Abstract
The importance of the F protein cytoplasmic tail (CT) for replication of human respiratory syncytial virus (HRSV) was examined by monitoring the behavior of viruses expressing F proteins with a modified COOH terminus. The F protein mutant viruses were recovered and amplified under conditions where F protein function was complemented by expression of a heterologous viral envelope protein. The effect of the F protein modifications was then examined in the context of a viral infection in standard cell types (Vero and HEp-2). The F protein modifications consisted of a deletion of the predicted CT or a replacement of the CT with the CT of the vesicular stomatitis virus (VSV) G protein. In addition, engineered HRSVs that lacked all homologous glycoprotein genes (SH, G, and F) and expressed instead either the authentic VSV G protein or a VSV G containing the HRSV F protein CT were examined. We found that deletion or replacement of the F protein CT seriously impaired the production of infectious progeny. Cells infected with viruses bearing CT modifications displayed increased F protein surface expression and increased syncytium formation. The distribution of F protein in the plasma membrane of infected cells was altered, resulting in an F protein that was evenly distributed rather than localized predominantly to virus-induced surface filaments. CT deletion or exchange also abrogated interaction of F protein with Triton-insoluble lipid rafts. Addition of the F protein CT to the VSV G protein, expressed as the only viral glycoprotein in an HRSV genome, had the opposite effects: the number of infectious progeny was higher, the surface distribution was changed from relatively even to localized, and the proportion of VSV G protein associated with lipid rafts was higher. Together, these results show that the HRSV F protein CT plays a critical role in F protein cellular localization and production of infectious virus and suggest that the function provided by the CT is independent of the F protein ectodomain and transmembrane domain and is mediated by F protein-lipid raft interaction.
Human respiratory syncytial virus (HRSV) is a major cause of severe lower respiratory tract disease in infants, children, immunosuppressed individuals, and the elderly (13, 22, 41). Prevention and treatment of HRSV disease remain a significant challenge. HRSV is a member of the Paramyxoviridae family and has a 15,222-nucleotide, negative-sense, single-stranded RNA genome. Eleven known viral proteins are expressed, three of which have been characterized as transmembrane glycoproteins: SH (small hydrophobic, unknown function), G (attachment), and F (fusion) (11, 25). The G and F proteins are required for optimal virus infectivity and contain important immunoprotective epitopes (12, 40, 57). In infected cells at late times postinfection, the G and F proteins are present in virus-induced filamentous structures of 2 to 10 μm in length at the cell surface (2, 18, 27, 38, 44, 54). It is not clear whether these filamentous structures represent single infectious virions or virion factories or whether they may play additional, yet-unidentified roles in the HRSV life cycle.
HRSV and the other pneumoviruses are unique within the Paramyxoviridae in that they express an attachment glycoprotein, G, that is highly O glycosylated as well as N glycosylated and has no homology to the other paramyxovirus attachment proteins (63, 64). The HRSV F protein resembles the prototypic paramyxovirus fusion protein (12). However, in contrast to most other paramyxovirus F proteins, the HRSV F protein can induce membrane fusion in the absence of the homotypic attachment protein and was shown to be a determinant of species-specific virus entry into host cells (23, 46).
Both G and F proteins are structural components of the HRSV virion, but little is known about the mechanisms by which they coalesce and incorporate into virus particles and about the HRSV assembly process in general. For several members of the Mononegavirales, the viral matrix protein is thought to be an organizer of viral assembly, and interactions between matrix and glycoproteins have been demonstrated or suggested (for reviews, see references 47 and 56). For some, such as measles, influenza, and Ebola viruses, Triton-insoluble membranes (lipid rafts) have been implicated in the assembly process (3, 30, 45, 59). Recent reports suggest that for HRSV too, lipid rafts appear to play a role in viral assembly. For example, HRSV virions were shown to contain lipid raft markers, and the HRSV matrix, SH, G, and F proteins were reported to associate with lipid rafts (6-8, 24, 27, 32-34, 43). Biochemical analyses of cells infected with a recombinant vaccinia virus expressing HRSV F indicate that the F protein, but not the matrix protein, can associate with lipid rafts in the absence of the other HRSV proteins (24). In addition, in previous work we found that F distribution in infected Vero cells is highly targeted relative to G distribution: while G is detected at the cell surface in an evenly distributed, and seemingly random, fashion, the F protein is observed predominantly localized to viral filaments (38). These findings suggest that F protein targeting may play an organizing role in viral assembly.
By analogy to other enveloped viruses, two domains likely to be involved in F protein functions in assembly are the transmembrane (TM) domain and the cytoplasmic tail (CT). A previous study in which the HRSV F protein was expressed in uninfected cells reported that the TM domain contains an orientation-independent apical sorting signal and that the CT was not a determinant of F protein targeting (4a). Experimental CT truncations in the viral context have been reported for other enveloped viruses and were shown in many cases to affect assembly-related processes such as glycoprotein incorporation, budding, or morphology (14, 17, 26, 28, 47, 48, 55, 56, 62). For influenza virus and simian virus 5 (SV5), viruses lacking one of the membrane glycoprotein CTs, but not both, could be generated with relative ease, indicative of a dually redundant role for the transmembrane glycoprotein CTs in viral assembly (26, 28, 62). We previously reported that a heterologous viral transmembrane glycoprotein (vesicular stomatitis virus [VSV] G protein) carrying the HRSV F protein CT concentrated in HRSV-induced surface filaments and colocalized with F when the two proteins were expressed simultaneously (38). In addition, providing a partial F CT to the baculovirus GP64 protein modestly increased the amount of GP64 incorporated into HRSV virions as determined by immunogold labeling (39). These observations suggested a role for the F protein CT in viral assembly.
In this study, we examined the importance of the F protein CT for replication of HRSV in cell culture. Due to the potential impact of F modifications on F protein function, we recovered viruses encoding tail-modified F proteins from cDNA in a complementing cell line (Vbac), which expresses a heterologous viral membrane fusion protein, the baculovirus GP64 protein with an HRSV F protein CT (named GP64/F) (40). The GP64/F protein provided in trans was previously shown to allow recovery and efficient amplification of HRSV independent of F protein function (40), thus enabling us to recover viruses with mutations that otherwise might be detrimental. After recovery and amplification of engineered viruses in Vbac cells, Vero and HEp-2 cells were used to examine growth in cell culture, surface expression, and lipid raft association in the presence or absence of the predicted F protein CT. In addition, engineered HRSVs expressing as their only transmembrane glycoprotein either the authentic VSV G protein or a VSV G protein carrying the F protein CT were compared. The results indicate that the F protein CT plays a key role in the cellular distribution of the F protein and production of infectious HRSV progeny.
MATERIALS AND METHODS
Cells and antibodies.
Vero 76 (Vero) and HEp-2 cells were acquired from the American Type Culture Collection (ATCC) and grown in standard growth medium containing 5% fetal bovine serum. Vbac cells (Vero cells expressing the baculovirus GP64 protein carrying F protein COOH-terminal residues 563 to 573) were described previously (40). Monoclonal antibodies (MAbs) 6 and 19 were provided by Geraldine Taylor (Institute for Animal Health, Compton, United Kingdom), MAb L9 by Ed Walsh (University of Rochester School of Medicine, Rochester, NY), and MAb AcV5 by Gary Blissard (Boyce Thompson Institute at Cornell University, Ithaca, NY). The humanized anti-F MAb (Synagis) and anti-VSV G antibody (mouse ascitic fluid) were acquired from MedImmune, Inc., and ATCC, respectively. Anti-caveolin-1 antibody was purchased from Santa Cruz Biotechnology, Inc., and the anti-human transferrin receptor antibody from Zymed Laboratories, Inc.
Virus infections.
Infections of cell monolayers were carried out by adsorbing virus to cells for 1.5 h at 37°C, unless indicated otherwise.
Construction of F and VSV G protein COOH-terminal modifications.
Through the use of PCR, an XbaI site was generated at the TM domain/CT border region of the F protein by changing nucleotides 1654 and 1656 of the F protein open reading frame (ORF) in a cDNA clone from G to T and from C to T, respectively. This resulted in a coding change of amino acid 552 from alanine to serine (F552S) (Fig. 1A). With the use of this XbaI site (spanning the coding sequence for F residues 552 [TCT] and 553 [AGA]) and an AscI site flanking the stop codon of the F protein ORF (38), the sequence encoding the native F protein CT was deleted and replaced with a sequence carrying a stop codon immediately following residue R553 (FΔCT) or with a sequence encoding amino acids 492 to 511 of the VSV G protein (FVSV-G). Construction of VSV-GRS-F (previously termed GVSV) is described elsewhere (38). All VSV G sequences represent the Indiana serotype.
