Abstract
Pseudorabies virus (PRV), an alphaherpesvirus related to herpes simplex virus type 1 and varicella-zoster virus, infects a broad host range of mammals. A striking characteristic of PRV infection is the different symptoms and outcomes of infection in natural and nonnatural hosts. Adult pigs, the natural hosts of PRV, survive infection with only mild respiratory symptoms, while nonnatural hosts, including rodents and cattle, invariably die after exhibiting neurological symptoms. Here, we show that the PRV EP0 protein is necessary to overcome an interferon-mediated antiviral response in primary cells from the natural host of PRV but is not necessary in nonnatural-host cells.
Pseudorabies virus (PRV) is a member of the alphaherpesvirus subfamily related to the human pathogens herpes simplex virus type 1 (HSV-1) and HSV-2 and varicella-zoster virus (17). Like many alphaherpesviruses, PRV infects a broad range of mammals, including rodents, cattle, dogs, and cats (15). Higher-order primates, including humans, are not susceptible to PRV infection. A striking characteristic of PRV infection is the different symptoms and outcomes of infection in natural and nonnatural hosts. Wild-type PRV infection causes respiratory and reproductive disease in its natural host, the adult pig, and mortality is rare (15). In contrast, nonnatural hosts infected with PRV exhibit neurological symptoms and die as a result of infection. The death of nonnatural hosts may be caused by a systemic inflammatory response (1). The type I interferon (IFN) system is the front line of host antiviral defense and is involved in establishing an inflammatory response (7, 19). Accordingly, the different outcomes of PRV infection in natural and nonnatural hosts may be linked to how PRV infection engages the IFN system in natural and nonnatural hosts.
The HSV-1 protein ICP0 prevents the transcriptional induction of IFN-stimulated genes (4). ICP0-null mutants are sensitive to IFN-α pretreatment in a cell line-specific manner (11). While PRV EP0 is considered a homolog of ICP0 because of its position in the genome and its function as a transcriptional activator, the two genes exhibit different temporal expression and have low sequence similarity outside their N-terminal domains (14, 15). In this report, we compared the growth of PRV EP0 deletion mutants with that of HSV-1 ICP0 deletion mutants in the presence and absence of IFN. We wanted to determine whether the growth levels of these two mutants were equally sensitive to IFN pretreatment in primary fibroblasts from natural and nonnatural hosts.
Monolayers of primary fibroblasts from natural and nonnatural hosts of PRV were treated with 1,000 U/ml of the species-appropriate IFN-α (PBL Biomedical Laboratories) and infected 24 h later with either PRV Be or the PRV EP0 deletion mutant (PRV EP0-1) (2) at 1 PFU per cell. The natural host cells of PRV were pig embryonic lung fibroblasts (PELF) and pig embryonic spleen fibroblasts (C. Jones, University of Nebraska). Nonnatural host cells for PRV were rat embryonic fibroblasts and mouse embryonic fibroblasts. IFN-α was chosen for this assay because it is commercially available for all animal species tested in this experiment. All IFNs used were active and capable of inducing STAT1 phosphorylation in their target cells (data not shown). Virus was collected 24 h postinfection (hpi) and viral yield determined by plaque assay. The results were expressed as the ratios of titers obtained from nontreated cells and the titers obtained from pretreated cells (Table 1). The yields of PRV Be were marginally reduced by IFN-α on both porcine and rodent cells. The titers of PRV EP0-1 were also marginally affected on rodent fibroblasts, but yields were 100- to 300-fold reduced on porcine fibroblasts pretreated with IFN-α.
TABLE 1.
Effects of IFN-α and cell type on viral yield
| Virus strain | Gene inactivated | −IFN/+IFN ratioa for indicated cell group
|
||||
|---|---|---|---|---|---|---|
| PELF | PESF | REF | MEF | HFF | ||
| PRV Becker | None | 2.4 | 2 | 2.3 | 2.9 | — |
| PRV EP0-1 | EP0 | 297 | 114 | 5.6 | 7.1 | — |
| HSV-1 KOS | None | — | — | — | 2.8 | 90 |
| HSV-1 n212 | ICP0 | — | — | — | 2.2 | 58 |
| HSV-1 d99 | ICP0 | — | — | — | 12 | 107 |
The ratio was determined by dividing the viral titer obtained from nontreated cells by the titer obtained from pretreated cells. Each titer measurement was an average obtained from three independent experiments. −IFN, nontreated cells; +IFN, pretreated cells; —, not done.
