Abstract
Human immunodeficiency virus type 1 can generally use CCR3 and CCR5 for cell entry. We show that envelopes with novel phenotypes arise during “coreceptor switch”: one loses the ability to use CCR3 (R5-only phenotype), and another gains use of CXCR4 in addition to CCR5 and CCR3 (R3/R5/X4-using phenotype). The envelope determinants for CCR3 use mapped to three amino acids. One, N356 in conserved region 3, is a potential glycosylation site and has not previously been associated with coreceptor use. The other two, R440 and N448 in conserved region 4, are proximal to but distinct from residues already identified as being important for CCR5 binding.
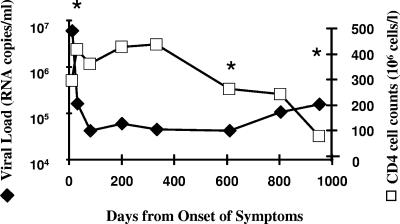
We have previously described that efficient CCR3 use is a general feature of human immunodeficiency virus type 1 (HIV-1) envelopes (Envs) from uncultured viruses (1). Most Envs from patient blood could use CCR3 as well as CCR5 (R3/R5). Potential targets for CCR3-mediated infection include microglial cells, macrophages, eosinophils, and T-helper 2 cells. Here the dynamics of coreceptor use, from acute infection to disease progression, was tested with one patient. Figure 1 shows a sharp decline in CD4 cell numbers and an elevation in viral load between days 608 and 957. Since this is characteristic of R5/X4 “coreceptor switch,” we determined coreceptor use by the prevailing quasispecies. Envs were cloned by reverse transcription-PCR and inserted into a replication-competent HIV-1 vector, and viruses were produced by transfection of 293T cells (1, 2). Coreceptor use on NP2/CD4 and U87/CD4 cells expressing various receptors was determined. Infection (focus-forming units [FFU])/ml) was detected by p24 immunostaining after 48 h of culture (1). As expected, Envs from acute (day 12 to 32) and asymptomatic (day 608) infection were R3/R5-tropic. Indeed, the efficiency of CCR3 use was similar to that of CCR5 use in the majority of Envs (Table 1). We have previously shown that the high level of CCR3 use observed is not an artifact of our system (1). Little or no use of CCR1, CCR2b, CCR8, and APJ was seen (data not shown). Envs isolated from day 957 revealed two different receptor phenotypes (Table 1). Both types of Env maintained CCR5 use, but two lost the use of CCR3 (8.9.I and 8.9.K). The other maintained CCR3 use but additionally gained CXCR4 use (8.9.B, 8.9.D, 8.9.H, and 8.9.J). Gain of CXCR4 use with time occurs in about 50% of HIV-1 patients, but as far as we are aware, the phenotypic switch (from R3/R5 to R5 only or to R3/R5/X4) described here has not been previously reported (5, 13, 14).
FIG. 1.
Virus load and CD4 cell counts over time. The patient's virus load (Chiron [Emeryville, Calif.] 3.0) (closed symbols) and CD4 cell numbers (open symbols) are plotted against the time in days from onset of symptoms characteristic of acute HIV infection. The time points from which HIV Envs were cloned are indicated with asterisks.
TABLE 1.