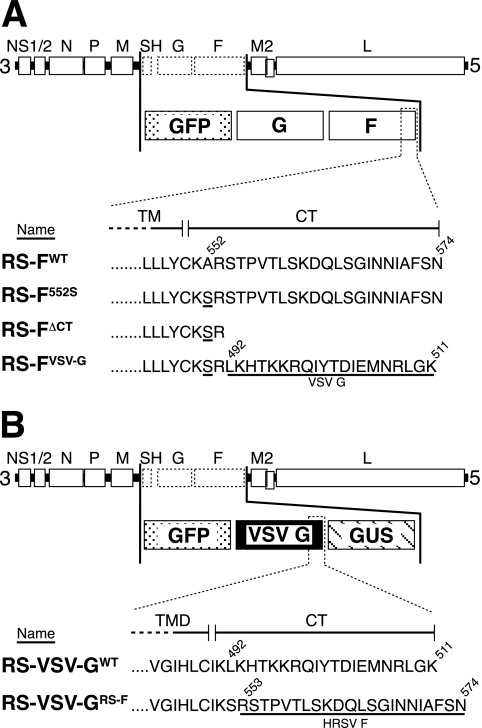
FIG. 1.
Composition of COOH-terminally modified F and VSV G proteins and viral cDNAs. (A) The F protein CT was either deleted (FΔCT) or replaced with that of the VSV G protein (FVSV-G). This was done with the use of a restriction site created at amino acid position 552, which resulted in a change from alanine to serine (A552S). An F protein of wt length but containing the sole change of A552S (F552S) was included in all experiments as a control. Non-HRSV sequences are underlined. Shuttle vectors containing the GFP ORF (in place of the nonessential SH ORF), the G protein ORF, and each of the modified F protein ORFs or a fully authentic F protein ORF (FWT), separated by authentic gene junctions, were inserted into a cDNA of the A2 strain of HRSV. (B) A VSV G ORF carrying the HRSV F protein CT in place of its own (VSV-GRS-F) was constructed and reported previously (38). Non-VSV sequences are underlined. The VSV-GRS-F ORF or the authentic VSV G protein ORF (VSV-GWT) was cloned into a shuttle vector already containing the GFP and β-glucuronidase ORFs, separated by authentic HRSV gene junctions. Shuttle vectors were inserted into a cDNA backbone of the A2 strain of HRSV, generating cDNAs that lacked the SH, G, and F ORFs and expressed a heterologous viral protein as their only transmembrane glycoprotein. (A and B) Viruses were recovered from all cDNAs shown by using the baculovirus GP64-complementing cell line Vbac, as previously described (40), and named as indicated.
Construction of cDNAs and recovery of infectious HRSVs by using the Vbac cell line.
Engineered cDNAs were based on the A2 strain of HRSV and were constructed as described previously (38). Briefly, using standard cloning techniques, shuttle vectors containing ORFs encoding green fluorescent protein (GFP), HRSV G, and HRSV F (wild type [wt] or COOH-terminally modified) were constructed. To facilitate future cloning, the nucleotide preceding the F start codon was changed from A to C to create an NcoI site, slightly altering the F protein translation context from ATAACA-ATG to ATAACC-ATG. The ORFs contained in the shuttle vectors, separated by authentic HRSV intergenic junctions and flanked by unique FseI and AscI restriction sites, were cloned into an SH/G/F-deleted cDNA backbone containing matching FseI and AscI restriction sites. The gene content of the resulting cDNAs thus varied from that of wt HRSV only at genome positions 6 (GFP) and 8 (modified F). Construction of the RS-VSV-GRS-F cDNA was previously reported, and this was termed RSΔSH,G,F/Gvsv (38). Virus RS-VSV-GWT was similarly constructed but contains the authentic VSV G protein ORF in place of the chimeric VSV-GRS-F ORF. All modified areas of engineered cDNAs were verified by nucleotide sequencing prior to virus recovery.
Infectious viruses were recovered from cDNA as described previously (40). Notably, to relieve selection pressure that might result from the effect of F modifications, a plasmid encoding a chimeric VSV G protein was included in the initial transfection, and Vbac cells were used for virus amplification. Viral RNAs were harvested from cells infected with the engineered viruses at passage 3 and amplified by reverse transcription-PCR, and selected areas (see Results) were verified by bulk nucleotide sequence analysis. No changes were found. Passage 3 stocks were used for the experiments described here.
Protein labeling and immunoprecipitation.
Vero cells were infected at a multiplicity of 5 and incubated at 37°C. At 20 to 21 h postinfection (p.i.), cells were starved for 30 min in medium without methionine and cysteine and then incubated in the presence of Tran35S-label (MP Biochemicals, Inc.) for 4 h. Cells were washed and subsequently lysed by incubation in cold immunoprecipitation (IP) buffer (150 mM NaCl, 50 mM Tris [pH 7.4], 1% NP-40, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS]) in the presence of protease inhibitors (Halt inhibitor cocktail; Pierce) for 30 min on ice. Cell lysates were clarified by two rounds of centrifugation for 5 min at 15,800 × g. To the clarified supernatants, anti-F antibodies MAb 19 and Synagis were added, and the lysate was then incubated for 45 min at 4°C. Protein G beads (Pierce) were added, and the lysates were incubated for an additional 2 h with agitation. Next, the beads were washed extensively with cold IP buffer, and the final pellet was boiled in SDS-polyacrylamide gel electrophoresis (SDS-PAGE) buffer.
Samples for the lipid raft experiment were immunoprecipitated as described above, with the following exceptions. Cells were metabolically labeled from 24 to 27 h p.i. Chase samples were washed with phosphate-buffered saline and incubated for an additional 1 h in standard growth medium before lysis.
Cell ELISA analysis.
Protein expression by the engineered viruses was examined by cell enzyme-linked immunosorbent assay (ELISA) as previously described (35, 39). Briefly, for each virus duplicate wells of infected Vero cells were fixed with 4% paraformaldehyde for 15 min at approximately 24 h p.i. and permeabilized by incubation with 0.2% Triton X-100 for 5 min at room temperature. Cells were blocked and then incubated with antibodies against HRSV F (MAb 19), HRSV G (MAb L9), GP64 (MAb AcV5), or HRSV nucleocapsid (N) protein (MAb 6) as a control; washed; and incubated with a horseradish peroxidase-conjugated secondary goat anti-mouse antibody (Pierce). Next, cells were washed with phosphate-buffered saline and incubated in 1 ml 3,3′,5,5′-tetramethylbenzidine substrate (Pierce). At various times after addition of substrate, 100-μl aliquots were collected and added to 2 M sulfuric acid in a 96-well plate to stop the reaction. The optical density at 450 nm was determined in an ELISA plate reader. To determine relative F protein surface levels (see Fig. 5A), parallel samples were not detergent permeabilized but were otherwise treated identically. The protein levels were normalized to the level of N protein expression (average of results from duplicate wells) measured in parallel samples that were permeabilized.
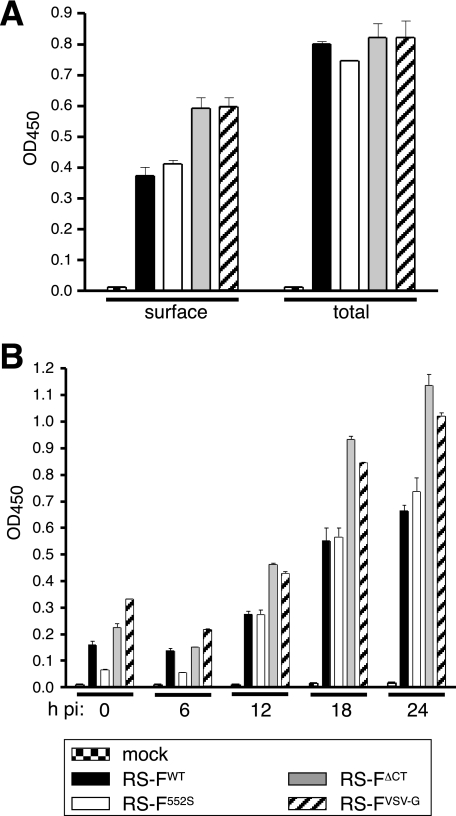
FIG. 5.
