0.01) in all four experiments. Some of these genes were still similarly regulated at day 4 and day 7 in blood, as shown in the left heat map (red font). Several of these genes shown to be regulated in blood were also regulated at multiple time points in the lungs (red arrows) as shown in the right heat map. Braces indicate genes whose profiles for blood must be interpreted with caution because the expression levels at day 0 in these individual blood samples differed by a factor of fourfold or more from those in all samples combined at day 0. All gene expression profiles for lung tissue are the results of comparing gene expression samples from individual experimental animals versus gene expression samples from mock-infected animals (pooled). All gene expression profiles for blood are the results of comparing gene expression samples from individual experimental animals at the indicated time points versus day 0 samples from the same animal.
Proteome analysis of infected and control lung samples yields results consistent with influenza virus infection.
Several observations point to the importance of monitoring the proteins expressed in a cell or tissue and relating these to mRNA expression measurements in order to maximize the chance of identifying biomarkers of the conditions being studied and uncovering cellular mechanistic pathways that contribute to disease processes. Perhaps most notable is the often poor to moderate correlation (e.g., <40% concordance) between the relative expression abundances of a gene and its biologically active protein product (12, 15, 27, 77), which can arise, for example, from differences in their rates of synthesis and turnover. Moreover, the limited presence of mRNA in body fluids restricts the identification of clinically relevant disease biomarkers to the measurement of secreted proteins in these samples. Additionally, phenotypic protein variations (e.g., modification, interaction with other proteins, subcellular distribution, and activity) that confer or modify protein function cannot be predicted from mRNA measurements. For these reasons, we have begun to expand upon our functional genomics efforts by constructing a macaque lung protein database and subsequently analyzing the protein abundance changes that occur in macaques infected with influenza virus. A total of 14,100 peptides and 3,548 proteins were identified for the entire study (Table 2). Among the proteins identified were many candidates of particular interest based on gene expression in the lung and peripheral blood (Fig. 2B and 4) as well as previous clinical, pathological, and gene expression data initially demonstrating the suitability of the nonhuman primate model for studying influenza virus pathogenesis (3). The proteins identified included several well-known interferon-induced proteins (e.g., MX1, IFI-, GBP-, and STAT1) and other noncellular mediators of the innate immune response (elements of the complement pathways, C7 and C5) (Fig. 5). Additional examples include proteins relevant to neutrophil and monocyte/macrophage function (e.g., MIF, MMP9, and MNDA).
TABLE 2.
Numbers of peptides and proteins identified in this studya
| Animal group | No. of:
|
||
|---|---|---|---|
| Peptidesa | Cys peptides (%)b | Proteinsc | |
| Mock infected | |||
| Non-Cys | 9,268 | 57 (0.6) | 2,885 |
| Cys | 1,943 | 1,849 (95) | 1,033 |
| Influenza infected | |||
| Non-Cys | 6,980 | 46 (0.6) | 2,322 |
| Cys | 1,670 | 1,596 (96) | 905 |
| Totald | 14,100 | 2,356 | 3,548 |
Total number of peptides detected in each sample.
Total number of cysteinyl-peptides detected in each sample.
Total number of proteins detected in each sample following ProteinProphet analysis.
Total number of unique peptides and proteins.
FIG. 5.
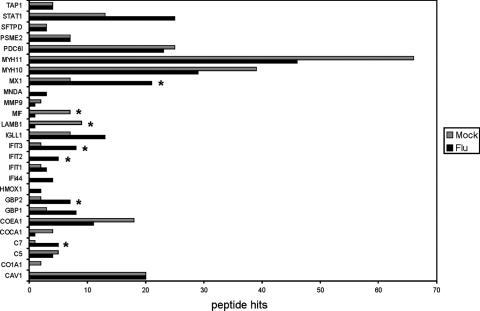
Bar graph showing the total number of peptide identifications for proteins having previously known associations with influenza (Flu) virus infection or acute respiratory distress. Those proteins exhibiting an apparent regulation during influenza virus infection are indicated with asterisks. Apparent regulation indicates that the protein was identified with at least five peptides and also showed at least a threefold increase/decrease between samples from influenza-infected and mock-infected animals.
The ability to globally profile changes in protein abundance is essential for elucidating the events that occur during cellular processes. The macaque lung protein database described here lays the foundation for subsequent comparative quantitative proteomic studies of nonhuman primate models of influenza virus infection by using stable isotope labeling strategies. While such a detailed proteomic characterization is beyond the scope of this study, we have taken advantage of the “semiquantitative” relationship between the total number of peptide identifications and the relative abundance of the corresponding protein in a sample (24, 47, 57) as a first-pass means of detecting changes in protein abundance when comparing uninfected and influenza virus-infected lung tissue samples. This approach has been verified in our laboratory, where we previously described perturbations in the abundances of a variety of proteins by using an in vitro model system for hepatitis C virus replication (36). We observed apparent upregulation of MX1, an interferon-induced protein involved in the innate immune response, and C7, a noncellular mediator of the innate immune response, which is consistent with the establishment of an antiviral state in the lungs of influenza virus-infected macaques. Additionally, by mining proteins having functional annotations of interest, we identified three proteins (MIF, C7, and GBP2) whose changes in relative abundance would not have been predicted from analysis at the mRNA level. These findings are consistent with previous studies demonstrating that mRNA and protein levels do not necessarily correlate, thus pointing to the complementarity of high-throughput proteomic studies for assisting in the determination of proteins/pathways affected by influenza virus infection.
