Abstract
Our aim was to isolate and characterize white spot syndrome virus (WSSV)-binding proteins from shrimp. After a blot of shrimp hemocyte membrane proteins was overlaid with a recombinant WSSV envelope protein (rVP28), the reactive bands on the blot were detected using anti-VP28 antibody. Among three membrane-associated molecules identified by liquid chromatography-tandem mass spectrometry, there was a 25-kDa protein that bound to both rVP28 and WSSV. Since it had a primary structure with high homology to the small GTP-binding protein Rab7, we named it Penaeus monodon Rab7 (PmRab7). The full-length PmRab7 cDNA was obtained, and results from a glutathione S-transferase pull-down assay confirmed specific binding to rVP28. Reverse transcriptase PCR analysis revealed PmRab7 expression in many tissues, and real-time PCR analysis revealed that expression was constitutive. Binding of PmRab7 to rVP28 or WSSV occurred in a dose-dependent manner and was inhibited by anti-Rab7 antibody. In an in vivo neutralization assay, the number of dead shrimp after challenge with WSSV plus PmRab7 (15%) or WSSV plus anti-Rab7 antibody (5%) was significantly lower than after challenge with WSSV alone (95%). In contrast to the WSSV-injected group, shrimp injected with WSSV plus PmRab7 or WSSV plus anti-Rab7 showed no WSSV-type histopathology. We conclude that PmRab7 is involved in WSSV infection in shrimp. This is the first study to identify a shrimp protein that binds directly to a major viral envelope protein of WSSV.
White spot syndrome virus (WSSV) is a viral pathogen that emerged in the early 1990s and has since spread throughout Asia and to the Americas. Diseased shrimp are lethargic and slow swimming and show reduced feed consumption. Histopathology has revealed that WSSV-infected shrimp tissues are of ectodermal and mesodermal origin (3, 6, 19, 39). WSSV is an ellipsoid to bacilliform, enveloped particle of about 275 nm in length and 120 nm in width, with a tail-like appendage at one end. It is the type species of the genus Whispovirus in the family Nimaviridae (36). It is unique, with an infection strategy that does not match infection models of any other known virus, and must therefore be investigated ab initio.
All three of the WSSV isolates that have been sequenced have a genome of about 300 kbp, and genetic comparisons have shown a high degree of genetic similarity (16). The availability of the complete WSSV sequence facilitates the global molecular characterization of the virus by genomic and proteomic approaches and has recently led to the discovery of many important WSSV genes, including latency-associated genes (10, 11), immediate-early genes (15), many other nonstructural genes (5, 29, 30, 33), and more than 39 structural genes (6, 13, 19, 31, 32, 35, 43). To date, however, little is known of the interaction between shrimp and WSSV at the cellular and molecular levels.
Neutralization experiments with a major WSSV envelope protein, VP28, have shown that it is involved in systemic infection of WSSV (34). It has further been shown that VP28 is able to bind to the surface of shrimp cells (41) and that feeding with recombinant VP28 can protect shrimp from WSSV infection (38). However, until now there have been no reports on the interaction of VP28 with a specific shrimp protein(s). Therefore, in the present study, to identify shrimp hemocyte membrane (SHM) proteins involved in WSSV binding, a virus overlay protein binding assay (VOPBA) was performed (9, 21). VP28 was selected as the WSSV target because it is the most abundant exposed protein in the WSSV envelope (32). One of the candidate proteins from this assay was further characterized, and its full-length sequence was analyzed. Its expression pattern in response to WSSV infection was investigated, and a glutathione S-transferase (GST) pull-down assay tested the specificity of its binding to recombinant VP28 (rVP28). An enzyme-linked immunosorbent assay (ELISA) and an in vivo neutralization assay were also conducted. This is the first study to describe the specific binding of a shrimp protein to the WSSV major structural protein VP28.
MATERIALS AND METHODS
Shrimp.
Shrimp (Penaeus monodon) were injected with WSSV stock in our laboratory to prepare WSSV-infected shrimp. Hemolymph was collected from Penaeus monodon brood stock (40 shrimp) received from the Thailand brood stock domestication program (BIOTEC, Bangkok, Thailand) and used to prepare a hemocyte membrane fraction. Domesticated white shrimp (Penaeus vannamei, also called Litopenaeus vannamei) that were specific pathogen free (meaning free of specifically listed pathogens, including WSSV) were obtained from SyAqua Thailand and used for neutralization tests.
Expression and purification of recombinant WSSV VP28 protein.
The WSSV VP28 gene was PCR amplified from WSSV genomic DNA by using the forward primer 5′-GGA TCT AAG CTTA (CAT)6 ATA ATG GAT CTT TCT TT-3′ and the reverse primer 5′-CAA TGA GCT CTT ACT CGG TCT CAG TG-3′. The forward primer contained recognition sequences for HindIII with a His6 tag, and the reverse primer had a recognition site for SacI. For the template, DNA was extracted from a WSSV-infected shrimp that had been diagnosed using a commercial WSSV detection kit (IQ2000 WSSV detection kit; Farming Intelligene Co., Ltd., Taiwan). The PCR amplicon was cloned into pET-17b vector (Novagen). The recombinant plasmid was transformed into Escherichia coli strain BL21, and the insert was confirmed by sequencing. The fusion recombinant protein (i.e., as rVP28) was purified by Ni-nitrilotriacetic acid-ribotriacetic acid affinity chromatography according to the manufacturer's protocol (QIAGEN). The purified rVP28 was stored at −20°C.
Preparation of shrimp hemocyte membrane protein.
Hemolymph from adult specific-pathogen-free shrimp was collected in AC-1 anticoagulant solution (27) at a hemolymph/AC-1 ratio of 1:2. The hemocyte pellet was collected, resuspended, and homogenized in 0.9% NaCl. This lysate was then sedimented by centrifugation, and the supernatant portion was collected and ultracentrifuged at 100,000 × g for 1 h at 4°C. After ultracentrifugation, the pellet was solubilized in NaCl/phosphate buffer (7) with 1% Triton X-100, 1× protease inhibitor mix (Amersham Biosciences). The suspension was ultracentrifuged, and the supernatant was collected and referred to as shrimp hemocyte membrane protein solution. The total protein concentration in SHM protein solution was determined using Bradford's reagent protein assay (Bio-Rad). To determine the membrane protein profile, the SHM fraction was subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue.
Western blot analysis of rVP28.
Purified rVP28 was separated by standard SDS-PAGE (12). For immunoblotting experiments, the purified rVP28 was electrophoresed and transferred to a nitrocellulose membrane (Amersham Biosciences). The membrane was immersed in blocking buffer (5% skim milk in 140 mM phosphate-buffered saline [PBS]) before incubation overnight at 4°C with a 1:1,000 dilution of mouse anti-VP28 antiserum (kindly provided by P. Sithikornkul, Srinakarinwirote University, Bangkok, Thailand). The blot was then washed twice and incubated for 2 h with a 1:2,000 dilution of goat anti-mouse immunoglobulin G conjugated with horseradish peroxidase (HRP) (Zymed). Subsequently, the blot was washed extensively and the color was developed with an AEC (red) substrate kit (Zymed).