Virus coreceptor use and sensitivity to neutralization by autologous sera
| Virus clonea | Isolation dayb | Coreceptor use
|
IC90 at time (days) from onset of acute infection symptomsd:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Infectivity (103 FFU/ml) for NP2/CD4:
|
Classificationc | |||||||||
| CD4 alone | CCR3 | CCR5 | CXCR4 | 20 | 81 | 608 | 957 | |||
| 8.2.50 | 12 | 0.3 | 1,200 | 1,400 | 0.3 | R3/R5 | <10f | 10 | 20 | 40 |
| 8.2.51 | 12 | —e | 150 | 90 | 0.3 | R3/R5 | <10 | 10 | 80 | 80 |
| 8.4.51 | 32 | 0.2 | 10 | 270 | 1 | R3/R5 | <10 | 10 | 40 | 40 |
| 8.8.3 | 608 | — | 80 | 120 | 0.1 | R3/R5 | <10 | 10 | 80 | 80 |
| 8.8.4 | 608 | — | 40 | 470 | 2 | R3/R5 | <10 | 10 | 20 | 20 |
| 8.8.8 | 608 | — | 6 | 150 | 0.4 | R3/R5 | <10 | 10 | 80 | 80 |
| 8.9.B | 957 | — | 110 | 280 | 230 | R3/R5/X4 | <10 | <10 | <10 | <10 |
| 8.9.D | 957 | — | 280 | 700 | 320 | R3/R5/X4 | <10 | <10 | <10 | <10 |
| 8.9.F | 957 | — | 0.2 | 1 | — | R3/R5 | <10 | <10 | 20 | 10 |
| 8.9.H | 957 | — | 140 | 340 | 590 | R3/R5/X4 | <10 | <10 | <10 | <10 |
| 8.9.I | 957 | — | — | 740 | 2 | R5 | <10 | <10 | <10 | <10 |
| 8.9.J | 957 | — | 70 | 140 | 150 | R3/R5/X4 | <10 | <10 | <10 | <10 |
| 8.9.K | 957 | — | — | 120 | — | R5 | <10 | <10 | <10 | <10 |
| YU2g | NAh | — | 1,700 | 4,800 | — | R3/R5 | NDi | ND | ND | ND |
| SDM4j | NA | — | 9 | 16 | — | R3/R5 | <10 | <10 | <10 | <10 |
Replication-competent molecular clone containing gp120 cloned directly from patient peripheral blood mononuclear cells (days 12 and 32) or plasma (days 608 and 957), and inserted into an HIV-1HXB2 based vector.
Time point from which the patient envelope was amplified, in days from onset of symptoms characteristic of primary HIV-1 infection.
Viruses were classified as dual-or triple-tropic if their titer on alternative coreceptors was within 1.5 log10 of the titer obtained on the dominant coreceptor cell line.
Titers are expressed as the reciprocal dilution of serum required to reduce infectivity by ≥90% (IC90), as measured by immunostaining of infected NP2/CD4/CCR5 cells against virus incubated with 10% (1:10) normal human serum.
—, ≤100 FFU/ml.
Less than 90% neutralization was observed at the lowest serum dilution (1:10) tested.
The titer the of prototypical CCR3-using HIV-1 virus YU2 on NP2 cells has been included for comparison.
NA, not applicable.
ND, not determined.
SDM4, site-directed mutant 4 (see Fig. 3>).
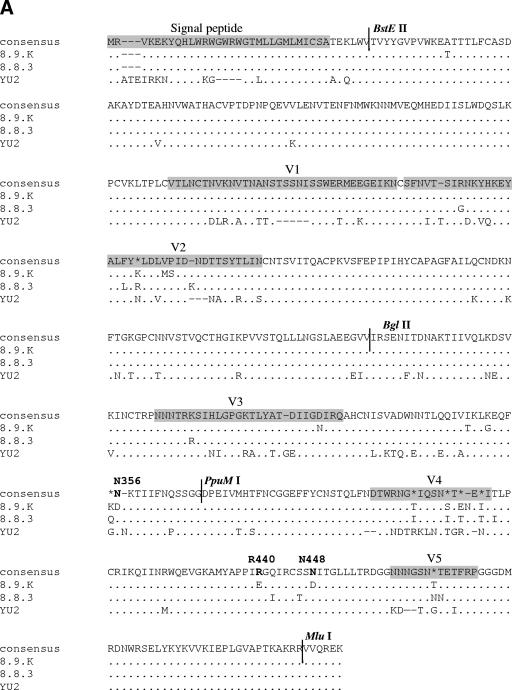
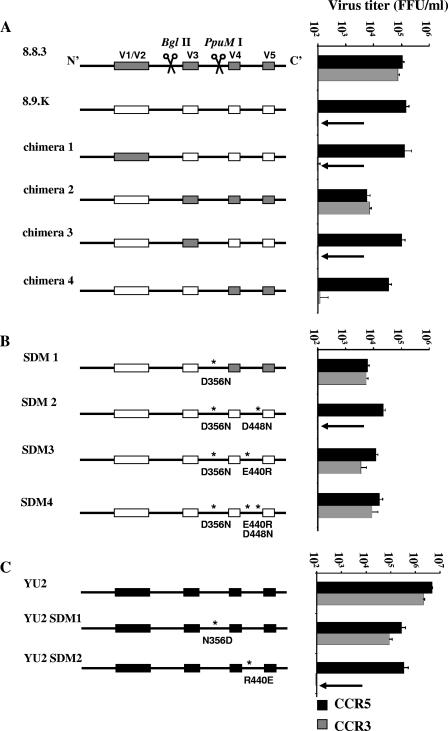