We next determined whether the HSV-1 ICP0 protein exhibits a similar type of species specificity in terms of IFN-α sensitivity. Therefore, human foreskin fibroblasts (HFF), cells from the natural host of HSV-1, and mouse embryonic fibroblasts, cells from a nonnatural host of HSV-1, were pretreated with a species-appropriate IFN-α, as described above. Cells were infected with the wild-type strain HSV-1 KOS or a HSV-1 KOS mutant bearing a null mutation in the ICP0 gene, n212 (3) or d99 (18). Viral yields were determined by plaque assays. In contrast to the relative insensitivity of PRV Be on porcine fibroblasts, HSV-1 KOS yields were 90-fold reduced on human foreskin fibroblasts (Table 1). This sensitivity of wild-type HSV-1 to IFN-α on cells from its natural host is consistent with previous results that showed that HSV-1 yields were reduced from 65% to 95% by various IFN-α subtypes on human cornea stromal cells (20). Unlike the PRV EP0-1 mutant, neither HSV-1 ICP0 mutant exhibited reduced yields on mouse cells pretreated with IFN.
The HSV-1 ICP0 protein did not function preferentially in human cells versus in rodent cells, since the viral yields of HSV-1 KOS and two ICP0-null mutants were similar in both cell types tested. Thus, despite being classified in the same virus subfamily, PRV and HSV-1 do not interact with the immune response in the same way. For example, wild-type PRV infection of mice is invariably fatal (1), while the lethality of wild-type HSV-1 infection depends on the strain of virus and the mouse (9, 21). Because of the rapid lethality of PRV infection of rodents, a latent infection is difficult to establish with virulent PRV strains. In fact, PRV latency in rodents has been possible only with the combination of passive immunization with high-titer neutralizing antibodies and infection with the attenuated strain PRV Bartha (13). HSV-1 latency, on the other hand, can be established in rodents with a variety of wild-type strains (5).
The different responses of the PRV EP0 mutant to IFN pretreatment in rodent and porcine cells may be caused by the adaptation of PRV to the IFN system of its natural host. The mouse (Mus musculus) and pig (Sus scrofa) protein sequences of several gene products involved in the type I IFN response were aligned with the rat (Rattus norvegicus) protein sequences by using DS Gene software (Accelrys) to analyze how closely related the IFN systems of the three species are (data not shown). The genes selected were those for IFN-α, signal transducer and activator of transcription 1 (STAT1), and double-stranded RNA-dependent protein kinase. In all three cases, the mouse and rat proteins had higher percentages of identical sequence than the pig and rat proteins. If EP0 needs to bind to some component of the IFN system to inhibit it efficiently and has evolved to interact structurally with the pig version of this component, the fact that the rodent versions of the protein are not very similar to the pig version may prevent efficient PRV EP0 function.
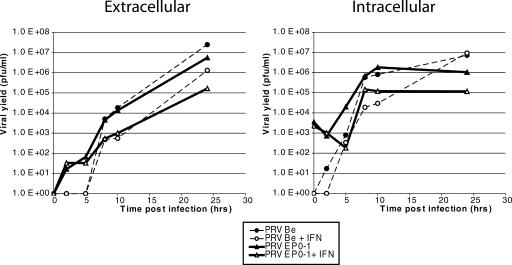
To determine the point in the viral replication cycle at which the EP0 protein functions, single-step growth curves were performed with both PRV Be and PRV EP0-1 on PELF pretreated or untreated with porcine IFN-α 24 h prior to infection. Extracellular and intracellular (Fig. 1) virus was collected over a 24-h period after infection, and the titers were measured by plaque assay. In IFN-treated cells, both PRV Be and PRV EP0-1 yields began to fall behind those from untreated cells between 5 and 10 hpi. PRV Be yields, however, caught up to those from untreated cells by 24 hpi, while PRV EP0-1 yields continued to lag behind. Both extracellular and intracellular yields exhibited similar patterns, suggesting that the defect linked to the absence of EP0 is not associated with viral egress.