Determination of WBPs by VOPBA.
To identify hemocyte membrane proteins involved in WSSV binding, a VOPBA was carried out (9, 21). SHM (50 μg) was separated by 12% SDS-PAGE and then transferred to a nitrocellulose membrane. Prior to the binding assay, the membrane was incubated with 5% skim milk in PBS buffer for 1 h. Following two washes, the membrane was equilibrated for 20 min with binding buffer (10 mM Tris-HCl [pH 6.5], 5 mM CaCl2, 10 mM MgCl2). Subsequently, the membrane was incubated with 0.8 mg of affinity-purified rVP28 (dialyzed against binding buffer at 4°C for 48 h) diluted in binding buffer with 0.02% skim milk and 1% Triton X-100. The membrane was extensively washed and WSSV-rVP28-binding proteins (WBPs) were detected by incubating with diluted mouse anti-VP28 antiserum (1:1,500). After washing, the blot was incubated with rabbit anti-mouse IgG-conjugated HRP (Roche). The hemocyte membrane proteins that interacted with rVP28 were detected by the addition of HRP substrate (0.003% [wt/vol] 3′,3′-diamino-benzenetetrahydrochloride [Sigma]-0.05% [wt/vol] H2O2).
Mass spectrometry analysis.
For internal sequence analysis, the WBPs were excised from the 12% SDS-polyacrylamide gel and separately digested overnight in gel with trypsin at 37°C. Liquid chromatography-electrospray ionization tandem mass spectrometry was performed, and the MASCOT program was used to analyze the results as described by Tsai et al. (31).
Expression of recombinant PmRab7.
The nucleotide sequence of shrimp Rab7 cDNA was identified from the information in a Taiwan expressed sequence tag library constructed from whole postlarval shrimp (C. F. Lo, unpublished data). The P. monodon Rab7 (PmRab7) sequence was analyzed and aligned with those of other Rab proteins from other microorganisms with BLAST 2.0 (http://www.ncbi.nlm.nih.gov/BLAST/) and Clustal W 1.7 multiple sequence alignment (http://www.ebi.ac.uk/clustalw/). Primers were designed to clone full-length PmRab7 in frame with both pET17b (Novagen) and pGEX4T-1 vector (Amersham Biosciences) for the production of recombinant PmRab7 fused with a His tag at its N terminus and GST at its C terminus, respectively. For pET17b vector, the forward primer 5′-CGA CGA TAG GTA CCC ATG GCA TCT TCG AA-3′ and the reverse primer 5′-TCG AGA CTC GAG GTG ATG GTG ATG GTG ATG TTA GCA AGA GCA TG-3′ were used. For GST fusion, the PmRab7 forward primer 5′-CGA CGA GAT GAA TTC ATG GCA TCT CGC AAG AA-3′ and the PmRab7 reverse primer 5′-CTA CTA GAG CGG CCG CGC AAG AGC ATG CAT-3′ were used. The protein was expressed in E. coli strain BL21 for the pET17b vector (as described above), whereas the protein with the GST tag was expressed in bacterial strain DH5α (Novagen). Protein production was accomplished by standard methods for bacterial growth, followed by induction with IPTG (isopropyl-β-d-thiogalactopyranoside). The His6-PmRab7 (rPmRab7) was used in ELISA and in vivo neutralization tests (K. Sritunyalucksana et al., 25 August 2006, Thai patent applications 0601004059 and 0601004060), whereas the GST-PmRab7 was used in GST pull-down assays. The rPmRab7 amino acid sequence was confirmed by liquid chromatography-electrospray ionization tandem mass spectrometry as described above. The specific binding of anti-Rab7 antibody to rPmRab7, as well as to Rab7 in SHM, was tested by Western blot analysis.
GST pull-down assay.
The GST fusion protein was purified using glutathione-Sepharose 4B resin (Amersham Biosciences). For the pull-down experiment, the interaction between rVP28 and GST-PmRab7 was examined by incubating purified GST-PmRab7 (1 μg) with a glutathione-Sepharose 4B resin (50 μl of a 50% bed slurry) for 1 h. Purified rVP28 was added, and incubation was continued for another 2 h at room temperature. Control reactions with GST plus rVP28, GST-PmRab7 plus bovine serum albumin (BSA), and GST-PmRab7 alone were included in the assay. After incubation, the beads were washed 10 times with PBS, pH 7.5. Fusion proteins were eluted by the addition of buffer containing reduced glutathione. The proteins were resolved by 12% SDS-PAGE and transferred onto nitrocellulose membranes. Blots were incubated with antihistidine antibody conjugated with HRP (Amersham Biosciences) and visualized using HRP substrate, as described above. To confirm the presence of GST-PmRab7, the blots were tested with anti-GST antibody conjugated with HRP (Amersham Biosciences).
Analysis of expression of the PmRab7 transcript.
Total RNAs were extracted from different organs (gill tissue, lymphoid tissue, hepatopancreas, stomach, heart, and hemocyte) of penaeid shrimp (WSSV-free P. monodon) by using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Reverse transcriptase PCRs (RT-PCRs) were carried out using the SuperScript III one-step RT-PCR kit (Invitrogen). The primer set used was the same as for GST-PmRab7 plasmid construction. The reactions were performed with the annealing temperature at 55°C. The amplified products were analyzed by electrophoresis on a 1.2% agarose gel.
For quantitation of WSSV, a TaqMan probe for WSSV detection was 6-carboxyfluorescein-5′-CGC TTC AGC CAT GCC AGC CG-3′-6-carboxytetramethylrhodamine. The primers were WSSV1 (5′-CCG ACG CCA AGG GAA CT-3′) and WSSV2 (5′-TTC AGA TTC GTT ACC GTT TCC A-3′). The TaqMan real-time PCR assay was carried out using TaqMan Universal PCR Master Mix (PE Applied Biosystems). A standard curve for WSSV was constructed as previously described (28). Quantitation of WSSV amplicons was accomplished by measuring the cycle threshold (CT) value. Since the plot of log of initial target copy number for the TaqMan assay was identical to that in WSSV DNA, it was considered that CT values obtained with infected shrimp DNA extracts could be converted to the numbers of viral genomic DNA targets by using the standard curve, and they are referred to here as viral titers. Relative amounts of PmRab7 expression were calculated from the CT value of PmRab7 divided by CT value of 18S rRNA.
WSSV purification.
The virus used in this study was isolated from WSSV-infected P. monodon shrimp from Thailand. WSSV purification followed the method described by Wang et al. (37). The viral titer (108 copies/μl) was determined by real-time PCR as described above. Viral preparations were stored at −20°C. The purity of the WSSV preparation was determined by transmission electron microscopy.