DISCUSSION
Suitability of nonhuman primate model for influenza studies.
This study represents the first complete investigation of a macaque model of influenza A virus infection, using both classical infection study protocols and the powerful technologies of functional genomics and proteomics. Our experimentally infected macaques showed signs of successful infection based on clinical features and pathology, as indicated in Table 1. Several of these features, such as rhinorrhea and conjunctivitis, are similar to symptoms in humans yet absent in mice infected with the same mildly pathogenic human virus. Likewise, the limited extents of pathology and viral replication in these nonhuman primates resemble what occurs in the human disease to a much greater extent. These findings emphasize that what occurs in inbred mice may not be as relevant to human host-virus interaction as an outbred mammalian model that can become naturally infected (38). For instance, ferrets have been successfully used to investigate the contribution of host-related factors to influenza virus pathogenesis, particularly H5N1 subtypes (26, 85). Unfortunately, limited genomic resources for ferrets render high-throughput analysis currently impossible with this species. By contrast, a macaque model combines a ose phylogenetic relationship and great similarities in immune response and physiology to humans. The size of macaques also makes them more amenable for discriminating response and pathology in different areas of the lungs, generating samples large enough for multiple analyses, and allowing serial sampling.
Consistently, pulmonary samples showed heterogeneity in gene expression, and the ability to compare signatures from samples very close anatomically but with or without the presence of influenza virus mRNA yielded new and valuable information about the effects of the virus on surrounding cells at the transcriptional level. At the same time, expression profiling suggested significant similarities in different lung samples from the same animal and even similarities in different experimental animals. Therefore, these findings contributed to alleviating the concern that transcriptional profiling may be too heavily influenced by individual variation to be of practical use in studying host-virus interaction. Commitment to this approach and systematic sampling are especially important when working with mildly pathogenic viruses because this graduated tissue response to virus may discern subtle yet telling differences. This understanding is important because it will help us characterize the constituents of a successful response to the insult at the tissue level or at the level of the entire host.
By analyzing individual samples rather than pooling them, gene expression patterns were not artificially diluted and samples with evidence of recently replicating virus could be used to study the direct effects of the infection while samples lacking viral mRNA helped further define the host response. For instance, the higher interferon pathway induction in tissues more directly affected by influenza virus was not surprising since secretion of interferon by infected cells induces interferon-sensitive genes in other cells whose protein synthesis machinery is not redirected toward making viral particles. On the other hand, the early and significant induction of T- and B-cell activation and proliferation pathways in tissues where influenza mRNA could be found was more intriguing in part because of its timing early in infection and because of its dependence on autocrine or paracrine effects of the virus. The bulk of T- and B-cell activation takes place in peripheral lymph nodes draining infected tissue, subsequent to antigen presentation. However, B cells are currently thought to also present exogenous antigens through still-incompletely characterized major histocompatibility complex class I pathways to cytotoxic T cells, a possibility that would increase the importance of their role during early influenza virus infection (64, 65, 80). Induction of T- and B-cell pathways in lung tissue at day 2 p.i. could indeed be compatible with activation of B cells through less-specific B-cell antigen receptor and antigen interactions. These interactions were shown to result in a lower activation threshold for B-cell intracellular signaling and antigen processing for presentation to T lymphocytes (5).
Transcriptional signature of pulmonary infection in whole blood.
Although blood work is typically unremarkable and viremia is undetectable during infection with a mildly pathogenic influenza virus (9), chemotaxis and activation of immune cells responding to a localized viral infection are detectable at the systemic level. It has been previously suggested that while viral replication is intrinsically capable of producing extensive damage to respiratory epithelium directly, the host immune cell response may significantly enhance pulmonary damage well beyond the immediate vicinity of infected cells (17). In line with this concept, we tried to detect early signatures in peripheral blood reflecting the effects of localized pulmonary influenza virus infection. There are currently only a few studies investigating the effect of pulmonary infection on gene expression in peripheral leukocytes in vivo (62, 84). Most often, transcriptional studies are performed on white blood cells after in vitro culture and infection. Our studies showed a number of genes consistently expressed within both peripheral white blood cells and lung samples positive for viral mRNA, particularly interferon-induced genes. Blood cells did not have viral mRNA detected by microarray, as the infection was local to pulmonary tissues, without viremia, indicating that changes do not reflect infection of blood cells but rather circulating cytokines and chemokines. Even with limited infiltration of white blood cells in lung tissue, highly activated immune cells in lung tissue account for at least part of the coregulation observed. The disproportionate involvement of interferon genes induced in peripheral white blood cells also raises the question of whether any acute respiratory viral infection would give similar transcriptional profiles. Current but very limited comparisons between severe acute respiratory syndrome and influenza in patients seem to suggest otherwise (62), indicating the possibility of unique signatures for respiratory infectious agents. Finding these unique signatures is possible if there is a continued commitment to using functional genomics as opposed to classical cytokines and antiviral protein assays.