Cell surface expression of the modified F proteins. (A) Proportion of total F protein expressed at the cell surface. Infected Vero cells were fixed with paraformaldehyde at 24 h p.i. and permeabilized with Triton X-100 for total cell protein detection (total) or left untreated for surface detection (surface). F protein levels were determined by cell ELISA as previously described (35, 39), using the broadly reactive anti-F antibody MAb 19. Bars represent the average values of ELISA signals, normalized to the amount of N protein. Error bars indicate the range for duplicate samples. (B) Timing of F protein cell surface expression. Infected Vero cells were fixed at 0, 6, 12, 18, or 24 h p.i., and relative F protein surface levels were determined using cell ELISA as described above, without Triton permeabilization.
Growth analysis.
Vero and HEp-2 cells were infected in six-well plates at a multiplicity of 0.2 for 1.5 h at 37°C, washed once, and then incubated in 1.5 ml normal growth medium at 37°C. Immediately following infection and at days 1, 2, 3, 4, and 5 postinfection, 50 mM HEPES was added and cells were scraped into the medium and transferred to −80°C. The titers of the virus suspensions were determined in Vero cells by a 50% tissue culture infective dose (TCID50) assay based on GFP expression from the viral genome as previously described (38, 39).
Syncytium formation analysis.
Vero and HEp-2 cells were plated in six-well plates at approximately 35% confluence, infected at a multiplicity of 1 or 0.1 for 1.5 h, and then washed and incubated at 37°C. Expression of GFP and syncytium formation were monitored at daily intervals. At 48 and 72 h p.i., cells were fixed in 4% paraformaldehyde for 15 min, incubated with Hoechst stain for 10 min (to visualize nuclei), and photographed on a Nikon Eclipse TE-2000E2 inverted fluorescence microscope at a magnification of ×100.
Confocal microscopy.
Confocal analysis was done as previously described with minor modifications (38). Briefly, Vero cells were plated onto glass coverslips and incubated overnight. Cells were infected at a multiplicity of 2 and incubated at 37°C. At 26 h p.i., cells were fixed with freshly prepared 4% paraformaldehyde for 15 min, washed, and blocked with 2% bovine serum albumin, 0.1% cold water fish skin gelatin (Sigma) for 30 min at room temperature. Next, cells were incubated simultaneously with anti-F (humanized) and anti-G (mouse) antibodies, or with an anti-VSV G antibody, for 1 h at room temperature, followed by secondary anti-human or anti-mouse antibodies carrying fluorescent conjugates (Alexa-594 and -350; Molecular Probes). Samples were examined on a Leica TCS NT confocal microscope system and photographed at a magnification of ×600.
Lipid raft analysis.
Triton X-100 solubility was examined based on previously described protocols for HRSV, with modifications (24, 33). Vero cells were infected at a multiplicity of 5 and incubated at 37°C. Part of the samples were radiolabeled with Tran35S-label (MP Biochemicals, Inc) from 24 to 27 h p.i. as described above. At 27 h p.i., cells were washed once in TNE buffer (50 mM Tris, 150 mM NaCl, 2 mM EDTA). After washing, ice-cold TNE buffer containing 1% Triton X-100 (TNE-T) and protease inhibitors (Halt inhibitor cocktail; Pierce) were added to the cells. Cells were harvested, collected in Eppendorf tubes, and placed on ice for 30 min. Selected samples were incubated at 22°C or 37°C for 30 min. To separate the soluble and insoluble fractions, samples were spun for 10 min at 15,800 × g. at 4°C, and the supernatant was transferred to new tubes. For Western blot analysis, pellet and supernatant fractions were adjusted to equal volumes and analyzed by electrophoresis on 12% polyacrylamide gels under reducing conditions. Western blots were generated and incubated with anti-caveolin-1 and anti-transferrin receptor antibodies and developed using ECL (Bio-Rad). For radioimmunoprecipitation analysis, cold TNE-T buffer including protease inhibitors (a volume equal to that of the supernatant fraction) was added to the pellet. Next, an equal volume of cold IP buffer at 2× concentration was added to each fraction, and samples were immunoprecipitated as described above. After electrophoresis, using the conditions described in the legend to Fig. 2A, gels were dried and then analyzed using the STORM system and ImageQuant software package (Molecular Dynamics).
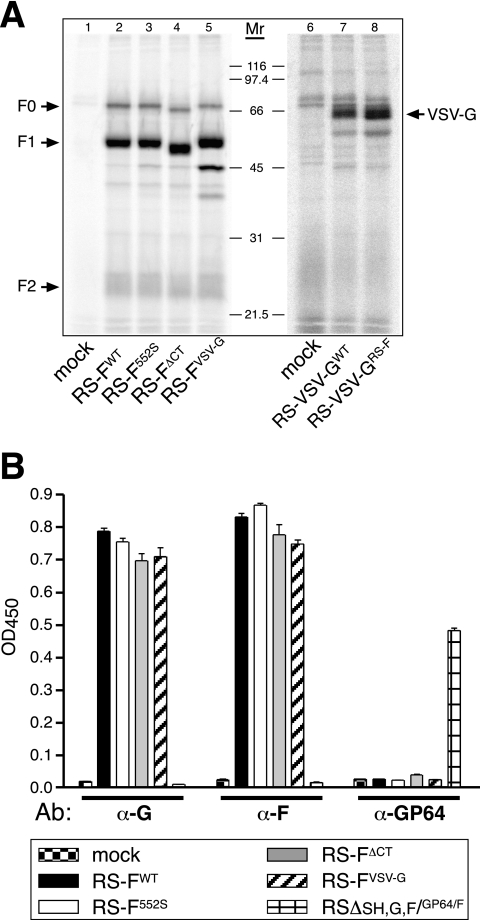
FIG. 2.
Transmembrane glycoprotein expression by the engineered HRSVs. (A) Immunoprecipitation analysis. Vero cells infected with the engineered viruses were exposed to [35S]methionine. Viral proteins were analyzed as described in Materials and Methods by immunoprecipitation with a combination of anti-F MAbs (MAb 19 and Synagis) or, in the case of VSV G-expressing viruses, with anti-VSV G antibody. Samples were electrophoresed on 12% polyacrylamide gels under reducing conditions. Molecular weights (in thousands) are indicated in between the panels. F0, uncleaved F protein precursor; F1 and F2, cleaved F protein subunits F1 and F2. (B) Cell ELISA analysis. Vero cells infected with viruses expressing modified F proteins were fixed at 25 h p.i. and analyzed by cell ELISA as previously described (39), using anti-F, anti-G, and anti-GP64 MAbs. A virus expressing the baculovirus GP64 protein (39) was included as a control for GP64 expression. Bars represent the average values of ELISA signals, normalized to the amount of N protein. Error bars indicate the range for duplicate samples.
RESULTS
Construction of cDNAs and recovery of engineered viruses.
To examine the importance of the F protein CT for HRSV replication in cell culture, viruses were recovered from engineered cDNAs that encoded COOH-terminally modified versions of the F protein. To facilitate modification of the F gene, an XbaI site was created at the TM domain/CT border region as described in Materials and Methods. Using the XbaI site, the sequence encoding the native F protein CT was deleted and replaced with a sequence encoding either a stop codon or amino acids 492 to 511 of the VSV G protein (Fig. 1A). These modifications resulted, respectively, in a tailless F protein (FΔCT) or an F protein carrying the VSV G protein CT in place of its own (FVSV-G). The creation of the XbaI site resulted in a change at amino acid 552 from alanine to serine. Therefore, an F construct containing only this alteration (F552S) (Fig. 1A) was included in all experiments to serve as a control for this change. Based on hydrophobicity profiles of the F protein, the predicted CT includes positively charged residues K551 and R553. However, because arginine- and lysine-rich sequences immediately following the TM domain can be part of translocation stop signals for membrane proteins (60), proteins FΔCT and FVSV-G were designed to retain the native residues K551 and R553 to prevent possible shedding into the endoplasmic reticulum lumen.
cDNAs representing the complete HRSV genome were engineered to contain the above-described modified F genes or a wt F gene (FWT), as shown in Fig. 1. The SH ORF, which is known to be dispensable for virus replication in cell culture (9), was replaced with the ORF of GFP to allow monitoring of virus spread. GFP expression from this location in the genome was previously shown to be an accurate indicator of infectivity that correlated with the number of PFU (38, 39).