Determination of binding specificity by ELISA.
Flat-bottomed 96-well ELISA plates (Nunc) were coated with 100 μg of either SHM, purified His6-PmRab7, or BSA (100 μg). The plates were incubated in a humid chamber at 4°C overnight and then blocked with 150 μl of 10% heat-inactivated fetal bovine serum (HyClone) diluted in PBS buffer for 2 h at room temperature. The plates were washed with PBS buffer containing 0.05% Tween 20, and various dilutions of rVP28 (0 to 200 μg) or WSSV titer (10 to 108 copies) were added. After incubation at room temperature for 1 h followed by extensive washing, mouse anti-VP28 antibody was added and the bound anti-VP28 antibody was detected with a 1:2,000 dilution of HRP-labeled goat anti-mouse antibody (Dako). The reaction was visualized using the HRP substrate 3,3,5,5′-tetramethylbenzidine (Sigma). The reaction was stopped by the addition of 2 N H2SO4, and the absorbance was immediately read at 450 nm using an ELISA reader (VersaMax).
For blocking the ELISA, coated plates were first blocked with various dilutions (from undiluted to 1:1,024) of rabbit anti-Rab7 antibody (anti-Rab7) (rabbit polyclonal IgG raised against epitopes corresponding to amino acids 158 to 207 of Rab7 of human origin [Santa Cruz]) or anti-shrimp molt-inhibiting hormone antibody (anti-MIH) (kindly provided by P. Wongthai and B. Withyachumnarnkul, Mahidol University, Thailand). This was followed by washing, reaction with 100 μg of rVP28 or 1,000 copies of WSSV, and detection of bound rVP28 or WSSV by anti-VP28 antibody as described above.
In vivo neutralization.
Penaeus vannamei shrimp (6 to 8 g [fresh weight]) were divided into five groups, with three replicates of 10 shrimp in each group. One replicate of each group was used to collect shrimp at intervals for histological examination. Shrimp were injected as follows: group 1 with WSSV (103 copies/shrimp) (positive control), group 2 with TN buffer (20 mM Tris-HCl, 400 mM NaCl [pH 7.4]) (buffer negative control), group 3 with WSSV plus PmRab7 (10 μg/shrimp) as the recombinant protein test, group 4 with WSSV plus anti-Rab7 antibody (diluted 1:10 in TN buffer) as the anti-Rab7 test, and group 5 with WSSV plus anti-MIH (diluted 1:50 in TN buffer) as the antibody negative control. The antibody or PmRab7-plus-WSSV preparations were mixed and immediately injected into the experimental shrimp. The experiment was carried out twice using the semipurified WSSV described above. Cumulative mortality at 13 days after WSSV injection was recorded and compared by one-way analysis of variance using Sigmastat software (Jandel Scientific Co., Ltd.). Differences were considered significant when the P value was <0.05.
Histological examination.
On day 4 postchallenge with WSSV, all shrimp survivors were sacrificed by stunning in ice water and fixed with Davidson's fixative, and cephalothoraxes were processed by standard methods (14) for histological detection of pathognomonic lesions of WSSV (14).
Nucleotide sequence accession number.
The full-length sequence of Rab7 in the shrimp P. monodon has been deposited in GenBank under accession number DQ231062.
RESULTS
Expression of WSSV rVP28 in bacteria.
For conducting the VOPBA, a recombinant viral envelope protein, rVP28, was produced by a bacterial expression system (Fig. 1, lane 2). A yield of 10 to 15 mg/liter of protein was obtained after affinity chromatography purification (Fig. 1, lane 3). The identity of rVP28 was confirmed by Western blotting with an anti-VP28 antibody (Fig. 1, lane 4). The rVP28 had an apparent molecular mass of approximately 29 kDa, including the His tag.
FIG. 1.
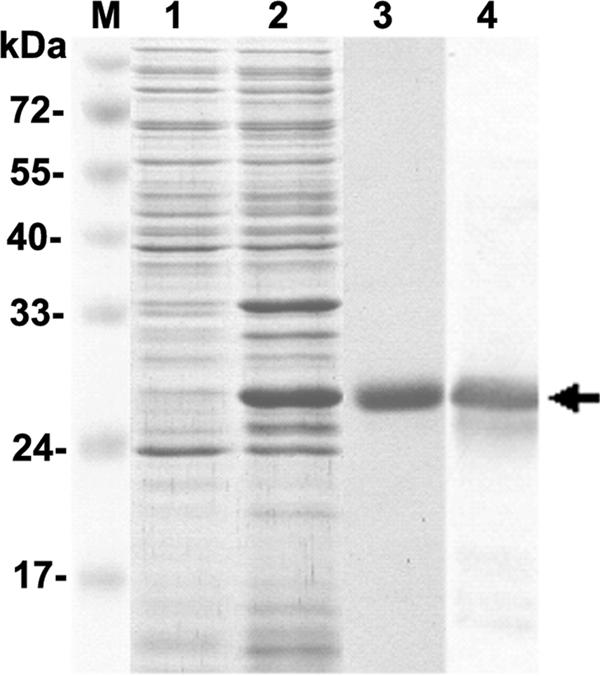
SDS-PAGE profile of recombinant WSSV envelope protein VP28 production. Lane 1, noninduced bacterial cell lysate; lane 2, induced bacterial cell lysate; lane 3, rVP28 purified by use of a Ni-nitrilotriacetic acid-ribotriacetic acid affinity chromatography column; lane 4, rVP28 detected with anti-VP28 antibody. The arrow shows a 29-kDa rVP28. Lane M, protein molecular mass markers.
Identification of WBPs.
By use of the VOPBA technique to identify host proteins that could bind with WSSV rVP28, anti-VP28 antibody revealed three distinct hemocyte membrane protein bands in SDS-PAGE, with apparent molecular masses of 14, 25, and 30 kDa (Fig. 2, lane 2). These three WBPs are referred to below as WBP14, WBP25, and WBP30. Analysis of these WBPs by liquid chromatography-tandem mass spectrometry showed that they were similar to proteins in the GenBank public database (Table 1). In particular, liquid chromatography-tandem mass spectrometry showed that WBP25 resembled a small GTP-binding protein, yptV5, from Volvox carteri (Table 1). The sequence of the algal protein yptV5 was similar to those of Rab7 proteins found in both plants and animals (65 to 71% identity). Since Rab7 is involved in the major routes of endocytosis used by viruses (26), WBP25 was investigated further.
FIG. 2.