The isolation of closely related Envs with different coreceptor preferences allowed us to map Env determinates of CCR3 use. Env fragments were swapped between 8.9.K (R5 only) and 8.8.3 (R3/R5). Figure 2A shows the amino acid alignment of these Envs and the prototypic R3/R5 Env YU2. Regions were swapped using conserved BglII and PpuMI restriction sites in combination with BstEII and MluI sites, which were incorporated in the primers used for Env cloning (Fig. 2A). Figure 3A shows the chimeric Envs generated and their titers on CCR3 and CCR5 cells. The parental Env 8.8.3 uses CCR3 and CCR5 with similar efficiencies (0.8 × 105 and 1.2 × 105 FFU/ml, respectively) whereas 8.9.K cannot use CCR3 (<1 FFU/ml, compared with 1.2 × 105 FFU/ml for CCR5). We excluded the involvement of regions N terminal of BglII, including the V1/V2 loops, since receptor preference followed the C-terminal end of the Envs (chimeras 1 and 2). Determinants for both CCR5 and CXCR4 binding and fusion have been mapped to amino acids in the V3 loop and to residues in the “bridging sheet” (3, 10-12). We sought to determine the contribution of the central V3 loop-containing region to CCR3 use. Insertion of the region flanked by BglII and PpuMI from CCR3-using Env 8.8.3 did not confer CCR3 use on 8.9.K (chimera 3), indicating that a region(s) C terminal of PpuMI is important. However, replacement of the region C terminal of PpuMI alone did not result in gain of CCR3 use (chimera 4). We concluded that determinants of CCR3 use are contained both in the central region and C-terminal end of Env.
FIG.2.
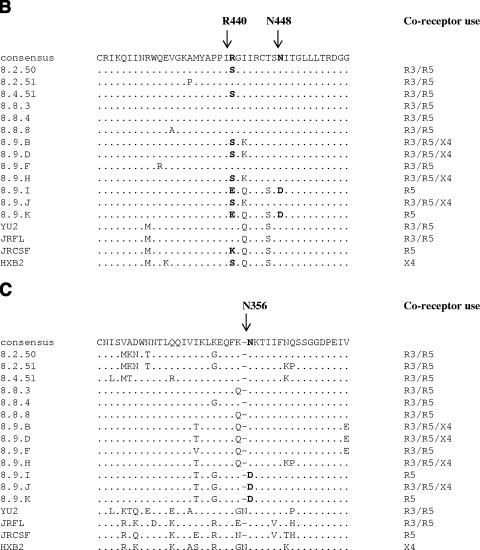
Env alignments. (A) Amino acid alignments (generated using ClustalW [http://www.ebi.ac.uk/clustralw]) of the R5-only-using Env 8.9.K (GenBank accession number DQ645384), the most closely related R3/R5-using Env 8.8.3 (GenBank accession number DQ425072), and the prototypical R3/R5-using Env YU2 (GenBank accession number M93258). The asterisks indicate lack of consensus between the three Envs. The dots indicate residues identical to those in the consensus sequence, and the dashes indicate gaps. The signal peptide and the gp120 variable loops are shaded gray. The locations of the sites for the restriction enzymes BglII, BstEII, MluI, and PpuMI used for the sequence swapping are also indicated. Residues subjected to site-directed mutagenesis are in boldface. (B) Alignments of a section of the C4 regions of all patient Envs (GenBank accession numbers AY295233, AY295235, AY295237, DQ425072 to -4, and DQ64378 to -84) and Envs YU2, JRFL, JRCSF, and HXB2 (GenBank accession numbers M93258, AY669728, M38429, and K03455, respectively). The dots indicate residues identical to those in the consensus sequence, and the dashes indicate gaps. The determinants for CCR3 use are in boldface. (C) Alignments of a section of the C3 Env regions (see description of panel B).
FIG. 3.