Additional engineered viruses were recovered to test whether the F protein CT possessed any function independent of the F protein ectodomain and TM domain. These viruses lacked the SH, G, and F ORFs and expressed as their only transmembrane glycoprotein either the authentic VSV G protein (RS-VSV-GWT) or a VSV G protein carrying the HRSV F protein CT (amino acids 553 to 574) in place of its own (RS-VSV-GRS-F) (Fig. 1B). Chimeric protein VSV-GRS-F and virus RS-VSV-GRS-F were previously reported (38) (see Materials and Methods). The VSV G-containing viruses expressed GFP from the same genome location as the F-modified viruses shown in Fig. 1A and had the F ORF replaced with that of the β-glucuronidase marker gene (Fig. 1B).
All cDNAs described above maintained the same number of genes as observed in wt HRSV and retained wild-type intergenic junctions throughout in order to maintain authentic transcription levels. Infectious viruses were recovered from cDNAs as described previously (40). Importantly, because a functional F protein is required for virus propagation and our experimental viruses were potentially F compromised, two modifications to the standard recovery protocol were employed: (i) to passage virus from the cells transfected with cDNA and support plasmids, an additional plasmid transiently expressing a VSV G protein containing the HRSV F CT (38) was included in the initial transfection mixture, and (ii) viral supernatants were passaged onto Vbac cells, which express a baculovirus-derived viral membrane fusion protein, GP64/F (see the introduction) (40). Vbac cells were previously shown to allow the recovery of HRSVs lacking the F protein gene (40). Viruses recovered from all engineered cDNAs were designated as shown in Fig. 1. At passage 3, virus stocks were verified by reverse transcription-PCR across the modified areas followed by sequence analysis (data not shown). Because interactions between viral glycoprotein CTs and matrix proteins have been proposed for other negative-strand RNA viruses, we similarly verified the matrix gene sequence to ensure that no potential compensatory mutations had accrued (data not shown). No nucleotide changes were found.
Characterization of glycoprotein expression by the engineered HRSVs.
Expression of the modified F and VSV G proteins from the engineered viruses was analyzed in Vero cells. Infected cells were labeled with [35S]methionine from 20 to 24 h p.i. and then lysed and immunoprecipitated with a combination of two previously described anti-F MAbs (MAb 19 and Synagis) or, in the case of VSV G, with anti-VSV G antibody (Fig. 2). Similar amounts of both the uncleaved F0 form and the cleaved fragments F1 and F2 (21) were detected for all F-expressing viruses after SDS-PAGE (Fig. 2A, lanes 2 to 5). Thus, the modified F proteins were expressed at similar levels and were similarly processed. Two additional bands of unknown identity were observed after immunoprecipitation from cells infected with virus RS-FVSV-G (Fig. 2A, lane 5). The expression level of VSV-GRS-F was slightly higher than that of VSV-GWT, but no significant differences in molecular mass were detected, as both proteins migrated on SDS-PAGE as reported previously (29) (Fig. 2A, lanes 7 and 8).
In addition to immunoprecipitation, we used a cell ELISA as previously described (35, 39) to examine F and G protein expression levels simultaneously in the F-modified engineered viruses (Fig. 2B). An anti-GP64 antibody (AcV5) was used to measure the level of GP64 from input virus. As a positive control for GP64 expression, a previously reported virus that expresses the baculovirus GP64 protein (RSΔSH,G,F/GP64/F) (39) was included. All viruses expressed GFP as well as the HRSV N protein (not shown), which was used to normalize the expression levels of the F, G, and GP64 proteins (Fig. 2B). All F-modified viruses expressed similar levels of both the G and F proteins, in agreement with the genome content shown in Fig. 1A. GP64 was not detected in cells infected with the F-modified viruses, confirming our previous observations that GP64 from the input virus (which was propagated on Vbac cells) is most likely degraded after entry (40).
Impact of CT modification on infectious progeny production and membrane fusion.
Amplification of the CT-modified viruses in Vbac cells yielded stock titers of approximately 106 to 107 PFU/ml, similar to titers generally reported for wt HRSV. To examine the ability of each of the engineered viruses to produce infectious progeny in standard cell types, i.e., in the absence of GP64/F complementation, growth analyses were carried out in Vero and HEp-2 cells infected at a multiplicity of 0.2 (Fig. 3). Viral titers were determined by TCID50 based on the expression of GFP as described in Materials and Methods. In both Vero and HEp-2 cells, control virus RS-F552S replicated as efficiently as virus RS-FWT, showing that the XbaI restriction site added to generate the F changes did not affect F function or the overall replication of the virus. Deletion of the F protein CT (RS-FΔCT), however, reduced the production of infectious progeny by 100- to 1,000-fold compared to that by a virus expressing the wt F protein. Replacement of the F protein CT with that of the VSV G protein had an even larger impact, resulting in little or no production of infectious virus. In contrast, when the F protein CT was added to VSV G, the ability of the VSV G protein to mediate HRSV infectivity improved. This effect was most pronounced in Vero cells, showing an approximately 10- to 50-fold increase in infectious progeny production compared to virus RS-VSV-GWT, which expressed the authentic VSV G protein. Together these results indicate that the F protein CT is important for infectious progeny production and suggest that the CT contains functions or signals which can operate independently of the F protein ectodomain and TM domain.
FIG. 3.
Analysis of viral growth in cell culture. Vero and HEp-2 cells were infected with the engineered viruses at a multiplicity of 0.2 and incubated at 37°C. Immediately after virus adsorption and at 1-day intervals thereafter, cells were scraped into the supernatants and stored at −80°C until viral titers were determined by TCID50 as previously described (38). Data points represent the averages from duplicate titrations.
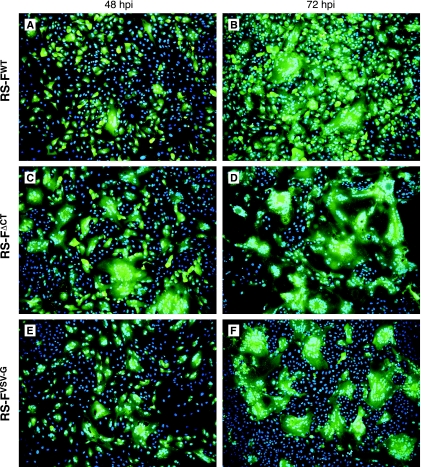
Observations made during the growth analyses suggested that cell-cell membrane fusion was affected by the F protein CT modifications. To compare the levels of cell-cell fusion induced by each of the F-modified viruses, Vero cells were infected at a multiplicity of 1 and syncytium formation was monitored for 72 h (Fig. 4). At 24 h p.i. equal numbers of individual cells expressing GFP were observed in each of the infected cell cultures, confirming equivalent multiplicities of infection (not shown). At this time, small syncytia (two to three nuclei) were observed occasionally only in cells infected with virus RS-FΔCT. By 48 h p.i. clear differences emerged in the level of virus-induced cell-cell fusion. In cells infected with virus RS-FWT (Fig. 4A) or virus RS-F552S (not shown), a small number of syncytia containing approximately five nuclei were present. The F-modified viruses RS-FΔCT and RS-FVSV-G induced syncytia that were both more abundant and larger (Fig. 4C and E). Particularly in the case of virus RS-FΔCT, the syncytia were large, containing approximately 10 to 40 nuclei each. By 72 h p.i., nearly the entire culture of RS-FΔCT-infected cells had fused to form massive syncytia (Fig. 4D). In the case of virus RS-FVSV-G, many large syncytia were observed, interspersed with uninfected cells (Fig. 4F). By comparison, virus RS-FWT had spread through the entire culture at 72 h p.i., displaying a mixture of single green cells as well as small and large syncytia (Fig. 4B). Similar results were obtained for Vero cells infected at lower multiplicity (0.1) and for HEp-2 cells (not shown). Thus, either deletion or replacement of the CT affected the extent of virus-induced cell-cell fusion.
FIG. 4.
F protein-mediated cell-cell fusion. Vero cells were infected with viruses RS-FWT (A and B), RS-FΔCT (C and D), or RS-FVSV-G (E and F) at a multiplicity of 1. Infected cells were incubated at 37°C, and GFP expression and syncytium formation were monitored. At 48 and 72 h p.i., infected cell cultures were fixed in 4% paraformaldehyde, incubated with Hoechst stain to visualize nuclei, and photographed on a fluorescence microscope (magnification, ×100) using green (GFP) and blue (nuclei) channels.
Steady-state cell surface levels of the modified F proteins.