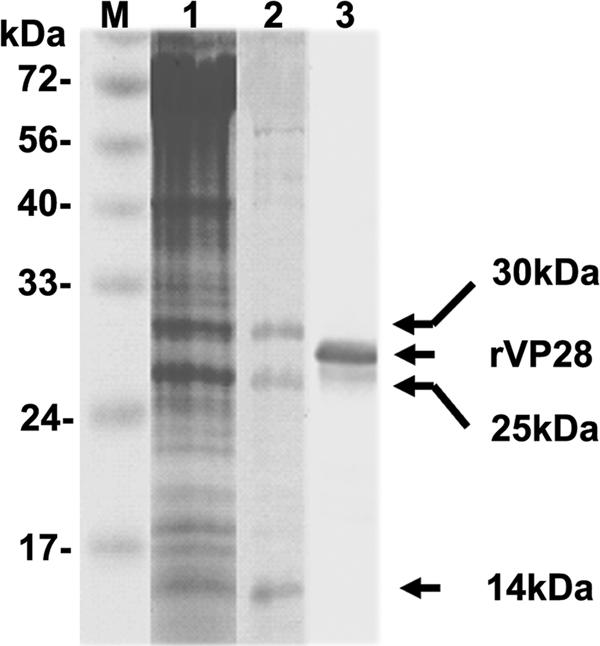
Results from VOPBA show the binding of rVP28 to SHM protein solution. Lane 1, Coomassie blue staining of shrimp hemocyte membrane protein; lane 2, blot of SHM incubated with rVP28 and then probed with anti-VP28 antibody reveals three rVP28-reactive bands; lane 3, blot of rVP28 (29 kDa) probed with anti-VP28 antibody. Lane M, protein molecular mass markers.
TABLE 1.
Analysis of WBPs by LC-MS/MS and MASCOT program
| WBP | Similar protein (protein name/organism/accession no.) | MASCOT score | Nominal mass (Da) | Sequence coverage (%) |
|---|---|---|---|---|
| WBP14 | Cyclic AMP-regulated protein-like protein/Marsupenaeus japonicus/BAB85575 | 205 | 17,052 | 24 |
| WBP25 | GTP-binding protein yptV5 (Rab7)/Volvox carteri/P36864 | 129 | 23,038 | 10 |
| WBP30 | Hemocyte kazal-type proteinase inhibitor/Penaeus monodon/ AAP92779 | 459 | 28,790 | 36 |
PmRab7 sequence analysis and tissue distribution.
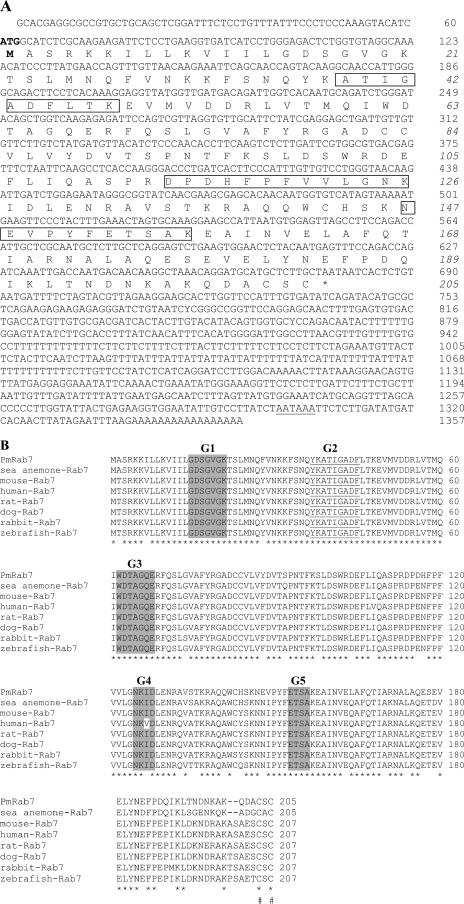
The full-length sequence of Rab7 in the shrimp P. monodon was identified in a Taiwan cDNA library (Fig. 3). The PmRab7 open reading frame has 1,357 bp encoding a polypeptide of 205 amino acids. Its estimated molecular mass is 21,930 Da, with a pI of 5.51. The widely spaced distribution of three WBP25 peptide sequences matching PmRab7 (Fig. 3A) further increased the reliability of the MASCOT results (MASCOT value of 128). The molecular mass calculated from the deduced amino acid sequence is slightly lower than that of the corresponding WBP25 protein observed by SDS-PAGE (∼25 kDa), suggesting that PmRab7 may undergo posttranslational modification. Consistent with this, Fig. 3B shows that the C terminus has two cysteine residues that may be isoprenylated, a crucial posttranslational modification that enables Rab proteins to associate and target the cell membrane (4). More importantly, PmRab7 has the four conserved GTP-binding or GTPase regions of the small G protein superfamily (G1 and G3 to G5), as well as an effector site (G2). These five regions are characteristic of Rab proteins (1). The sequence analysis suggests that PmRab7 may be an active GTPase that could cycle between the GDP- and GTP-bound states. PmRab7 is the first Rab homologue from crustaceans to be identified and characterized.
FIG. 3.
Primary structure of PmRab7 and sequence comparison. (A) Primary structure of PmRab7 cDNA and deduced amino acid sequence. The long 3′ untranslated sequence is included, and the polyadenylation signal is underlined. The start codon and the initiation methionine are in boldface. The stop codon is shown by an asterisk. The three matched peptide sequences from the MASCOT search are boxed. Ion scores for these three sequences were 52, 40, and 46, respectively. (B) Comparison of Rab7 amino acid sequences from different organisms. The putative PmRab7 (accession no. DQ231062) is shown aligned with sea anemone Rab7 (Aiptasia pulchella; AAQ23388), mouse Rab7 (Mus musculus; CAJ18560.1), human Rab7 (Homo sapiens; AAA86640), rat Rab7 (Rattus norvegicus; NP_076440), dog Rab7 (Canis familiaris; NP_001003316), rabbit Rab7 (Oryctolagus cuniculus; AAD02564), and zebrafish Rab7 (Danio rerio; AAH54602). Identical amino acids found in all sequences are indicated by asterisks; gaps were introduced to allow the best alignment. The putative effector binding domain (G2) is underlined. Conserved domains G1 and G3 to G5 are indicated by shaded boxes. Potential isoprenylation sites are indicated (#).
The tissue distribution of PmRab7-mRNA was studied by RT-PCR. The results revealed that PmRab7 is commonly expressed in various normal shrimp tissues, including hepatopancreas, hemocytes, stomach, lymphoid organ, gills, and heart (Fig. 4). Real-time PCR analysis showed that there was no significant change in the level of the PmRab7 transcript during the course of WSSV infection (data not shown). This result suggested that PmRab7 is a constitutively expressed gene.
FIG. 4.
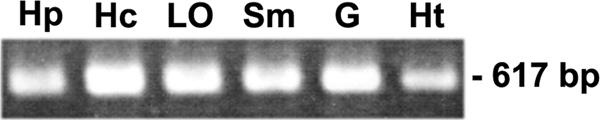
Tissue expression of PmRab7. The transcript of PmRab7 (617-bp product) was detected by RT-PCR in different tissues. Hp, hepatopancreas; Hc, hemocytes; Lo, lymphoid organ; Sm, stomach; G, gill; Ht, heart.