Relationship to human infections.
Classical cytokine studies have previously examined protein levels by using enzyme-linked immunosorbent assays with nasal lavage fluid, plasma, and serum samples from volunteers experimentally infected with influenza A/Texas/36/91 virus (22, 31). IL-6, TNF-α, IL-8, IFN-α, IFN-γ, IL-10, CCL3 (macrophage inflammatory protein 1α [MIP-1α]), CCL4 (MIP-1β), and CCL2 (monocyte chemoattractant protein 1) were elevated in nasal lavage fluid, and IL-6 and IFN-γ were elevated to a lesser extent in plasma. Our macaque studies concurred with these findings at the transcriptional level for TNF-α and IL-6 in pulmonary tissue. Additional cytokines not detected in the human studies were expressed as a result of the macaque infection in peripheral leukocytes (IL-1β and transforming growth factor βI) and in pulmonary tissue (transforming growth factor βI). Of note, only IL-6 was induced in pulmonary tissue of mice infected with influenza A/Texas/36/91 virus (unpublished data). CCL2 (monocyte chemoattractant protein 1) expression was also elevated in macaque pulmonary samples but differed from the results of the human study in that the response was early as opposed to sustained, perhaps a reflection of the fact that our data measured transcription as opposed to protein levels. What we have learned from our macaque model and functional genomics approach of cytokine and chemokine responses during influenza virus infection could be implemented in the context of human studies. Our examination of cytokines and chemokines expressed in macaque pulmonary tissue revealed CCL19 (MIP-3β), CCL11, CXCL11 (IP-9), CXCL13, CXCL10 (IP-10), and IL4I1 as attractive candidates for future investigation. For instance, CCL19, CXCL13, CXCL10, and CCL11 were highly induced by infection and have known roles in recruiting leukocytes to sites of inflammation. These could potentially be detected to a lesser extent at sites of infection in humans infected intranasally with influenza virus. Likewise, CXCL11 and IL4I1, although previously lacking a known association with influenza virus infection, had sufficiently significant responses to the presence of viral mRNA in pulmonary samples of macaques to warrant inclusion in future cytokine/chemokine lavage assays.
Proteomics as a complement to genomics.
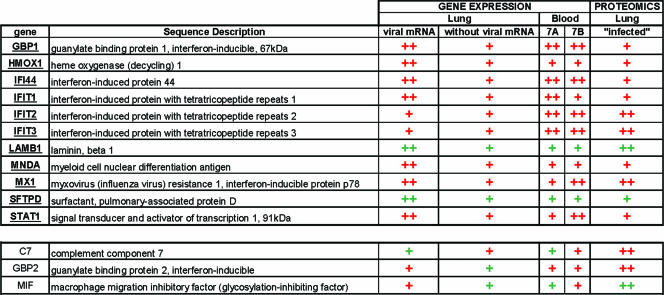
With the advent of a 22,000-macaque-oligonucleotide array, we have achieved superior genomic coverage and minimal standard error with our probe signals. We have supplemented our genomic coverage with proteomics data in an effort to move our study into the realm of a true functional global host response investigation. The utilization of refined, high-resolution multidimensional chromatographic separations that reduce sample complexity and the range of relative protein abundances enabled the identification of >3,000 proteins in macaque lung tissue. To our knowledge, this work represents the first comprehensive proteomic characterization yet reported for a nonhuman primate model system, providing a baseline for characterization of influenza virus-induced perturbations in the cellular environment, identification of potential targets for future antiviral treatment, and future comparative studies involving antiviral drug screening and evaluation for therapeutic intervention. To this end, we have used a semiquantitative approach (described above) to take a first look at lung protein abundance changes associated with influenza virus infection. Consistent with gene expression data demonstrating the establishment of an antiviral state in the lungs of influenza virus-infected macaques, our proteomic analyses also revealed an increase in the abundance of proteins in the lungs involved in the innate immune response. Furthermore, the complementary nature of proteomic studies was evidenced by the identification of changes in relative protein abundance that would not have been predicted from gene expression measurement, thus demonstrating the potential of proteomics for assisting in the determination of novel as well as previously described pathways affected by virus infection. To get a truly integrative view of the host molecular signature in response to influenza virus infection, we compared genomics profiles for blood and lung to proteomic profiles for lung in order to start identifying potential biomarkers indicative of influenza virus infection. Our method shows that the host response measured by pulmonary protein profiling is consistent with trends observed with genomic profiling, both in the lungs and in peripheral blood (Fig. 6). Statistical significance will be gained as refinements are made to the protocols for processing macaque protein samples and with the implementation of a macaque proteomics database. Still, this work has shown that even when screening macaque peptide hits to a human protein database, proteomics data augment robust genomics data to give a more complete picture of the host response to influenza virus infection.
FIG. 6.