FIG. 1.
Growth kinetics of PRV Be and PRV EP0-1 in PELF cells untreated and pretreated with porcine IFN-α. Cells were either untreated or treated with IFN-α for 24 h prior to infection with either PRV Be or PRV EP0-1 at a multiplicity of infection of 10. Extracellular and intracellular virus was collected at the indicated times postinfection, and the titers were determined by plaque assay (n = 2).
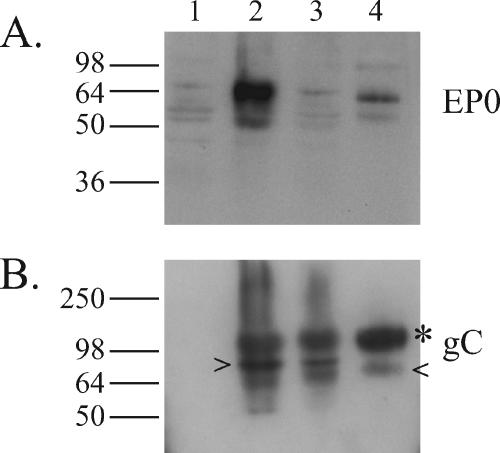
The finding that the EP0 gene product is necessary to overcome a previously established IFN-α-induced antiviral state suggests that the EP0 protein must act soon after a virus infects a cell. Previous results have shown that EP0 is packaged in the virions of the PRV YS-81 strain (12). Western blots were used to verify that the EP0 protein also is packaged in PRV Be virions. The anti-EP0 polyserum failed to recognize EP0 in the mock- and PRV EP0-1-infected cell lysates but recognized EP0 in the PRV Be-infected cell lysates and in PRV Be purified virions (Fig. 2A). The cell lysates and virions were also blotted for PRV glycoprotein C (gC) to demonstrate the purity of the virion preparations (Fig. 2B).
FIG. 2.
The EP0 protein is packaged in wild-type PRV virions. Whole-cell lysates of PK15 cells mock infected (lane 1), PRV Be infected (lane 2), or PRV EP0-1 infected (lane 3) and purified PRV-Be virions (lane 4) collected at 16 h postinfection were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted for EP0 (A) and gC (B). The mature (*), immature (>), and degradation (<) forms of gC are indicated (16). Positions of molecular mass markers are shown on the left in kilodaltons.
PRV is capable of preventing the establishment of an IFN-β-induced antiviral state by interfering with IFN signal transduction (2). Here, we have demonstrated that PRV infection can overcome a previously established antiviral state through the function of the EP0 protein. Because the proteins required for both modes of blocking the IFN response are carried in the virion, most likely the tegument, they are available soon after viral entry into cells. While the protein(s) that interacts with IFN signaling is active in nonnatural host cells, EP0 appears to function only in natural host cells. We propose the following model for the necessity of two modes of counteracting the IFN response. The protein(s) that blocks IFN signaling allows wild-type PRV to replicate normally in both kinds of hosts by preventing the expression of IFN-stimulated genes that affect viral macromolecule synthesis and that may cause killing of PRV-infected cells before viral replication is complete (2). The EP0 protein keeps the natural host of PRV from dying from a systemic inflammatory response activated by the IFN system because most successful viruses do not kill their natural hosts rapidly (6). We have shown that the EP0 deletion renders PRV more sensitive to IFN treatment in terms of viral replication (Table 1). It is likely that the lack of EP0 would also make the virus unable to control other aspects of the IFN system, such as the induction of an inflammatory response. This model can be tested by comparing PRV Be and PRV EP0-1 infection of adult pigs. Blood cytokine levels of proinflammatory cytokines, such as interleukin-8 and tumor necrosis factor alpha, can be measured, and infiltration of inflammatory cells into tissues can be monitored. If the model is correct, pigs infected with PRV EP0-1 are expected to have a more robust inflammatory response than pigs infected with PRV Be. While it is possible that the pathway targeted by EP0 is not found in rodents, it is unlikely that the absence of a pathway in nonnatural hosts would lead to a more lethal outcome.