GST pull-down assay.
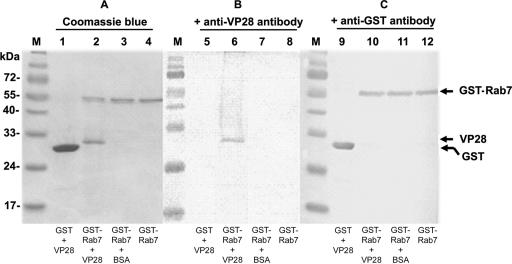
A GST pull-down experiment was used to confirm the functionality of PmRab7 and specifically its interaction with rVP28 (Fig. 5). Glutathione beads effectively pulled down both GST and GST-PmRab7 (Fig. 5), although the pull-down efficiency was higher with GST (Fig. 5, lanes 1 and 9) than with GST-PmRab7 (Fig. 5, lanes 4 and 12). When the same membrane was probed with antihistidine antibody, VP28 was present only in the pull-down product of GST-PmRab7 (Fig. 5, lanes 2 and 6). These data suggest that GST-PmRab7, not GST alone, is able to coprecipitate rVP28 and confirmed the specific binding of PmRab7 and viral envelope protein. The BSA control showed no coprecipitation of BSA with GST-PmRab7 (lane 3).
FIG. 5.
Results from GST pull-down assays show the specific binding of rVP28 to PmRab7. (A) Coomassie brilliant blue staining; (B) immunoblotting using antihistidine antibody; (C) immunoblotting using anti-GST antibody. Reaction components for each Sepharose 4B resin are shown at the bottom of each lane.Recombinant VP28 was coprecipitated with GST-Rab7 (lanes 2 and 6). Lane M, protein molecular mass markers.
Specificity of PmRab7 binding as determined by ELISA.
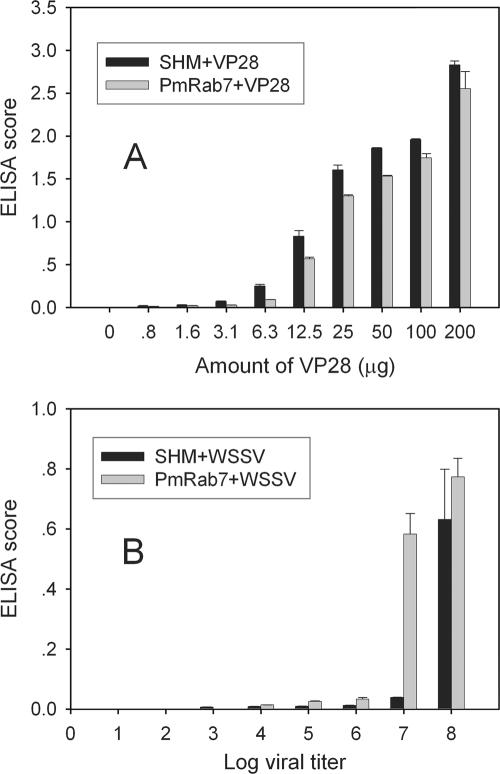
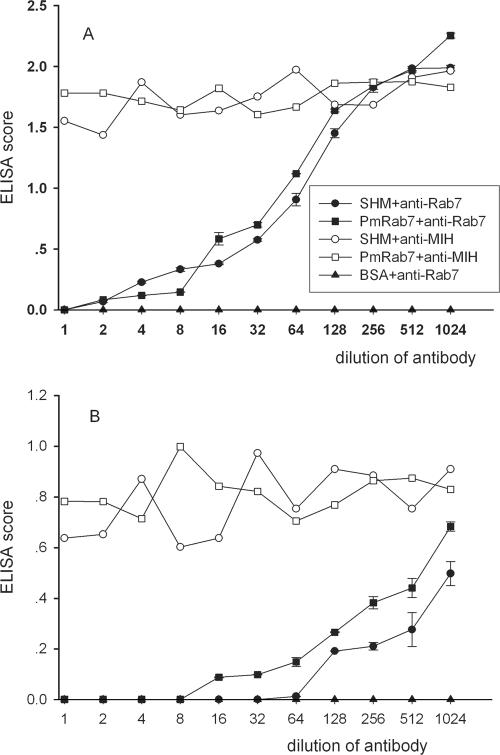
We have developed ELISA tests to determine the specificity of PmRab7 binding to rVP28 or WSSV particles. ELISA tests with SHM, purified rPmRab7, and BSA (control) showed that binding of SHM and rPmRab7 to rVP28 and WSSV was dose dependent (Fig. 6). No binding was observed with BSA. Binding of higher concentrations of virus would clarify whether the binding was truly saturable, but the required amount of virus with the excess to test for binding specificity was practically unattainable by current methodology. The interaction between SHM or rPmRab7 and WSSV could be detected when a viral titer of 10,000 was used. The amount of binding increased as the titer increased. However, at 107, binding to SHM was much less than it was to PmRab7. The difference was not as great at a titer of 108. The reason for this is unclear but may be related to differences in the dynamics of WSSV binding to purified PmRab7 as opposed to PmRab7 associated with other molecules within cell membranes. These other molecules may modulate the PmRab7 interaction with VP28, or they may interact directly with other WSSV proteins in a manner that affects PmRab7-VP28 binding.
FIG. 6.
Binding of rVP28 or WSSV particles to purified PmRab7. Purified rPmRab7 (100 μg) or SHM (100 μg) was applied to a microwell plate incubated with different amounts of rVP28 (0 to 200 μg) (A) or purified WSSV (viral titer of 10 to 108 copies) (B). A test with coated BSA was also performed as negative control. The binding specificity was determined with anti-VP28 antibody and secondary antibody conjugated with HRP. Error bars indicate standard deviations.
Binding of PmRab7 to WSSV is blocked by anti-Rab7 antibody.
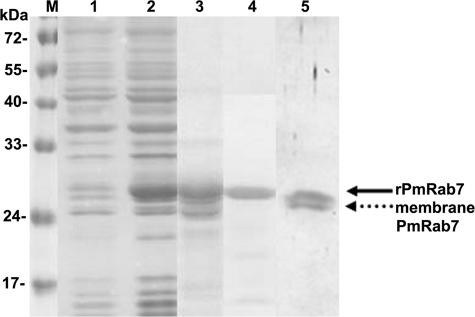
In this study, we tested whether anti-Rab7 antibody could block the binding of SHM or rPmRab7 to rVP28 or to WSSV particles. After Western blotting confirmed that the commercial rabbit anti-human Rab7 antibody could detect both recombinant PmRab7 (Fig. 7, lane 4) and hemocyte membrane Rab7 (Fig. 7, lane 5), ELISA results showed that binding was reduced as the amount of anti-Rab7 antibody was increased (Fig. 8). The control antibody (anti-MIH) had no effect on the binding. Similarly, there was no color detected when BSA was applied to plates instead of SHM or rPmRab7.