Diagram showing the overlap between proteins detected by mass spectrometry in pulmonary samples and mRNA detected by gene expression in pulmonary and peripheral blood samples. Red indicates induction, and green repression. We have chosen to highlight genes/proteins that showed concordance in the top panel. In addition, we show three proteins that were not previously identified using genomics alone. Gene expression: “++” indicates genes meeting the criteria of a >2-fold change and a P value of <0.01; “+” indicates genes that showed a consistent trend as far as induction or repression but did not make the array statistical cutoff. Gene expression results for determined trends were either combined in silico (Lung) or averaged from the time course data (Blood). Proteomics: “++” indicates that the protein was identified with at least five peptides and also showed a >3-fold change between samples from influenza-infected and mock-infected animals. “+” indicates that the protein was identified.
Conclusions.
By integrating macaque-based functional genomics, using both microarrays and mass spectrometry, we have refined an integrated, multifaceted macaque model of influenza virus infection. The field of virus-host bioinformatics has expanded greatly in the past few years, and efforts are being undertaken to improve methodologies for compendium analyses, including software tools for visualizing genomic and proteomic data simultaneously, beginning with established human databases and progressing to establish macaque genomic and proteomic databases. As more wild-type and mutant viruses showing differences in pathogenicity are studied in the macaque model, host-related factors leading to a successful outcome and viral gene combinations leading to host demise will be better understood. This strategy, combined with the concurrent profiling of markers in peripheral blood, and with the investigation of a molecular basis for pathogenesis in cell culture, may ultimately lead to effective early prognostic tools for influenza.
.
Acknowledgments
We acknowledge Rosalind Billharz, Olivia Perwitasari, Jim Wallace, and Doug Chan from the Katze laboratory and Leon R. Flanary from the Washington National Primate Research Center for their intellectual contributions and technical assistance.
Portions of this research were supported by the NIH National Center for Research Resources (grants RR018522 to R.D.S. and RR016354 to M.G.K.), the National Institute of Allergy and Infectious Diseases (grant P01 AI058113 to A.G.-S. and M.G.K.), the National Institute on Drug Abuse (grant 1P30DA01562501 to M.G.K.), and the Environmental Molecular Sciences Laboratory at PNNL, Richland, WA, which provided the instrumentation applied in this research. The Environmental Molecular Sciences Laboratory, a national scientific user facility, is sponsored by the Department of Energy's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory. Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U.S. Department of Energy under contract no. DE-AC06-76RLO 1830.
Footnotes
Published ahead of print on 6 September 2006.
REFERENCES
- 1.Aggarwal, S., S. Gollapudi, and S. Gupta. 1999. Increased TNF-alpha-induced apoptosis in lymphocytes from aged humans: changes in TNF-alpha receptor expression and activation of caspases. J. Immunol. 162:2154-2161. [PubMed] [Google Scholar]
- 2.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 3.Baskin, C. R., A. García-Sastre, T. M. Tumpey, H. Bielefeldt-Ohmann, V. S. Carter, E. Nistal-Villán, and M. G. Katze. 2004. Integration of clinical data, pathology, and cDNA microarrays in influenza virus-infected pigtailed macaques (Macaca nemestrina). J. Virol. 78:10420-10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batista, F. D., and M. S. Neuberger. 2000. B cells extract and present immobilized antigen: implications for affinity discrimination. EMBO J. 19:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bin, L. H., L. D. Nielson, X. Liu, R. J. Mason, and H. B. Shu. 2003. Identification of uteroglobin-related protein 1 and macrophage scavenger receptor with collagenous structure as a lung-specific ligand-receptor pair. J. Immunol. 171:924-930. [DOI] [PubMed] [Google Scholar]
- 7.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. P. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME) toward standards for microarray data. Nat. Genet. 29:365-371. [DOI] [PubMed] [Google Scholar]
- 8.Burnet, F. 1941. Influenza virus “A” infections of cynomolgus monkeys. Aust. J. Exp. Biol. Med. 19:281-290. [Google Scholar]
- 9.Cate, T. R. 1987. Clinical manifestations and consequences of influenza. Am. J. Med. 82(6A):15-19. [DOI] [PubMed] [Google Scholar]
- 10.Cenciarelli, C., D. S. Chiaur, D. Guardavaccaro, W. Parks, M. Vidal, and M. Pagano. 1999. Identification of a family of human F-box proteins. Curr. Biol. 9:1177-1179. [DOI] [PubMed] [Google Scholar]
- 11.Chai, V., A. Vassilakos, Y. Lee, J. A. Wright, and A. H. Young. 2005. Optimization of the PAXgene blood RNA extraction system for gene expression analysis of clinical samples. J. Clin. Lab. Anal. 19:182-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, G., T. G. Gharib, C. C. Huang, J. M. Taylor, D. E. Misek, S. L. Kardia, T. J. Giordano, M. D. Iannettoni, M. B. Orringer, S. M. Hanash, and D. G. Beer. 2002. Discordant protein and mRNA expression in lung adenocarcinomas. Mol. Cell. Proteomics 1:304-313. [DOI] [PubMed] [Google Scholar]
- 13.Chipman, H., T. J. Hastie, and R. Tibshirani. 2003. Clustering microarray data, p. 159-200. In T. Speed (ed.), Statistical analysis of gene expression microarray data. CRC Press Company, New York, N.Y.