One possible explanation for the enhanced cell-cell fusion was increased F expression at the cell surface. However, the data presented in Fig. 2 showed no differences in total F protein expression between the engineered viruses. To examine the relative levels of F at the cell surface, we compared surface and total cell F protein levels by cell ELISA. Infected Vero cells were briefly fixed with paraformaldehyde at 24 h p.i., permeabilized with Triton X-100 or left untreated, and then incubated with a broadly reactive anti-F antibody (Fig. 5A). Previous immunofluorescence observations showed that this fixation regimen, in the absence of permeabilization, detects only F protein at or near the cell surface, while permeabilization with Triton X-100 allows for recognition of both surface and intracellular F protein. F protein levels were normalized to the level of N protein expression in permeabilized samples (not shown). While total F protein levels were similar for the different viruses, both in the absence of the CT and when the CT was replaced with that of the VSV G protein, cell surface levels of F were elevated (Fig. 5A). Thus, absence of the CT increased the proportion of F protein expressed at the cell surface.
To determine whether the timing of F surface expression was affected, infected cells were examined for F protein surface expression as described above, at 6-h intervals (Fig. 5B). At 12, 18, and 24 h p.i., surface levels of FΔCT and FVSV-G were elevated relative to those of wt F protein, but no significant differences in timing were detected. Thus, manipulation of the CT affected the relative cell surface levels but not the timing of F surface expression.
Deletion or replacement of the CT alters F protein surface distribution.
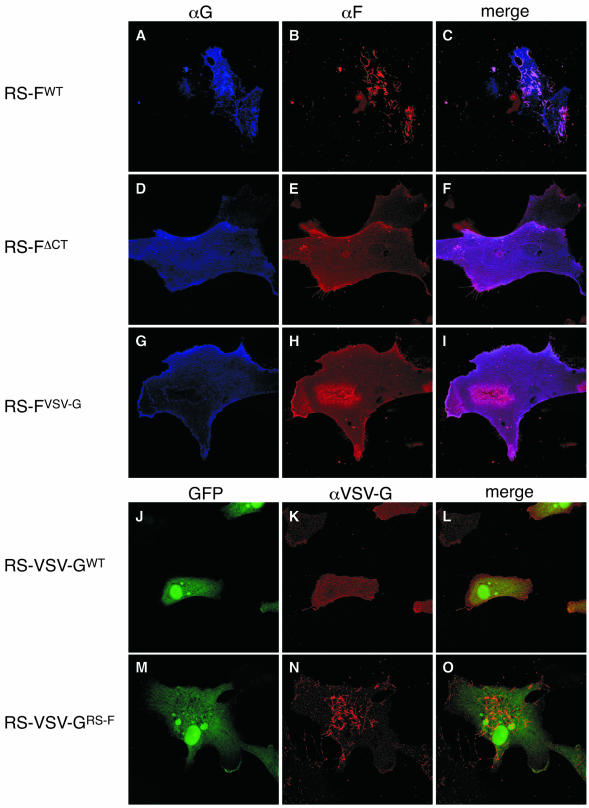
Because the F proteins expressed from viruses RS-FΔCT and RS-FVSV-G were present at the cell surface and were fusion competent yet largely failed to support infectious progeny production, we examined the surface expression of the various F proteins by using confocal microscopy (Fig. 6). Infected Vero cells were fixed with paraformaldehyde at 26 h p.i. and incubated simultaneously with anti-F (humanized) and anti-G (mouse origin) antibodies. Anti-F and anti-G antibodies were detected with secondary anti-human and anti-mouse antibodies carrying fluorescent conjugates, as described in Materials and Methods. In agreement with our previous observations, a virus expressing wt G and F proteins (virus RS-FWT) displayed a G protein that is expressed across the entire cell surface but is also present in virus-induced cell surface filaments (Fig. 6A), where it colocalized with the F protein (Fig. 6C). In contrast to G, the wt F protein was observed almost exclusively in association with filamentous structures (Fig. 6B). No differences were observed between viruses RS-FWT and RS-F552S (not shown). However, deletion or replacement of the CT dramatically altered the plasma membrane distribution of the F protein: proteins FΔCT and FVSV-G were evenly spread across the plasma membrane relative to the wt F protein (Fig. 6E and H). In addition, filamentous structures were rarely observed in the absence of the F protein CT. No filaments were detected by incubation with anti-G or anti-F antibodies in samples infected with virus RS-FVSV-G. We also noted that in the absence of the F protein CT, no G protein was found localized to filamentous structures (Fig. 6D and G). At this time the mechanism by which G protein distribution may have been affected is unclear.
FIG. 6.
Plasma membrane distribution of the modified F proteins. Infected Vero cells were fixed with freshly dissolved paraformaldehyde at 26 h p.i. Cells were then incubated simultaneously with anti-F (Synagis, humanized) and anti-G (L9, mouse origin) antibodies, followed by secondary anti-human and anti-mouse antibodies carrying red and blue fluorescent conjugates, respectively. Samples of the F protein-expressing viruses were scanned sequentially for GFP expression (not shown), F expression (B, E, and H), and G expression (A, D, and G). Samples of the VSV G protein-expressing viruses were examined by confocal microscopy sequentially for GFP expression (J and M) and VSV G expression (K and N). For each virus, the left and middle panels were overlaid (merge; panels C, F, I, L, and O). Magnifications, ×600.
We previously reported that a VSV G protein carrying the F protein CT localized predominantly to virus-induced filamentous structures at the cell surface (38). Examination of cells infected with virus RS-VSV-GRS-F (Fig. 6N) confirmed these prior observations. In contrast, the authentic VSV G protein expressed from virus RS-VSV-GWT did not localize to filaments but instead was distributed evenly across the plasma membrane (Fig. 6K). Thus, having the VSV G protein CT replaced with the CT of the HRSV F protein changed the cell surface distribution of the heterologous VSV G membrane protein from even to predominantly localized to filamentous structures.
A role for the CT in F protein-lipid raft association.
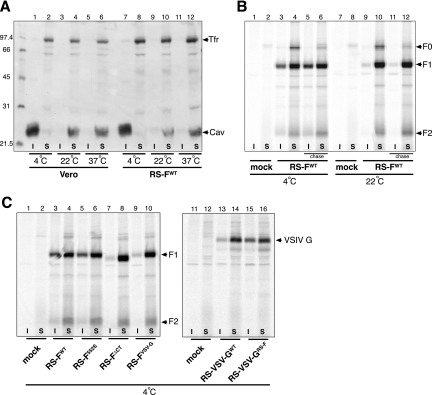
Given the reported interaction between F protein and lipid rafts (24) and the altered distribution of proteins FΔCT and FVSV-G compared to wt F protein (Fig. 6), we asked whether the CT may play a role in targeting of F protein to lipid rafts. To compare lipid raft targeting in the presence or absence of the F protein CT, Triton X-100 solubilization conditions were established using previously described methods (24, 31), with the use of a known lipid raft and non-lipid raft marker, caveolin-1 and transferrin receptor, respectively (Fig. 7A). Uninfected or RS-FWT-infected Vero cells were lysed in Triton X-100-containing buffer at 27 h p.i. at 4, 22, or 37°C. Different temperatures were examined because resistance to solubilization in Triton X-100 is a temperature-dependent phenomenon, occurring at 4°C but not at elevated temperatures (5, 51). After Triton solubilization at the various temperatures, samples were spun to separate soluble and insoluble fractions, as described in Materials and Methods. Pellets and supernatants were adjusted to equal volumes and analyzed by SDS-PAGE under reducing conditions. Western blots were generated and incubated with anti-caveolin-1 and -transferrin receptor antibodies and developed using ECL. Consistent with the literature, caveolin-1 was insoluble in Triton X-100 at 4°C, and this was apparent in both uninfected and HRSV-infected cells (Fig, 7A, lanes 1 and 7). In contrast, transferrin receptor, a nonraft marker, was soluble under the same conditions (Fig. 7A, lanes 2 and 8). As predicted, Triton extraction at 22 and 37°C resulted in a shift from the insoluble to the soluble fraction for caveolin-1 (Fig. 7A, lanes 4, 6, 10, and 12).
FIG. 7.