Mapping of CCR3 determinants. (A) Sections of the env gene were swapped between env 8.8.3 (gray boxes) and env 8.9.K (white boxes) by restriction enzyme digestion with BglII and/or PpuMI. Only chimera 2 (8.9.K-BglII-8.8.3) was able to infect NP2/CD4/CCR3 cells efficiently. (B) Amino acid substitutions, indicated with asterisks, were introduced into chimera 4 (SDM1) and env 8.9.K (SDM2, -3, and -4) by site-directed mutagenesis. Mutation of D356 to N in combination with mutation of E440 to R was sufficient to transform the R5-only-using Env 8.9.K to an efficient R3 user (SDM3). The titer of SDM3 on R3 cells was ∼30% of that on R5 cells. Additional mutation of D448 to N (SDM4) increased the virus titer on CCR3 cells to ∼60% of the titer on CCR5 cells. (C) Mutation of amino acids N356 in YU2 to aspartic acid (D) did not affect CCR3 use by YU2. However, mutation of R440 in YU2 to glutamic acid (E) resulted in an almost-complete abolition of CCR3 use (from 40% to less than 0.1% of the titer on R5 cells).
Site directed mutagenesis identified specific determinates of CCR3 use. We initially used chimera 4 as a template and replaced aspartic acid 356 with asparagine (D356N [residue numbering is according to the HXBc2 sequence]), since the CCR3-using Envs YU2 and 8.8.3 both have an asparagine at this position (Fig. 2A). The resulting Env fully gained CCR3 use (SDM1) (Fig. 3B). Further mutagenesis defined the CCR3 determinants needed in addition to N356 and located C terminal of the PpuMI site. We introduced the D356N substitution into 8.9.K and substituted D448N and E440R, residues that also differ between the R5-only-using (8.9.K) and R3/R5-using (8.8.3 and YU2) Envs. The combination of D356N and D448N was not sufficient for CCR3 use (SDM2) (Fig. 3B), but substitution of E440R in combination with D356N resulted in a near-complete reconstitution of CCR3 use (SDM3) (Fig. 3B). The titer of SDM3 on R3 cells was ∼30% of that on R5 cells. The joint substitution of E440R and D448N with N356 resulted in CCR3 use that was 60% of that on R5 cells (SDM4) (Fig. 3B). Thus, amino acid N356 in conserved region 3 (C3), together with R440 and N448 in C4, determines CCR3 use. R440 and N448 are proximal to residues that mediate binding to CCR5 (10, 11). N448 was shared by all R3-using patient Envs, but serine replaced R440 in the R3/R5/X4-using Envs (Fig. 2B). The switch to negatively charged residues at positions 440 and 448 may affect electrostatic interactions between Env and CCR3. N356, however, is located on the outer domain of Env, distal from the bridging-sheet region implicated in CCR5 and CXCR4 binding (7, 8). The resultant loss of a potential N-linked glycosylation site could result in a global conformational change, with indirect effects on the bridging sheet specifically affecting CCR3 usage. Interestingly, Env 8.9.J could use CCR3 efficiently despite having D356, suggesting that other regions may compensate for this CCR3 determinant (Fig. 2C).
The Envs used in the experiment described above were closely related. To ascertain the influence of the determinants for CCR3 use in a broader context, we used the R3/R5 Env YU2. Substitution of N356D did not result in loss of CCR3 use (Fig. 3C), again suggesting that the influence of this CCR3 determinant is context dependent. However, maintaining R440 was crucial for CCR3-mediated entry, confirming the association of this residue with CCR3 use (Fig. 3C). The identified residues had minimal effects on CCR5 use, and thus use of CCR3 is not merely a fortuitous extension of CCR5 use.
CCR3 use has previously been associated with V1/V2 and V3 (6). Here we have mapped additional determinants in C3 and C4. Determinants N356 and N448 are highly conserved among subtype B viruses (Fig. 2B and C). Residue 440 has previously been implicated in T-cell tropism (4). An arginine or serine, but not glutamic acid or lysine, at this position appears to be compatible with CCR3 use (Fig. 2C).
It has previously been reported that producer cells influence the type of N-linked carbohydrate structures present on HIV Env (9). Since the identified CCR3 determinants included putative targets for N-glycans, we tested whether the ability to use CCR3 was altered by growth through an in vivo relevant cell type. The parental R5-only-using virus 8.9.K and the R3/R5-using virus SDM3 (which contain N356, a potential glycosylation site, and the R440 determinants) were cultured in T cells (SupT1/CCR5). The level of CCR3 use remained the same as for virus produced in 293T cells (data not shown).