- 14.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 15.Cox, B., T. Kislinger, and A. Emili. 2005. Integrating gene and protein expression data: pattern analysis and profile mining. Methods 35:303-314. [DOI] [PubMed] [Google Scholar]
- 16.Debey, S., T. Zander, B. Brors, A. Popov, R. Eils, and J. L. Schultze. 2006. A highly standardized, robust, and cost-effective method for genome-wide transcriptome analysis of peripheral blood applicable to large-scale clinical trials. Genomics 87:653-654. [DOI] [PubMed] [Google Scholar]
- 17.Enelow, R. I., A. Z. Mohammed, M. H. Stoler, A. N. Liu, J. S. Young, Y. H. Lou, and T. J. Braciale. 1998. Structural and functional consequences of alveolar cell recognition by CD8(+) T lymphocytes in experimental lung disease. J. Clin. Investig. 102:1653-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan, J., X. Liang, M. S. Horton, H. C. Perry, M. P. Citron, G. J. Heidecker, T. M. Fu, J. Joyce, C. T. Przysiecki, P. M. Keller, V. M. Garsky, R. Ionescu, Y. Rippeon, L. Shi, M. A. Chastain, J. H. Condra, M. E. Davies, J. Liao, E. A. Emini, and J. W. Shiver. 2004. Preclinical study of influenza virus A M2 peptide conjugate vaccines in mice, ferrets, and rhesus monkeys. Vaccine 22:2993-3003. [DOI] [PubMed] [Google Scholar]
- 19.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franitza, S., R. Hershkoviz, N. Kam, N. Lichtenstein, G. G. Vaday, R. Alon, and O. Lider. 2000. TNF-alpha associated with extracellular matrix fibronectin provides a stop signal for chemotactically migrating T cells. J. Immunol. 165:2738-2747. [DOI] [PubMed] [Google Scholar]
- 22.Fritz, R. S., F. G. Hayden, D. P. Calfee, L. M. Cass, A. W. Peng, W. G. Alvord, W. Strober, and S. E. Straus. 1999. Nasal cytokine and chemokine responses in experimental influenza A virus infection: results of a placebo-controlled trial of intravenous zanamivir treatment. J. Infect. Dis. 180:586-593. [DOI] [PubMed] [Google Scholar]
- 23.Gabay, C., and I. Kushner. 1999. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340:448-454. [DOI] [PubMed] [Google Scholar]
- 24.Gao, J., G. J. Opiteck, M. S. Friedrichs, A. R. Dongre, and S. A. Hefta. 2003. Changes in the protein expression of yeast as a function of carbon source. J. Proteome Res. 2:643-649. [DOI] [PubMed] [Google Scholar]
- 25.Gao, W., A. Tamin, A. Soloff, L. D'Aiuto, E. Nwanegbo, P. D. Robbins, W. J. Bellini, S. Barratt-Boyes, and A. Gambotto. 2003. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet 362:1895-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Govorkova, E. A., J. E. Rehg, S. Krauss, H. L. Yen, Y. Guan, M. Peiris, T. D. Nguyen, T. H. Hanh, P. Puthavathana, H. T. Long, C. Buranathai, W. Lim, R. G. Webster, and E. Hoffmann. 2005. Lethality to ferrets of H5N1 influenza viruses isolated from humans and poultry in 2004. J. Virol. 79:2191-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haagmans, B. L., T. Kuiken, B. E. Martina, R. A. Fouchier, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, T. de Jong, S. Itamura, K. H. Chan, M. Tashiro, and A. D. Osterhaus. 2004. Pegylated interferon-alpha protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nat. Med. 10:290-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harris, M. A., J. Clark, A. Ireland, J. Lomax, M. Ashburner, R. Foulger, K. Eilbeck, S. Lewis, B. Marshall, C. Mungall, J. Richter, G. M. Rubin, J. A. Blake, C. Bult, M. Dolan, H. Drabkin, J. T. Eppig, D. P. Hill, L. Ni, M. Ringwald, R. Balakrishnan, J. M. Cherry, K. R. Christie, M. C. Costanzo, S. S. Dwight, S. Engel, D. G. Fisk, J. E. Hirschman, E. L. Hong, R. S. Nash, A. Sethuraman, C. L. Theesfeld, D. Botstein, K. Dolinski, B. Feierbach, T. Berardini, S. Mundodi, S. Y. Rhee, R. Apweiler, D. Barrell, E. Camon, E. Dimmer, V. Lee, R. Chisholm, P. Gaudet, W. Kibbe, R. Kishore, E. M. Schwarz, P. Sternberg, M. Gwinn, L. Hannick, J. Wortman, M. Berriman, V. Wood, N. de la Cruz, P. Tonellato, P. Jaiswal, T. Seigfried, and R. White. 2004. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32:D258-D261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 31.Hayden, F. G., R. Fritz, M. C. Lobo, W. Alvord, W. Strober, and S. E. Straus. 1998. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J. Clin. Investig. 101:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann, E., S. Krauss, D. Perez, R. Webby, and R. G. Webster. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165-3170. [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann, E., and R. G. Webster. 2000. Unidirectional RNA polymerase I-polymerase II transcription system for the generation of influenza A virus from eight plasmids. J. Gen. Virol. 81:2843-2847. [DOI] [PubMed] [Google Scholar]