The mechanism by which EP0 acts intracellularly is not known. HSV-1 ICP0 mediates the inhibition of activated interferon regulatory factor 3 (IRF-3) and IRF-7 and prevents nuclear translocation of IRF-3 induced by Sendai virus (8). IRF-3 has been linked to induction of RANTES, a monocyte-recruiting chemokine (10). PRV EP0 may also affect promyelocytic leukemia protein nuclear bodies, which have been implicated in immune surveillance and transcription regulation (22).
Acknowledgments
We thank Christina Paulus for generating and providing the PRV EP0-1 mutant and rat embryonic fibroblasts. We are grateful to Priscilla Schaffer, Neal DeLuca, and Clinton Jones for providing HSV-1 strains and pig primary fibroblasts. All members of the Enquist laboratory provided helpful suggestions and insightful comments.
This research was supported by NIH grant 5P01 CA87661 to L.W.E. A.B. was supported by an NSF Predoctoral Fellowship and a Princeton Graduate School Centennial Fellowship.
Footnotes
Published ahead of print on 23 August 2006.
REFERENCES
- 1.Brittle, E. E., A. E. Reynolds, and L. W. Enquist. 2004. Two modes of pseudorabies virus neuroinvasion and lethality in mice. J. Virol. 78:12951-12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brukman, A., and L. W. Enquist. 2006. Suppression of the interferon-mediated innate immune response by pseudorabies virus. J. Virol. 80:6345-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, W. Z., and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral infection. J. Virol. 76:2180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellison, A. R., L. Yang, C. Voytek, and T. P. Margolis. 2000. Establishment of latent herpes simplex virus type I infection in resistant, sensitive, and immunodeficient mouse strains. Virology 268:17-28. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Sastre, A., and C. A. Biron. 2006. Type I interferons and the virus-host relationship: a lesson in detente. Science 312:879-882. [DOI] [PubMed] [Google Scholar]
- 7.Grob, P., V. E. Schijns, M. F. van den Broek, S. P. Cox, M. Ackermann, and M. Suter. 1999. Role of the individual interferon systems and specific immunity in mice in controlling systemic dissemination of attenuated pseudorabies virus infection. J. Virol. 73:4748-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin, R., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 78:1675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez, C. 1975. Genetics of natural resistance to herpesvirus infections in mice. Nature 258:152-153. [DOI] [PubMed] [Google Scholar]
- 10.Melchjorsen, J., and S. R. Paludan. 2003. Induction of RANTES/CCL5 by herpes simplex virus is regulated by nuclear factor kappa B and interferon regulatory factor 3. J. Gen. Virol. 84:2491-2495. [DOI] [PubMed] [Google Scholar]
- 11.Mossman, K. L., and J. R. Smiley. 2002. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J. Virol. 76:1995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ono, E., S. Watanabe, H. Nikami, T. Tasaki, and H. Kida. 1998. Pseudorabies virus (PRV) early protein 0 activates PRV gene transcription in combination with the immediate-early protein IE180 and enhances the infectivity of PRV genomic DNA. Vet. Microbiol. 63:99-107. [DOI] [PubMed] [Google Scholar]
- 13.Osorio, F. A., and D. L. Rock. 1992. A murine model of pseudorabies virus latency. Microb. Pathog. 12:39-46. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type I ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pomeranz, L. E., A. E. Reynolds, and C. J. Hengartner. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol. Mol. Biol. Rev. 69:462-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robbins, A. K., R. J. Watson, M. E. Whealy, W. W. Hays, and L. W. Enquist. 1986. Characterization of a pseudorabies virus glycoprotein gene with homology to herpes simplex type 1 and type 2 glycoprotein C. J. Virol. 58:339-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roizman, B., and P. E. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 18.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 20.Taylor, J. L., S. D. Little, and W. J. O'Brien. 1998. The comparative anti-herpes simplex virus effects of human interferons. J. Interferon Cytokine Res. 18:159-165. [DOI] [PubMed] [Google Scholar]
- 21.Wagner, E. K., and D. C. Bloom. 1997. Experimental investigation of herpes simplex virus latency. Clin. Microbiol. Rev. 10:419-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]