Effect of CT modification on F protein-lipid raft association. (A) Confirmation of Triton X-100 solubilization conditions. Vero cells, either uninfected (lanes 1 to 6) or HRSV infected (lanes 7 to 12), were solubilized in Triton X-100-containing buffer at 4°C, 22°C, or 37°C. Next, pellet and supernatant fractions were separated by centrifugation for 10 min at 15,800 × g, adjusted to equal volumes, and analyzed by SDS-PAGE under reducing conditions. Western blots were generated and incubated with anti-caveolin-1 (Cav) and anti-transferrin receptor (Tfr) antibodies and developed using ECL. Molecular weight markers (in thousands) are indicated on the left. I, Triton-insoluble fraction; S, Triton-soluble fraction. (B) Triton X-100 solubility of wt F protein. RS-FWT- or mock-infected Vero cells were metabolically labeled with [35S]methionine from 24 to 27 h p.i. A subset of samples (indicated as chase; lanes 5, 6, 11, and 12) were washed and incubated for an additional 1 h in standard growth medium. All samples were detergent extracted as described above at either 4 or 22°C (lanes 1 to 6 and lanes 7 to 12, respectively). Pellet and supernatant fractions were adjusted to equal volumes and then lysed, immunoprecipitated, and electrophoresed using the conditions described in the legend to Fig. 2A. Dried gels were scanned and analyzed using the STORM system and the ImageQuant software package (Molecular Dynamics). F0, uncleaved F protein precursor; F1 and F2, cleaved F protein subunits F1 and F2. (C) Effect of CT modifications on Triton X-100 solubility of F and VSV G proteins. Vero cells were infected with viruses RS-FWT (lanes 3 and 4), RS-F552S (lanes 5 and 6), RS-FΔCT (lanes 7 and 8), RS-FVSV-G (lanes 9 and 10), RS-VSV-GWT (lanes 13 and 14), or RS-VSV-GRS-F (lanes 15 and 16), and Triton solubility was examined at 4°C as indicated for the chase samples in Fig. 7B.
Using the detergent solubilization conditions established above, we investigated the association of wt F protein with lipid rafts (Fig. 7B). For optimal visualization of F, radioimmunoprecipitation was used in place of Western blotting. Cells infected with virus RS-FWT were metabolically labeled with [35S]methionine from 24 to 27 h p.i. A subset of samples (indicated as chase) were washed and incubated for an additional 1 h in standard growth medium. All samples were detergent extracted as described above at either 4 or 22°C. Next, pellet and supernatant fractions were immunoprecipitated and electrophoresed using the conditions described in the legend to Fig. 2A. Dried gels were analyzed using the STORM system and the ImageQuant software package (Molecular Dynamics). Consistent with previous reports, a proportion of the wt F protein was resistant to Triton extraction at 4°C, as evidenced by the presence of the F1 and F2 subunits in the insoluble fraction (Fig. 7B, lane 3). When radiolabel was replaced with standard growth medium and cells incubated for an additional 1 h at 37°C (chase), the labeled uncleaved precursor F0 was detected at very low levels (presumably due to cleavage into its subunits), and the proportion of Triton-resistant F protein had increased (Fig. 7B, lanes 5 and 6). In contrast, at 22°C the amount of wt F protein that was Triton resistant was negligible (Fig. 7B, lanes 9 and 11), in agreement with the temperature-dependent nature of resistance to solubility in Triton X-100. To examine Triton X-100 solubility in the absence of the CT, cells were infected with the various engineered viruses and processed like the chase samples described above, at 4°C (Fig. 7C). At this temperature, approximately 30% of the FWT and F552S proteins was found associated with the Triton-insoluble fraction. In contrast, in the absence of a CT (FΔCT) or when the F CT was replaced with the CT of VSV G (FVSV-G), only approximately 1.5% or 2%, respectively, of the total F protein was Triton resistant (Fig. 7C, lanes 7 and 9). Moreover, when the F CT was used to replace the CT of the VSV G protein (VSV-GRS-F), the proportion of Triton-resistant VSV G protein increased from approximately 7.5% to 34% (Fig. 7C, lanes 13 and 15). Together, these results indicate that the homologous CT is required for association of F protein with Triton-insoluble lipid rafts and suggest that the F protein CT contains a lipid raft sorting signal that is functional in the absence of the F ectodomain and TM domain.
DISCUSSION
Cytoplasmic tails of viral attachment or membrane fusion proteins have been implicated in a range of assembly-related processes (see the introduction) and have been shown in many cases to modulate virus-induced membrane fusion (15, 36, 42, 49, 58, 61, 62). For measles virus, absence of the F protein CT resulted in enhanced cell-cell fusion but had only a moderate impact on virus propagation in Vero cells (10). For another paramyxovirus, SV5, the F protein CT was reported to be dispensable for normal virus replication and budding (48). In the work presented here we found that either deletion or replacement of the HRSV F protein CT reduced the amount of infectious progeny after low-multiplicity infection by approximately 100- to 1,000-fold in different cell types, while the F protein remained abundant and fusion competent but displayed an altered distribution at the plasma membrane.
Impact of CT modification on membrane fusion.
Modulation of membrane fusion capacity is one possible mechanism by which transmembrane glycoprotein CTs can affect or regulate the ability of a virus to spread to neighboring cells. Experimental CT truncations have been reported for a number of viruses. In most cases of COOH-terminal truncation of F proteins, surface expression is not or is only minimally affected but the truncations differ in their impact on cell-cell fusion. For example, cell-cell fusion levels were unaffected or enhanced, respectively, after CT truncation of the F proteins of human parainfluenza virus type 2 and measles virus (10, 65). For some retro- and paramyxoviruses, truncation of the Env or F protein CT both enhanced fusion and induced conformational changes, indicating an ability of the CT to modulate cell-cell fusion by influencing ectodomain conformation (16, 50, 53, 61). In the case of SV5, the F protein CT was hypothesized to affect the energy threshold required to initiate fusion via conformational changes in the ectodomain (61). Using a broadly reactive anti-F antibody, we found that modifications of the HRSV F protein CT increased the level of syncytium formation as well as the proportion of F protein expressed at the cell surface. As higher F surface levels may explain the increase in syncytium formation, it remains unclear whether a mechanism such as the one proposed for SV5 applies. It is possible, however, that in addition to its impact on protein surface levels, absence of the CT may affect the ability of the HRSV F protein to remain in the proposed prefusion or metastable energy state, resulting in extensive but premature membrane fusion.
Previous work, in which a 26-amino-acid deletion of the HRSV F protein COOH terminus was studied outside the viral context, showed a smaller increase in surface expression and syncytium formation than observed here (4). This may indicate that additional HRSV factors affect the level of F expression and fusion function. Alternatively, a proportion of the F protein lacking the 26 COOH-terminal amino acids may have shed into the endoplasmic reticulum lumen, as no basic residues were left at the TM domain/CT border region and a cysteine residue known to be palmitylated (C550) was absent (1, 4). Those residues were retained in the constructs reported here.
How does F protein CT modification affect the level of infectious progeny?
Either deletion or replacement of the HRSV F protein CT severely decreased HRSV growth in cell culture. This argues that the CTs of the HRSV fusion and attachment proteins (F and G) are not dually redundant, as has been proposed for the CTs of the fusion and attachment proteins of paramyxovirus SV5, as well as for the CTs of influenza virus hemagglutinin and neuraminidase proteins (28, 62).
One explanation for the strong effect of F protein CT deletion on HRSV progeny production may be premature fusion in the absence of the CT, as this might prevent the virus from assembling virions containing F protein in the proper energy state. Additional explanations include interaction and/or targeting defects. By analogy to other enveloped viruses, the CT may mediate interactions between F protein and other viral components during assembly. For example, in the case of the measles virus F and human immunodeficiency virus type 1 Env proteins, the CT is thought to interact with the viral matrix protein (37, 52). It was recently reported that the HRSV G protein interacts with the matrix protein via its CT (19), but an interaction between matrix and F proteins has not been reported. The CT may also contain targeting signals that direct the F protein to the proper subcellular location, presumably sites where a stage of HRSV assembly occurs. Although, in the absence of viral infection, deletion of CT amino acids A552 to N574 was previously reported to have no effect on F protein distribution (4a), we found a marked change in plasma membrane distribution and lipid raft association of the HRSV F protein upon CT modification (Fig. 6 and 7), supporting a targeting function for this domain. No canonical targeting motifs are present in the CT, but residues 561 to 564 (KDQL) (Fig. 1A) bear similarity to the previously described endoplasmic reticulum retention signal KDEL.
HRSV virion morphology, F-lipid raft interaction, and the role of the CT.