- 34.Horimoto, T., and Y. Kawaoka. 2005. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 3:591-600. [DOI] [PubMed] [Google Scholar]
- 35.Hwu, P., M. X. Du, R. Lapointe, M. Do, M. W. Taylor, and H. A. Young. 2000. Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J. Immunol. 164:3596-3599. [DOI] [PubMed] [Google Scholar]
- 36.Jacobs, J. M., D. L. Diamond, E. Y. Chan, M. A. Gritsenko, W. Qian, M. Stastna, T. Baas, D. G. Camp, R. L. Carithers, Jr., R. D. Smith, and M. G. Katze. 2005. Proteome analysis of liver cells expressing a full-length hepatitis C virus (HCV) replicon and biopsy specimens of posttransplantation liver from HCV-infected patients. J. Virol. 79:7558-7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs, J. M., H. M. Mottaz, L. R. Yu, D. J. Anderson, R. J. Moore, W. N. Chen, K. J. Auberry, E. F. Stritmatter, M. E. Monroe, B. D. Thrall, I. I. Camp, and R. D. Smith. 2004. Multidimensional proteome analysis of human mammary epithelial cells. J. Proteome Res. 3:68-75. [DOI] [PubMed] [Google Scholar]
- 38.Jones-Engel, L., G. A. Engel, M. A. Schillaci, R. Babo, and J. Froehlich. 2001. Detection of antibodies to selected human pathogens among wild and pet macaques (Macaca tonkeana) in Sulawesi, Indonesia. Am. J. Primatol. 54:171-178. [DOI] [PubMed] [Google Scholar]
- 39.Kash, J. C., C. F. Basler, A. García-Sastre, V. Carter, R. Billharz, D. E. Swayne, R. M. Przygodzki, J. K. Taubenberger, M. G. Katze, and T. M. Tumpey. 2004. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J. Virol. 78:9499-9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerr, M. K., and G. A. Churchill. 2001. Statistical design and the analysis of gene expression microarray data. Genet. Res. 77:123-128. [DOI] [PubMed] [Google Scholar]
- 41.Kobasa, D., A. Takada, K. Shinya, M. Hatta, P. Halfmann, S. Theriault, H. Suzuki, H. Nishimura, K. Mitamura, N. Sugaya, T. Usui, T. Murata, Y. Maeda, S. Watanabe, M. Suresh, T. Suzuki, Y. Suzuki, H. Feldmann, and Y. Kawaoka. 2004. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431:703-707. [DOI] [PubMed] [Google Scholar]
- 42.Kuiken, T., G. F. Rimmelzwaan, G. van Amerongen, and A. D. Osterhaus. 2003. Pathology of human influenza A (H5N1) virus infection in cynomolgus macaques (Macaca fascicularis). Vet. Pathol. 40:304-310. [DOI] [PubMed] [Google Scholar]
- 43.Kuiken, T., B. G. van den Hoogen, D. A. van Riel, J. D. Laman, G. van Amerongen, L. Sprong, R. A. Fouchier, and A. D. Osterhaus. 2004. Experimental human metapneumovirus infection of cynomolgus macaques (Macaca fascicularis) results in virus replication in ciliated epithelial cells and pneumocytes with associated lesions throughout the respiratory tract. Am. J. Pathol. 164:1893-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawler, J. V., T. P. Endy, L. E. Hensley, A. Garrison, E. A. Fritz, M. Lesar, R. S. Baric, D. A. Kulesh, D. A. Norwood, L. P. Wasieloski, M. P. Ulrich, T. R. Slezak, E. Vitalis, J. W. Huggins, P. B. Jahrling, and J. Paragas. 2006. Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med. 3:e149. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, B. J., Q. Tang, D. Cheng, C. Qin, F. Y. Xie, Q. Wei, J. Xu, Y. Liu, B. J. Zheng, M. C. Woodle, N. Zhong, and P. Y. Lu. 2005. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in rhesus macaque. Nat. Med. 11:944-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, Z., H. Chen, P. Jiao, G. Deng, G. Tian, Y. Li, E. Hoffmann, R. G. Webster, Y. Matsuoka, and K. Yu. 2005. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 79:12058-12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, H., R. G. Sadygov, and J. R. Yates III. 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76:4193-4201. [DOI] [PubMed] [Google Scholar]