The observation that two new phenotypes arose at “coreceptor switch,” one which gained CXCR4 while maintaining CCR3 use and another which lost CCR3 use, is intriguing. We determined if selection by neutralizing antibodies is involved by testing the viruses for susceptibility to autologous sequential serum (Table 1). Virus was incubated with serially diluted sera and plated onto NP2/CD4/CCR5 cells (2). In contrast to the earlier R3/R5 Envs, the R5-only- and R3/R5/X4-using Envs were resistant to neutralization. The day 957 R3/R5 Env (8.9.F) retained some sensitivity to autologous neutralization. However, we found no association between CCR3 determinants and neutralization sensitivity, since SDM4 also was neutralization resistant (Table 1). It is therefore unlikely that neutralizing antibodies were an important selective pressure for coreceptor switch in this case.
In summary we describe a novel “coreceptor switch” phenotype, from R3/R5-using to R5-only- or R3/R5/X4-using Envs. The determinants for CCR3 use were amino acid N356, which has not previously been associated with coreceptor use, and R440 and N448, which are proximal to but distinct from residues previously shown to be important for CCR5 use. Therefore, CCR3 use is not merely a fortuitous event that occurs because of CCR5 use. It will be important to determine if CCR3 use can be correlated with a specific niche or tropism in vivo.
Acknowledgments
We thank the medical staff at the Centre for Sexual Health and HIV Research (University College London) for their help with the clinical side of the study, Paul Kellam for advice, and Suzy Willey for critical reading of the manuscript.
The Wellcome Trust, the MRC-UK, and core funding from the Edward Jenner Institute for Vaccine Research supported this work.
REFERENCES
- 1.Aasa-Chapman, M. M. I., K. Aubin, I. Williams, and A. McKnight. 2006. Primary CCR5 only using HIV-1 isolates does not accurately represent the in vivo replicating quasi-species. Virology 351:489-496. [DOI] [PubMed] [Google Scholar]
- 2.Aasa-Chapman, M. M. I., A. Hayman, P. Newton, D. Cornforth, I. Williams, P. Borrow, P. Balfe, and A. McKnight. 2004. Development of the antibody response in acute HIV-1 infection. AIDS 18:371-381. [DOI] [PubMed] [Google Scholar]
- 3.Basmaciogullari, S., G. J. Babcock, D. Van Ryk, W. Wojtowicz, and J. Sodroski. 2002. Identification of conserved and variable structures in the human immunodeficiency virus gp120 glycoprotein of importance for CXCR4 binding. J. Virol. 76:10791-10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrillo, A., and L. Ratner. 1996. Human immunodeficiency virus type 1 tropism for T-lymphoid cell lines: role of the V3 loop and C4 envelope determinants. J. Virol. 70:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiore, J. R., G. Buccoliero, P. Pezzotti, G. Rezza, A. Saracino, G. Pastore, E. M. Fenyo, and G. Angarano. 1999. HIV-1 disease progression in women: role of the viral phenotype of the HIV-positive steady partner. AIDS 13:1801-1802. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman, T. L., E. B. Stephens, O. Narayan, and R. W. Doms. 1998. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc. Natl. Acad. Sci. USA 95:11360-11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, G., G. Simmons, S. Pohlmann, F. Baribaud, H. Ni, G. J. Leslie, B. S. Haggarty, P. Bates, D. Weissman, J. A. Hoxie, and R. W. Doms. 2003. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J. Virol. 77:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeves, J. D., J. L. Miamidian, M. J. Biscone, F. H. Lee, N. Ahmad, T. C. Pierson, and R. W. Doms. 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J. Virol. 78:5476-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retroviruses 16:741-749. [DOI] [PubMed] [Google Scholar]
- 12.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 13.Scarlatti, G., E. Tresoldi, A. Bjorndal, R. Fredriksson, C. Colognesi, H. K. Deng, M. S. Malnati, A. Plebani, A. G. Siccardi, D. R. Littman, E. M. Fenyo, and P. Lusso. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 3:1259-1265. [DOI] [PubMed] [Google Scholar]
- 14.Shankarappa, R., P. Gupta, G. H. Learn, Jr., A. G. Rodrigo, C. R. Rinaldo, Jr., M. C. Gorry, J. I. Mullins, P. L. Nara, and G. D. Ehrlich. 1998. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology 241:251-259. [DOI] [PubMed] [Google Scholar]