FIG. 7.
Protein profile of rPmRab7 production. Lane 1, noninduced bacterial cell lysate; lane 2, induced bacterial cell lysate; lane 3, affinity-purified rPmRab7; lane 4, rPmRab7 probed with anti-Rab7 antibody (rPmRab7 is shown by the solid arrow); lane 5, Rab7 in shrimp hemocyte membrane probed with anti-Rab7 antibody (membrane PmRab7 is shown by the dotted arrow). Lane M, protein molecular mass markers.
FIG. 8.
Anti-Rab7 antibody inhibits the binding of rVP28 or WSSV to PmRab7. SHM or PmRab7 (100 μg) was applied to a microwell plate and incubated with different dilutions of anti-Rab7 antibody or anti-MIH antibody (positive control). After removal of excess antibodies, rVP28 (100 μg) (A) or purified WSSV (103 copies) (B) was added to each well. The amount of rVP28 or WSSV binding to PmRab7 were determined with anti-VP28 antibody and secondary antibody conjugated with HRP. The binding was determined by spectrophotometry after addition of HRP substrate (ELISA score). Only one replicate was done for SHM plus anti-MIH and PmRab7 plus anti-MIH. Error bars indicate standard deviations.
In vivo neutralization.
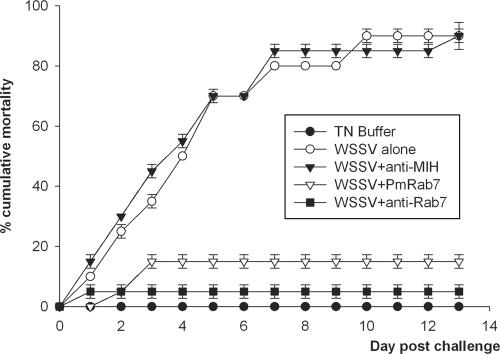
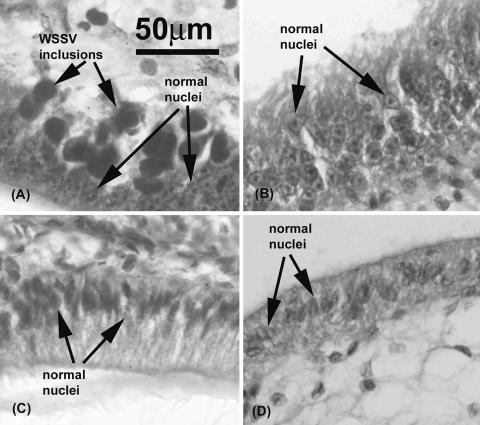
The in vivo assay was developed to test whether PmRab7 or anti-Rab7 antibody could block WSSV infection in shrimp. Shrimp mortality increased steadily from day 1 to day 7, ending at 95% for both shrimp groups injected with WSSV alone (positive control) and groups injected with WSSV plus anti-MIH antibody (antibody negative control) (Fig. 9). By contrast, there was no shrimp mortality in the buffer-injected group (negative control group) and low shrimp mortality in groups injected with WSSV plus anti-Rab7 (5%) or WSSV plus PmRab7 (15%). The tests were done twice, and similar results were observed. Thus, the results indicated that both PmRab7 and anti-Rab7 antibody could block white spot disease in shrimp. Histological examination of surviving shrimp in each group on day 4 postchallenge confirmed typical WSSV histopathology (i.e., large basophilic, intranuclear viral inclusions in subcuticular epithelial cells) in shrimp injected with WSSV alone (Fig. 10A) or with WSSV plus anti-MIH antibody (results were similar to those in Fig. 10A). These inclusions were not found in tissues of the surviving shrimp injected with WSSV plus anti-Rab7 antibody (Fig. 10B), with WSSV plus PmRab7 (Fig. 10C), or with buffer (Fig. 10D).
FIG. 9.
Neutralization of WSSV with anti-Rab7 antibody and PmRab7. On day 0, shrimp were injected as follows: group 1, WSSV (103 copies/shrimp); group 2, TN buffer; group 3, WSSV plus PmRab7 (10 μg/shrimp); group 4, WSSV plus anti-Rab7 antibody; group 5, WSSV plus anti-MIH antibody. Cumulative mortality data represent the pooled results for two replications (n = 10 for each group). The experiment was done two times. Error bars indicate standard deviations.
FIG. 10.
Histological examination (magnification, ×400) of experimental shrimp on day 4 postchallenge. (A) Shrimp injected with WSSV (group 1); (B) shrimp injected with WSSV plus anti-Rab7 antibody (group 4); (C) shrimp injected with WSSV plus PmRab7 (group 3); (D) shrimp injected with buffer (group 2).
DISCUSSION
We have shown that the GTP-binding protein Rab7 may be a receptor for VP28 envelope protein of WSSV in shrimp. Rab proteins are known to be regulators of vesicle budding and fusion events and represent a family of over 30 proteins that are localized on the surfaces of distinct membrane-enclosed compartments of exocytic and endocytic pathways. They are found in all eukaryotes, including yeasts, plants, insects, and mammals (22, 24, 42). Pan et al. (20) studied differential gene expression in WSSV-resistant shrimp (Penaeus japonicus) by subtractive hybridization and showed that genes for small GTPases are up-regulated in virus-resistant shrimp. These smallGTP-binding proteins comprise products of the Ras, Rab/YPT, and Rho gene families and are involved in diverse cellular functions, including growth, differentiation, and vesicular traffic (42). In mammalian cells, Rab7 protein is associated with late endosomes and regulates the traffic from early to late endosomes and/or from late endosomes to lysosomes (4, 8). Rab7 function appears to be required for efficient phagocytosis (2, 17). In its primary structure, every Rab protein contains five characteristic regions (1). Four of these are GTP-binding or GTPase regions that are conserved in all members of the small G protein superfamily. The fifth is termed an effector site, which interacts with accessory protein and contains amino acids especially conserved in the Rab family (Fig. 3B). In agreement with this, the PmRab7 gene sequence codes for a deduced polypeptide containing five extremely conserved motifs involved in GTP-binding or GTPase activity and an isoprenylation site, suggesting that PmRab7 is an active GTPase that is able to cycle between GDP- and GTP-bound states.