- 48.Luke, C. J., and K. Subbarao. 2006. Vaccines for pandemic influenza. Emerg. Infect. Dis. 12:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magness, C. L., P. C. Fellin, M. J. Thomas, M. J. Korth, M. B. Agy, S. C. Proll, M. Fitzgibbon, C. A. Scherer, D. G. Miner, M. G. Katze, and S. P. Iadonato. 2005. Analysis of the Macaca mulatta transcriptome and the sequence divergence between Macaca and human. Genome Biol. 6:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAuliffe, J., L. Vogel, A. Roberts, G. Fahle, S. Fischer, W. J. Shieh, E. Butler, S. Zaki, M. St. Claire, B. Murphy, and K. Subbarao. 2004. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology 330:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naschberger, E., M. Bauer, and M. Sturzl. 2005. Human guanylate binding protein-1 (hGBP-1) characterizes and establishes a non-angiogenic endothelial cell activation phenotype in inflammatory diseases. Adv. Enzyme Regul. 45:215-227. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 52.Nesvizhskii, A. I., A. Keller, E. Kolker, and R. Aebersold. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646-4658. [DOI] [PubMed] [Google Scholar]
- 53.Neumann, G., K. Fujii, Y. Kino, and Y. Kawaoka. 2005. An improved reverse genetics system for influenza A virus generation and its implications for vaccine production. Proc. Natl. Acad. Sci. USA 102:16825-16829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palese, P. 2004. Influenza: old and new threats. Nat. Med. 10:S82-S87. [DOI] [PubMed] [Google Scholar]
- 56.Perdue, M. L. 2003. Molecular diagnostics in an insecure world. Avian Dis. 47:1063-1068. [DOI] [PubMed] [Google Scholar]
- 57.Qian, W. J., J. M. Jacobs, D. G. Camp, M. E. Monroe, R. J. Moore, M. A. Gritsenko, S. E. Calvano, S. F. Lowry, W. Xiao, L. L. Moldawer, R. W. Davis, R. G. Tompkins, and R. D. Smith. 2005. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics 5:572-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian, W. J., T. Liu, M. E. Monroe, E. F. Strittmatter, J. M. Jacobs, L. J. Kangas, K. Petritis, D. G. Camp, and R. D. Smith. 2005. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Proteome Res. 4:53-62. [DOI] [PubMed] [Google Scholar]
- 59.Qin, C., J. Wang, Q. Wei, M. She, W. A. Marasco, H. Jiang, X. Tu, H. Zhu, L. Ren, H. Gao, L. Guo, L. Huang, R. Yang, Z. Cong, L. Guo, Y. Wang, Y. Liu, Y. Sun, S. Duan, J. Qu, L. Chen, W. Tong, L. Ruan, P. Liu, H. Zhang, J. Zhang, H. Zhang, D. Liu, Q. Liu, T. Hong, and W. He. 2005. An animal model of SARS produced by infection of Macaca mulatta with SARS coronavirus. J. Pathol. 206:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin, E., H. Shi, L. Tang, C. Wang, G. Chang, Z. Ding, K. Zhao, J. Wang, Z. Chen, M. Yu, B. Si, J. Liu, D. Wu, X. Cheng, B. Yang, W. Peng, Q. Meng, B. Liu, W. Han, X. Yin, H. Duan, D. Zhan, L. Tian, S. Li, J. Wu, G. Tan, Y. Li, Y. Li, Y. Liu, H. Liu, F. Lv, Y. Zhang, X. Kong, B. Fan, T. Jiang, S. Xu, X. Wang, C. Li, X. Wu, Y. Deng, M. Zhao, and Q. Zhu. 2006. Immunogenicity and protective efficacy in monkeys of purified inactivated Vero-cell SARS vaccine. Vaccine 24:1028-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rainen, L., U. Oelmueller, S. Jurgensen, R. Wyrich, C. Ballas, J. Schram, C. Herdman, D. Bankaitis-Davis, N. Nicholls, D. Trollinger, and V. Tryon. 2002. Stabilization of mRNA expression in whole blood samples. Clin. Chem. 48:1883-1890. [PubMed] [Google Scholar]
- 62.Reghunathan, R., M. Jayapal, L. Y. Hsu, H. H. Chng, D. Tai, B. P. Leung, and A. J. Melendez. 2005. Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reid, A. H., and J. K. Taubenberger. 2003. The origin of the 1918 pandemic influenza virus: a continuing enigma. J. Gen. Virol. 84:2285-2292. [DOI] [PubMed] [Google Scholar]
- 64.Reimann, J., W. Bohm, and R. Schirmbeck. 1994. Alternative processing pathways for MHC class I-restricted epitope presentation to CD8+ cytotoxic T lymphocytes. Biol. Chem. Hoppe Seyler 375:731-736. [DOI] [PubMed] [Google Scholar]
- 65.Reimann, J., and R. Schirmbeck. 1999. Alternative pathways for processing exogenous and endogenous antigens that can generate peptides for MHC class I-restricted presentation. Immunol. Rev. 172:131-152. [DOI] [PubMed] [Google Scholar]
- 66.Rennels, M. B., and H. C. Meissner. 2002. Technical report: reduction of the influenza burden in children. Pediatrics 110:e80. [DOI] [PubMed] [Google Scholar]
- 67.Rimmelzwaan, G. F., M. Baars, R. van Beek, G. van Amerongen, K. Lovgren-Bengtsson, E. C. Claas, and A. D. Osterhaus. 1997. Induction of protective immunity against influenza virus in a macaque model: comparison of conventional and iscom vaccines. J. Gen. Virol. 78:757-765. [DOI] [PubMed] [Google Scholar]