Previous work has shown that the F protein concentrates in HRSV-induced cell surface filaments and interacts with lipid rafts (see the introduction). In addition, based on a reported reduction in number and length of filaments after chemical disruption of rafts, it was suggested that lipid rafts are required for the formation or maintenance of these filaments (20, 33). Because modification of the CT altered F distribution from filament associated to even (Fig. 6), we asked whether the F protein CT might also affect F protein-lipid raft interaction. Our experiments showed that the F protein CT is required for both localization of F protein to filaments and association of F protein with lipid rafts (Fig. 7). A role for the F protein CT in raft interaction is consistent with a recent paper in which mass spectrometry was used to predict protein domains that may be important for HRSV-raft interactions. Five peptides that may be raft-interacting domains were identified within the F protein, one of which was comprised of the COOH-terminal 13 amino acids of the F protein CT (34).
Combined with previous reports, our findings suggest that the interaction between F protein and lipid rafts is a critical step, required for F incorporation into filaments. Whether this is a direct or indirect interaction remains to be established; However, F protein-lipid raft interaction does not require additional HRSV proteins (24). It is not clear whether filaments are truly absent after F protein CT deletion or are no longer visualized due to absence of the F protein from filaments. Thus, it is possible that F-lipid raft interaction is also required for the formation or maintenance of cell surface filaments.
The CT provides an independent function.
Providing the COOH-terminal 22 amino acids of the HRSV F protein to VSV G had a significant effect on replication of RS-VSV-GRS-F, a virus engineered to express VSV-G as its only glycoprotein (38). In both Vero and HEp-2 cells, virus RS-VSV-GRS-F replicated to higher titers than virus RS-VSV-GWT. Moreover, adding the F protein CT to the VSV G protein changed its plasma membrane distribution from predominantly even to predominantly filament associated (Fig. 6). The exact mechanism by which the F protein CT imparts these changes is not yet clear. However, lipid rafts may be involved, as the proportion of the VSV G protein associated with Triton-insoluble rafts increased when its homologous CT was replaced with that of the HRSV F protein (Fig. 7). The results observed with the VSV G protein-expressing viruses further suggest that the mechanisms underlying the function(s) provided by the CT can operate independently from the F protein TM domain and ectodomain.
Conclusion.
For some members of the Paramyxoviridae, CT deletions of a single glycoprotein have shown only a moderate impact on infectious virus production and overall virus viability. In some cases this may be explained by a redundancy in function of the attachment and fusion protein CT, as proposed for paramyxovirus SV5 and influenza virus (see above). In contrast to the case for SV5, the cytoplasmic domains of the HRSV F and G protein do not appear to be dually redundant, as our data show that HRSV replication is critically dependent upon the F protein CT.
The critical nature of the F protein CT may represent another feature that sets apart HRSV and/or the pneumovirus genus from the other paramyxoviruses. It is important, however, to keep in mind that the engineered viruses recovered here were amplified in a cell line expressing a complementing heterologous viral protein (GP64/F) previously shown to be highly efficient in mediating HRSV infectivity (40). As a result, selection pressure on the CT-modified viruses to accrue compensatory mutations should have been alleviated (no nucleotide changes were found in the F or matrix gene at the passage used for this study, passage 3). It is possible that in some previous reports concerning CT deletions, the consequences of these deletions may have been moderated due to compensating mutations occurring during recovery and amplification of viruses.
The most likely mechanisms to explain the high impact of F protein CT modification on the amount of HRSV infectious progeny are defective F protein targeting and/or F protein conformational changes that may lead to premature membrane fusion. The latter we have not yet investigated, while the former is supported by confocal microscopy and lipid raft association analysis (Fig. 6 and 7). In either case, we can conclude that the F protein CT plays a role that involves Triton-insoluble lipid rafts and that determines both the distribution of the F protein in an infected cell and the efficiency with which infectious progeny are produced.
Acknowledgments
We thank the members of the Wertz laboratory for helpful discussions during the preparation of the manuscript.
This work was supported by Public Health Service grant AI20181 to G.W.W. from the National Institutes of Health.
Footnotes
Published ahead of print on 23 August 2006.
REFERENCES
- 1.Arumugham, R. G., R. C. Seid, Jr., S. Doyle, S. W. Hildreth, and P. R. Paradiso. 1989. Fatty acid acylation of the fusion glycoprotein of human respiratory syncytial virus. J. Biol. Chem. 264:10339-10342. [PubMed] [Google Scholar]
- 2.Bachi, T., and C. Howe. 1973. Morphogenesis and ultrastructure of respiratory syncytial virus. J. Virol. 12:1173-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branigan, P. J., N. D. Day, C. Liu, L. L. Gutshall, J. A. Melero, R. T. Sarisky, and A. M. Del Vecchio. 2006. The cytoplasmic domain of the F protein of human respiratory syncytial virus is not required for cell fusion. J. Gen. Virol. 87:395-398. [DOI] [PubMed] [Google Scholar]
- 4a.Brock, S. C., J. M. Heck, P. A. McGraw, and J. E. Crowe, Jr. 2005. The transmembrane domain of the respiratory syncytial virus F protein is an orientation-independent apical plasma membrane sorting sequence. J. Virol. 79:12528-12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. A., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111-136. [DOI] [PubMed] [Google Scholar]
- 6.Brown, G., J. Aitken, H. W. Rixon, and R. J. Sugrue. 2002. Caveolin-1 is incorporated into mature respiratory syncytial virus particles during virus assembly on the surface of virus-infected cells. J. Gen. Virol. 83:611-621. [DOI] [PubMed] [Google Scholar]
- 7.Brown, G., C. E. Jeffree, T. McDonald, H. W. Rixon, J. D. Aitken, and R. J. Sugrue. 2004. Analysis of the interaction between respiratory syncytial virus and lipid-rafts in Hep2 cells during infection. Virology 327:175-185. [DOI] [PubMed] [Google Scholar]
- 8.Brown, G., H. W. Rixon, and R. J. Sugrue. 2002. Respiratory syncytial virus assembly occurs in GM1-rich regions of the host-cell membrane and alters the cellular distribution of tyrosine phosphorylated caveolin-1. J. Gen. Virol. 83:1841-1850. [DOI] [PubMed] [Google Scholar]
- 9.Bukreyev, A., S. S. Whitehead, B. R. Murphy, and P. L. Collins. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, P. L., Y. T. Huang, and G. W. Wertz. 1984. Identification of a tenth mRNA of respiratory syncytial virus and assignment of polypeptides to the 10 viral genes. J. Virol. 49:572-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, P. L., K. McIntosh, and R. M. Chanock. 1996. Respiratory syncytial virus. Lippincott-Raven, Philadelphia, Pa.