It is well known that many viruses use endocytosis as a route of entry into host cells (23, 25, 26). According to this model, a virus binds to cell surface receptors and is then internalized into the endocytic network in a manner that is generally considered to be calthrin dependent. For example, adenovirus subgroup B (Ad7) undergoes high-affinity interaction with its receptor Ad7 that is colocalized with Rab7 as it is trafficked to late endosomes (18). Rabs act as molecular switches to control trafficking of endocytic vesicles within cells, as well as their subsequent fusion to endosomes However, our work now shows that an enveloped virus can bind directly to a Rab protein (Fig. 2, 5, 6, 8, and 9). In particular, Fig. 6 shows that PmRab7 binds to WSSV virions and to the recombinant WSSV envelope protein or VP28 in a dose-dependent manner, Fig. 8 shows that anti-Rab7 antibody can inhibit these interactions, and Fig. 5 confirms the specificity of this binding. Further tests, such as immunofluorescence by confocal microscopy or immunogold labeling for transmission electron microscopy, would be necessary to establish whether or not PmRab7 is localized at the hemocyte cell surface. If it is, then simple receptor interference would explain the protective effect of PmRab7 and anti-PmRab7 in our WSSV challenge tests (see below). If, on the other hand, it is exclusively localized in cytoplasmic membranes as previously suggested (1), the mechanism for this protection would be more complex. For example, prior binding of PmRab7 to WSSV via VP28 might interfere with viral binding to the cell surface via a different receptor, but an explanation for protection by anti-PmRab7 would be more difficult. Thus, we believe that the balance of our results leans in favor of at least some presence of PmRab7 at the cell surface, despite previous information suggesting that it is exclusively cytoplasmic (1).
In vivo neutralization experiments have been widely used for many vertebrate viruses and have led to the development of passive immunization strategies. In shrimp, use of an anti-VP28 antibody resulted in a low cumulative mortality in immersion or challenge tests with WSSV (34). Witteveldt et al. (38) also showed that injection of recombinant VP28 could reduce the mortality caused by WSSV in shrimp. Similarly, a reduction and delay in mortality were observed upon the use of antibodies to other WSSV envelope proteins, such as VP68, VP281, and VP466 (40). Using an alternative strategy for the first time in shrimp, we have shown that either PmRab7 or an antibody to it can reduce and delay mortality upon WSSV challenge. These results may open the way for the design of novel methods to prevent shrimp death from WSSV infection.
Acknowledgments
We thank Paisarn Sithigorngul for providing mouse anti-VP28 antibody and Printhip Wongthai and Boonsirm Withyachumnarnkul for providing anti-molt-inhibiting hormone antibody.
This investigation was supported financially by National Center for Genetic Engineering and Biotechnology (BIOTEC) grant BT-B-07-SG-B7-4512 and by a National Research Council of Thailand grant.
REFERENCES
- 1.Bourne, H. R., D. A. Sanders, and F. McCormick. 1990. The GTPase superfamily: a conserved switch for diverse cell functions. Nature 38:125-132. [DOI] [PubMed] [Google Scholar]
- 2.Bucci, C., P. Thomsen, P. Nicoziani, J. McCarthy, and B. van Deurs. 2000. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell 11:467-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, P., C. F. Lo, Y. Wang, and G. H. Kou. 1996. Identification of white spot syndrome associated baculovirus (WSBV) target organs in the shrimp Penaeus monodon by in situ hybridization. Dis. Aquat. Org. 27:131-139. [Google Scholar]
- 4.Chavrier, P., R. G. Parton, P. H. Hauri, K. Simons, and M. Zerial. 1990. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell 62:317-329. [DOI] [PubMed] [Google Scholar]
- 5.Chen, L. L., J. H. Leu, C. J. Huang, C. M. Chou, S. M. Chen, C. H. Wang, C. F. Lo, and G. H. Kou. 2002. Identification of a nucleocapsid protein (VP35) gene of shrimp white spot syndrome virus and characterization of the motif important for targeting VP35 to the nuclei of transfected insect cells. Virology 293:44-53. [DOI] [PubMed] [Google Scholar]
- 6.Durand, S., D. V. Lightner, R. M. Redman, and J. R. Bonami. 1997. Ultrastructure and morphogenesis of white spot syndrome baculovirus (WSSV). Dis. Aquat. Org. 29:205-211. [Google Scholar]
- 7.Duvic, B., and K. Söderhäll. 1992. Purification and partial characterization of a beta-1,3-glucan-binding-protein membrane receptor from blood cells of the crayfish Pacifastacus leniusculus. Eur. J. Biochem. 207:223-228. [DOI] [PubMed] [Google Scholar]
- 8.Feng, Y., B. Press, and A. Wandinger-Ness. 1995. Rab7: an important regulator of late endocytic membrane traffic. J. Cell Biol. 131:1435-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gastka, M., J. Horvath, and T. L. Lentz. 1996. Rabies virus binding to the nicotinic acetylcholine receptor α subunit demonstrated by virus overlay protein binding assay. J. Gen. Virol. 77:2437-2440. [DOI] [PubMed] [Google Scholar]
- 10.Hossain, M. S., S. Khadijah, and J. Kwang. 2004. Characterization of ORF89, a latency-related gene of white spot syndrome virus. Virology 325:106-115. [DOI] [PubMed] [Google Scholar]
- 11.Khadijah, S., S. Y. Neo, M. S. Hossain, L. D. Miller, S. Mathavan, and J. Kwang. 2003. Identification of white spot syndrome virus latency-related genes in specific-pathogen-free shrimps by use of a microarray. J. Virol. 77:10162-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Leu, J. H., J. M. Tsai, H. C. Wang, A. H. Wang, C. H. Wang, G. H. Kou, and C. F. Lo. 2005. The unique stacked rings in the nucleocapsid of the white spot syndrome virus virion are formed by the major structural protein VP664, the largest viral structural protein ever found. J. Virol. 79:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lightner, D. V. 1996. A handbook of shrimp pathology and diagnostic procedures for diseases of cultured penaeid shrimp. World Aquaculture Society, Baton Rouge, La.
- 15.Liu, W. J., Y. S. Chang, C. H. Wang, G. H. Kou, and C. F. Lo. 2005. Microarray and RT-PCR screening for white spot syndrome virus immediate-early genes in cycloheximide-treated shrimp. Virology 334:327-341. [DOI] [PubMed] [Google Scholar]
- 16.Marks, H., R. W. Goldbach, J. M. Vlak, and M. C. W. van Hulten. 2004. Genetic variation among isolates of white spot syndrome virus. Arch. Virol. 149:673-697. [DOI] [PubMed] [Google Scholar]
- 17.Meresse, S., J. P. Gorvel, and P. Chavrier. 1995. The rab7 GTPase resides on a vesicular compartment connected to lysosmes. J. Cell Sci. 108:3349-3358. [DOI] [PubMed] [Google Scholar]
- 18.Miyazawa, N., R. G. Crystal, and P. L. Leopold. 2001. Adenovirus serotype 7 retention in a late endosomal compartment prior to cytosol escape is modulated by fiber protein. J. Virol. 75:1387-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nadala, E. C. B., L. M. Tapay, and P. C. Loh. 1998. Characterization of a non-occluded baculovirus-like agent pathogenic to penaeid shrimp. Dis. Aquat. Org. 33:221-229. [DOI] [PubMed] [Google Scholar]
- 20.Pan, D., N. He, Z. Yang, H. Liu, and X. Xu. 2005. Differential expression profile in hepatopancreas of WSSV-resistant shrimp (Penaeus japonicus) by suppression substractive hybridization. Dev. Comp. Immunol. 29:103-112. [DOI] [PubMed] [Google Scholar]
- 21.Pandya, J., A. Chakraborti, Y. Chawla, J. B. Dilawari, S. Sehgal, and N. K. Ganguly. 2002. Identification of human hepatocyte protein(s), which binds specifically to the recombinant envelope-2/non-structural-1 protein of hepatitis C virus. Virus Res. 87:135-143. [DOI] [PubMed] [Google Scholar]
- 22.Pereira-Leal, J. B., and M. C. Seabra. 2000. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J. Mol. Biol. 301:1077-1087. [DOI] [PubMed] [Google Scholar]
- 23.Russell, D. G., and M. Marsh. 2001. Endocytosis in pathogen entry and replication, p. 247-280. In M. Marsh (ed.), Endocytosis. Oxford University Press, Oxford, United Kingdom.