- 68.Rimmelzwaan, G. F., T. Kuiken, G. van Amerongen, T. M. Bestebroer, R. A. Fouchier, and A. D. Osterhaus. 2001. Pathogenesis of influenza A (H5N1) virus infection in a primate model. J. Virol. 75:6687-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rimmelzwaan, G. F., T. Kuiken, G. van Amerongen, T. M. Bestebroer, R. A. Fouchier, and A. D. Osterhaus. 2003. A primate model to study the pathogenesis of influenza A (H5N1) virus infection. Avian Dis. 47:931-933. [DOI] [PubMed] [Google Scholar]
- 70.Rowe, T., G. Gao, R. J. Hogan, R. G. Crystal, T. G. Voss, R. L. Grant, P. Bell, G. P. Kobinger, N. A. Wivel, and J. M. Wilson. 2004. Macaque model for severe acute respiratory syndrome. J. Virol. 78:11401-11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saslaw, S., H. E. Wilson, C. A. Doan, O. C. Woolpert, and J. L. Schwab. 1946. Reactions of monkeys to experimentally induced influenza virus A infection. Exp. Med. 84:113-125. [PMC free article] [PubMed] [Google Scholar]
- 72.Saslaw, S., and H. N. Carlisle. 1965. Aerosol exposure of monkeys to influenza virus. Proc. Soc. Exp. Biol. Med. 119:838-843. [DOI] [PubMed] [Google Scholar]
- 73.Shannon, W., R. Culverhouse, and J. Duncan. 2003. Analyzing microarray data using cluster analysis. Pharmacogenomics 4:41-52. [DOI] [PubMed] [Google Scholar]
- 74.Shen, Y., R. Zhao, M. E. Belov, T. P. Conrads, G. A. Anderson, K. Tang, L. Pasa-Tolic, T. D. Veenstra, M. S. Lipton, and R. D. Smith. 2001. Packed capillary reversed-phase liquid chromatography with high-performance electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry for proteomics. Anal. Chem. 73:1766-1775. [DOI] [PubMed] [Google Scholar]
- 75.Sidwell, R. W. 1999. The mouse model of influenza virus infection, p. 981-988. In O. Zak and M. A. Sande (ed.), Handbook of animal models of infection: experimental models in antimicrobial therapy. Academic Press, London, United Kingdom.
- 76.Sintchenko, V., G. L. Gilbert, E. Coiera, and D. Dwyer. 2002. Treat or test first? Decision analysis of empirical antiviral treatment of influenza virus infection versus treatment based on rapid test results. J. Clin. Virol. 25:15-21. [DOI] [PubMed] [Google Scholar]
- 77.Tian, Q., S. B. Stepaniants, M. Mao, L. Weng, M. C. Feetham, M. J. Doyle, E. C. Yi, H. Dai, V. Thorsson, J. Eng, D. Goodlett, J. P. Berger, B. Gunter, P. S. Linseley, R. B. Stoughton, R. Aebersold, S. J. Collins, W. A. Hanlon, and L. E. Hood. 2004. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell. Proteomics 3:960-969. [DOI] [PubMed] [Google Scholar]
- 78.Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solorzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. Garcia-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77-80. [DOI] [PubMed] [Google Scholar]
- 79.Tumpey, T. M., A. Garcia-Sastre, J. K. Taubenberger, P. Palese, D. E. Swayne, and C. F. Basler. 2004. Pathogenicity and immunogenicity of influenza viruses with genes from the 1918 pandemic virus. Proc. Natl. Acad. Sci. USA 101:3166-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Voeten, J. T., G. F. Rimmelzwaan, N. J. Nieuwkoop, R. A. Fouchier, and A. D. Osterhaus. 2001. Antigen processing for MHC class I restricted presentation of exogenous influenza A virus nucleoprotein by B-lymphoblastoid cells. Clin. Exp. Immunol. 125:423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Webby, R. J., and R. G. Webster. 2003. Are we ready for pandemic influenza? Science 302:1519-1522. [DOI] [PubMed] [Google Scholar]
- 82.Webster, R. G. 1997. Influenza virus: transmission between species and relevance to emergence of the next human pandemic. Arch. Virol. Suppl. 13:105-113. [DOI] [PubMed] [Google Scholar]
- 83.Yewdell, J., and A. Garcia-Sastre. 2002. Influenza virus still surprises. Curr. Opin. Microbiol. 5:414-418. [DOI] [PubMed] [Google Scholar]
- 84.Yu, S. Y., Y. W. Hu, X. Y. Liu, W. Xiong, Z. T. Zhou, and Z. H. Yuan. 2005. Gene expression profiles in peripheral blood mononuclear cells of SARS patients. World J. Gastroenterol. 11:5037-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zitzow, L. A., T. Rowe, T. Morken, W. J. Shieh, S. Zaki, and J. M. Katz. 2002. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J. Virol. 76:4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]