- 13.Couch, R. B., J. A. Englund, and E. Whimbey. 1997. Respiratory viral infections in immunocompetent and immunocompromised persons. Am. J. Med. 102:2-9, 25-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutch, R. E., and R. A. Lamb. 2001. Deletion of the cytoplasmic tail of the fusion protein of the paramyxovirus simian virus 5 affects fusion pore enlargement. J. Virol. 75:5363-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fouillot-Coriou, N., and L. Roux. 2000. Structure-function analysis of the Sendai virus F and HN cytoplasmic domain: different role for the two proteins in the production of virus particle. Virology 270:464-475. [DOI] [PubMed] [Google Scholar]
- 18.Fuchs, H., and T. Bachi. 1975. Scanning electron microscopical demonstration of respiratory syncytial virus antigens by immunological markers. J. Ultrastruct. Res. 52:114-119. [DOI] [PubMed] [Google Scholar]
- 19.Ghildyal, R., D. Li, I. Peroulis, B. Shields, P. G. Bardin, M. N. Teng, P. L. Collins, J. Meanger, and J. Mills. 2005. Interaction between the respiratory syncytial virus G glycoprotein cytoplasmic domain and the matrix protein. J. Gen. Virol. 86:1879-1884. [DOI] [PubMed] [Google Scholar]
- 20.Gower, T. L., M. K. Pastey, M. E. Peeples, P. L. Collins, L. H. McCurdy, T. K. Hart, A. Guth, T. R. Johnson, and B. S. Graham. 2005. RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J. Virol. 79:5326-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruber, C., and S. Levine. 1983. Respiratory syncytial virus polypeptides. III. The envelope-associated proteins. J. Gen. Virol. 64:825-832. [DOI] [PubMed] [Google Scholar]
- 22.Han, L. L., J. P. Alexander, and L. J. Anderson. 1999. Respiratory syncytial virus pneumonia among the elderly: an assessment of disease burden. J. Infect. Dis. 179:25-30. [DOI] [PubMed] [Google Scholar]
- 23.Heminway, B. R., Y. Yu, Y. Tanaka, K. G. Perrine, E. Gustafson, J. M. Bernstein, and M. S. Galinski. 1994. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology 200:801-805. [DOI] [PubMed] [Google Scholar]
- 24.Henderson, G., J. Murray, and R. Yeo. 2002. Sorting of the respiratory syncytial virus matrix protein into detergent-resistant structures is dependent on cell-surface expression of the glycoproteins. Virology 300:244. [DOI] [PubMed] [Google Scholar]
- 25.Huang, Y. T., P. L. Collins, and G. W. Wertz. 1985. Characterization of the 10 proteins of human respiratory syncytial virus: identification of a fourth envelope-associated protein. Virus Res. 2:157-173. [DOI] [PubMed] [Google Scholar]
- 26.Iwatsuki-Horimoto, K., T. Horimoto, T. Noda, M. Kiso, J. Maeda, S. Watanabe, Y. Muramoto, K. Fujii, and Y. Kawaoka. 2006. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 80:5233-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffree, C. E., H. W. Rixon, G. Brown, J. Aitken, and R. J. Sugrue. 2003. Distribution of the attachment (G) glycoprotein and GM1 within the envelope of mature respiratory syncytial virus filaments revealed using field emission scanning electron microscopy. Virology 306:254-267. [DOI] [PubMed] [Google Scholar]
- 28.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knipe, D., J. K. Rose, and H. F. Lodish. 1975. Translation of individual species of vesicular stomatitis viral mRNA. J. Virol. 15:1004-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leser, G. P., and R. A. Lamb. 2005. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342:215-227. [DOI] [PubMed] [Google Scholar]
- 31.Manie, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marty, A., J. Meanger, J. Mills, B. Shields, and R. Ghildyal. 2004. Association of matrix protein of respiratory syncytial virus with the host cell membrane of infected cells. Arch. Virol. 149:199-210. [DOI] [PubMed] [Google Scholar]
- 33.McCurdy, L. H., and B. S. Graham. 2003. Role of plasma membrane lipid microdomains in respiratory syncytial virus filament formation. J. Virol. 77:1747-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald, T. P., A. R. Pitt, G. Brown, H. W. Rixon, and R. J. Sugrue. 2004. Evidence that the respiratory syncytial virus polymerase complex associates with lipid rafts in virus-infected cells: a proteomic analysis. Virology 330:147-157. [DOI] [PubMed] [Google Scholar]
- 35.Monsma, S. A., and G. W. Blissard. 1995. Identification of a membrane fusion domain and an oligomerization domain in the baculovirus GP64 envelope fusion protein. J. Virol. 69:2583-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulligan, M. J., G. V. Yamshchikov, G. D. Ritter, Jr., F. Gao, M. J. Jin, C. D. Nail, C. P. Spies, B. H. Hahn, and R. W. Compans. 1992. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J. Virol. 66:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oomens, A. G., A. G. Megaw, and G. W. Wertz. 2003. Infectivity of a human respiratory syncytial virus lacking the SH, G, and F proteins is efficiently mediated by the vesicular stomatitis virus G protein. J. Virol. 77:3785-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oomens, A. G., and G. W. Wertz. 2004. The baculovirus GP64 protein mediates highly stable infectivity of a human respiratory syncytial virus lacking its homologous transmembrane glycoproteins. J. Virol. 78:124-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oomens, A. G., and G. W. Wertz. 2004. trans-complementation allows recovery of human respiratory syncytial viruses that are infectious but deficient in cell-to-cell transmission. J. Virol. 78:9064-9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pringle, C. R. 1987. Progress towards control of the acute respiratory viral diseases of childhood. Bull. W. H. O. 65:133-137. [PMC free article] [PubMed] [Google Scholar]
- 42.Ritter, G. D., Jr., M. J. Mulligan, S. L. Lydy, and R. W. Compans. 1993. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology 197:255-264. [DOI] [PubMed] [Google Scholar]
- 43.Rixon, H. W., G. Brown, J. Aitken, T. McDonald, S. Graham, and R. J. Sugrue. 2004. The small hydrophobic (SH) protein accumulates within lipid-raft structures of the Golgi complex during respiratory syncytial virus infection. J. Gen. Virol. 85:1153-1165. [DOI] [PubMed] [Google Scholar]
- 44.Roberts, S. R., D. Lichtenstein, L. A. Ball, and G. W. Wertz. 1994. The membrane-associated and secreted forms of the respiratory syncytial virus attachment glycoprotein G are synthesized from alternative initiation codons. J. Virol. 68:4538-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheiffele, P., A. Rietveld, T. Wilk, and K. Simons. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274:2038-2044. [DOI] [PubMed] [Google Scholar]
- 46.Schlender, J., G. Zimmer, G. Herrler, and K. K. Conzelmann. 2003. Respiratory syncytial virus (RSV) fusion protein subunit F2, not attachment protein G, determines the specificity of RSV infection. J. Virol. 77:4609-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt, A. P., and R. A. Lamb. 2004. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:145-196. [DOI] [PubMed] [Google Scholar]
- 48.Schmitt, A. P., G. P. Leser, D. L. Waning, and R. A. Lamb. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sergel, T., and T. G. Morrison. 1995. Mutations in the cytoplasmic domain of the fusion glycoprotein of Newcastle disease virus depress syncytia formation. Virology 210:264-272. [DOI] [PubMed] [Google Scholar]
- 50.Seth, S., A. Vincent, and R. W. Compans. 2003. Activation of fusion by the SER virus F protein: a low-pH-dependent paramyxovirus entry process. J. Virol. 77:6520-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 52.Spielhofer, P., T. Bachi, T. Fehr, G. Christiansen, R. Cattaneo, K. Kaelin, M. A. Billeter, and H. Y. Naim. 1998. Chimeric measles viruses with a foreign envelope. J. Virol. 72:2150-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spies, C. P., G. D. Ritter, Jr., M. J. Mulligan, and R. W. Compans. 1994. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J. Virol. 68:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stope, M. B., A. Karger, U. Schmidt, and U. J. Buchholz. 2001. Chimeric bovine respiratory syncytial virus with attachment and fusion glycoproteins replaced by bovine parainfluenza virus type 3 hemagglutinin-neuraminidase and fusion proteins. J. Virol. 75:9367-9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takimoto, T., T. Bousse, E. C. Coronel, R. A. Scroggs, and A. Portner. 1998. Cytoplasmic domain of Sendai virus HN protein contains a specific sequence required for its incorporation into virions. J. Virol. 72:9747-9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takimoto, T., and A. Portner. 2004. Molecular mechanism of paramyxovirus budding. Virus Res. 106:133-145. [DOI] [PubMed] [Google Scholar]
- 57.Teng, M. N., S. S. Whitehead, and P. L. Collins. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289:283-296. [DOI] [PubMed] [Google Scholar]
- 58.Tong, S., M. Li, A. Vincent, R. W. Compans, E. Fritsch, R. Beier, C. Klenk, M. Ohuchi, and H. D. Klenk. 2002. Regulation of fusion activity by the cytoplasmic domain of a paramyxovirus F protein. Virology 301:322-333. [DOI] [PubMed] [Google Scholar]
- 59.Vincent, S., D. Gerlier, and S. N. Manie. 2000. Measles virus assembly within membrane rafts. J. Virol. 74:9911-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Heijne, G., and Y. Gavel. 1988. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174:671-678. [DOI] [PubMed] [Google Scholar]
- 61.Waning, D. L., C. J. Russell, T. S. Jardetzky, and R. A. Lamb. 2004. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc. Natl. Acad. Sci. USA 101:9217-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waning, D. L., A. P. Schmitt, G. P. Leser, and R. A. Lamb. 2002. Roles for the cytoplasmic tails of the fusion and hemagglutinin-neuraminidase proteins in budding of the paramyxovirus simian virus 5. J. Virol. 76:9284-9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wertz, G. W., P. L. Collins, Y. Huang, C. Gruber, S. Levine, and L. A. Ball. 1985. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. USA 82:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wertz, G. W., M. Krieger, and L. A. Ball. 1989. Structure and cell surface maturation of the attachment glycoprotein of human respiratory syncytial virus in a cell line deficient in O glycosylation. J. Virol. 63:4767-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao, Q., and R. W. Compans. 1995. Differences in the role of the cytoplasmic domain of human parainfluenza virus fusion proteins. J. Virol. 69:7045-7053. [DOI] [PMC free article] [PubMed] [Google Scholar]