- 24.Segev, N. 2001. Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 13:500-511. [DOI] [PubMed] [Google Scholar]
- 25.Sieczkarski, S. B., and G. R. Whittaker. 2002. Dissecting virus entry via endocytosis. J. Gen. Virol. 83:1535-1545. [DOI] [PubMed] [Google Scholar]
- 26.Sieczkarski, S. B., and G. R. Whittaker. 2003. Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 4:333-343. [DOI] [PubMed] [Google Scholar]
- 27.Söderhäll, K., and V. J. Smith. 1983. Separation of the haemocyte populations of Carcinus maenas and other marine decapods, and prophenoloxidase distribution. Dev. Comp. Immunol. 7:229-239. [DOI] [PubMed] [Google Scholar]
- 28.Sritunyalucksana, K., J. Srisala, K. McColl, L. Nielsen, and T. W. Flegel. 2006. Comparison of PCR testing methods for white spot syndrome virus (WSSV) infections in penaeid shrimp. Aquaculture 255:95-104.
- 29.Tsai, M. F., C. F. Lo, M. C. W. van Hulten, H. F. Tzeng, C. M. Chou, C. J. Huang, C. H. Wang, J. Y. Lin, J. M. Vlak, and G. H. Kou. 2000. Transcriptional analysis of the ribonucleotide reductase genes of shrimp white spot syndrome virus. Virology 277:92-99. [DOI] [PubMed] [Google Scholar]
- 30.Tsai, M. F., H. T. Yu, H. F. Tzeng, J. H. Leu, C. M. Chou, C. J. Huang, C. H. Wang, J. Y. Lin, G. H. Kou, and C. F. Lo. 2000. Identification and characterization of a shrimp white spot syndrome virus (WSSV) gene that encodes a novel chimeric polypeptide of cellular-type thymidine kinase and thymidylate kinase. Virology 277:100-110. [DOI] [PubMed] [Google Scholar]
- 31.Tsai, J. M., H. C. Wang, J. H. Leu, H. H. Hsiao, A. H. Wang, G. H. Kou, and C. F. Lo. 2004. Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus. J. Virol. 78:11360-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai, J. M., H. C. Wang, J. H. Leu, A. H. Wang, Y. Zhung, P. J. Walker, G. H. Kou, and C. F. Lo. 2006. Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion. J. Virol. 80:3021-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzeng, H. F., Z. F. Chang, S. E. Peng, C. H. Wang, J. Y. Lin, G. H. Kou, and C. F. Lo. 2002. Chimeric polypeptide of thymidine kinase and thymidylate kinase of shrimp white spot syndrome virus: thymidine kinase activity of the recombinant protein expressed in a baculovirus/insect cell system. Virology 299:248-255. [DOI] [PubMed] [Google Scholar]
- 34.Van Hulten, M. C. W., J. Witteveldt, M. Snippe, and J. M. Vlak. 2001. White spot syndrome virus envelope protein VP28 is involved in the systemic infection of shrimp. Virology 285:228-233. [DOI] [PubMed] [Google Scholar]
- 35.Van Hulten, M. C. W., M. Reijus, A. M. G. Vermeesch, F. Zandbergen, and J. M. Vlak. 2002. Identification of VP19 and VP15 of white spot syndrome virus (WSSV) and glycosylation of the WSSV major structural proteins. J. Gen. Virol. 83:257-265. [DOI] [PubMed] [Google Scholar]
- 36.Vlak, J. M., J. R. Bonami, T. W. Flegel, G. H. Kou, D. V. Lightner, C. F. Lo, P. C. Loh, and P. W. Walker. 2004. Nimaviridae, p. 187-192. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.) VIIIth report of the International Committee on Taxonomy of Viruses. Elsevier, Amsterdam, The Netherlands.
- 37.Wang, C. H., C. F. Lo, J. H. Leu, C. M. Chou, P. Y. Yeh, H. Y. Chou, M. C. Tung, C. F. Chang, M. S. Su, and G. H. Kou. 1995. Purification and genomic analysis of baculovirus associated with white spot syndrome (WSBV) of Penaeus monodon. Dis. Aquat. Org. 23:239-242. [Google Scholar]
- 38.Witteveldt, J., J. M. Vlak, and M. C. W. van Hulten. 2004. Protection of Penaeus monodon against white spot syndrome virus using a WSSV subunit vaccine. Fish Shellfish Immunol. 16:571-579. [DOI] [PubMed] [Google Scholar]
- 39.Wongteerasupaya, C., J. E. Vickers, S. Sriurairatana, G. L. Nash, A. Akarajamorn, V. Boonsaeng, S. Panyim, A. Tassanakajon, B. Withyachumnarnkul, and T. W. Flegel. 1995. A non-occluded, systemic baculovirus that occurs in the cells of ectodermal and mesodermal origin and causes high mortality in the black tiger prawn Penaeus monodon. Dis. Aquat. Org. 21:69-77. [Google Scholar]
- 40.Wu, W., L. Wang, and X. Zhang. 2005. Identification of white spot syndrome virus (WSSV) envelope proteins involved in shrimp infection. Virology 332:578-583. [DOI] [PubMed] [Google Scholar]
- 41.Yi, G., Z. Wang, Y. Qi, L. Yao, J. Qian, and L. Hu. 2004. VP28 of shrimp white spot syndrome virus is involved in the attachment and penetration into shrimp cells. J. Biochem. Mol. Biol. 37:726-734. [DOI] [PubMed] [Google Scholar]
- 42.Zerial, M., and H. MacBride. 2001. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2:107-117. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, X., C. Huang, X. Tang, Y. Zhuang, and C. L. Hew. 2004. Identification of structural proteins from shrimp white spot syndrome virus (WSSV) by 2DE-MS. Proteins 55:229-235. [DOI] [PubMed] [Google Scholar]