Abstract
White spot syndrome virus (WSSV) virions were purified from the tissues of infected Procambarus clarkii (crayfish) isolates. Pure WSSV preparations were subjected to Triton X-100 treatment to separate into the envelope and nucleocapsid fractions, which were subsequently separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The major envelope and nucleocapsid proteins were identified by either matrix-assisted laser desorption ionization-time of flight mass spectrometry or defined antibody. A total of 30 structural proteins of WSSV were identified in this study; 22 of these were detected in the envelope fraction, 7 in the nucleocapsid fraction, and 1 in both the envelope and the nucleocapsid fractions. With the aid of specific antibodies, the localizations of eight proteins were further studied. The analysis of posttranslational modifications revealed that none of the WSSV structural proteins was glycosylated and that VP28 and VP19 were threonine phosphorylated. In addition, far-Western and coimmunoprecipitation experiments showed that VP28 interacted with both VP26 and VP24. In summary, the data obtained in this study should provide an important reference for future molecular studies of WSSV morphogenesis.
White spot syndrome virus (WSSV) is a major pathogen in the cultured penaeid shrimp and can also infect most species of crustaceans (2, 4, 6, 10, 22). Electron microscopy studies revealed that the WSSV virion is an enveloped, nonoccluded, and rod-shaped particle of approximately 275 by 120 nm in size (37, 39). The virus contains a double-stranded circular DNA of about 300 kb, which has been completely sequenced on three WSSV isolates (30, 43). Subsequent analysis revealed that the WSSV genome includes about 180 open reading flames (ORFs). However, so far, only about 30% of these ORFs were functionally annotated, including structural proteins and a variety of enzymes involved in DNA replication and repair, gene transcription, and protein modification, and the other potential gene products are known only as hypothetical proteins. On the basis of phylogenetic analysis, WSSV has been classified in a novel virus genus, Whispovirus, and family, Nimaviridae, as a sole member (23, 24).
With the elucidation of the WSSV genome, at present more attention has been paid to the identification and functional analysis of viral structural proteins, because they are considered to be the key molecules that interact first with the host. In previous studies, the major proteins of WSSV, such as VP28, VP26, VP24, VP19, and VP15, have been analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and identified by N-terminal sequencing (31-33). The recent attempts to identify the minor WSSV structural proteins relied on the combination of 1D or 2D gel electrophoresis separation and mass spectrometry (MS) followed by database searches, which is proven to be a fast and sensitive method for the identification of genes at the protein level (26). And so far, more than 30 polypeptides matching WSSV ORFs were identified with a molecular mass range of 7 to 660 kDa by using this approach (11, 29, 44). However, because of the lack of cell culture systems capable of supporting WSSV replication, many questions concerning viral structure, nature, and function remain to be elucidated.
For enveloped viruses, the envelope proteins are particularly important because these proteins often play vital roles in virus entry, assembly, and budding (3). In previous studies, intensive efforts have been undertaken for characterization of the envelope proteins of WSSV, but these previous approaches were focused on the analysis of individual proteins or a limited number of proteins and the overall biochemical composition of the envelope has for the most part remained elusive. Although the nucleocapsid of WSSV contains relatively few proteins compared with the envelope (42), the characterization of the nucleocapsid proteins is also poorly studied. At present, only two proteins (VP15 and VP664) are confirmed to be nucleocapsid associated (19, 31). Ultrathin section electron microscopy analysis revealed that the replication and assembly of WSSV take place in the nucleus (16, 21), but little is known about the molecular events occurring during the assembly. With this in mind, we decided to make a more comprehensive proteomic analysis, the goal of which was to identify and further characterize the major envelope and nucleocapsid proteins of WSSV. By systematically defining the protein composition of the envelope and nucleocapsid, a functional protein map of WSSV may be realized.
In this study, we have separated the envelope and nucleocapsid proteins of purified WSSV virions by 1D gel electrophoresis. The major envelope and nucleocapsid proteins were identified by using either matrix-assisted laser desorption ionization (MALDI) MS or defined antibodies. The structural proteins of WSSV were further characterized by analysis of glycosylation and phosphorylation. Finally, far-Western and coimmunoprecipitation experiments were conducted to identify the interaction between VP28 and other structural proteins.
MATERIALS AND METHODS
Purification of WSSV virion.
The purification of WSSV was performed as described previously with slight modification on ice or at 4°C (42). Briefly, five to six moribund crayfish (20 to 25 g each) were collected at 1 week postinfection; all tissues excluding hepatopancreas were homogenized for 2 min using a mechanical homogenizer (IKA T-25) in 1,200 ml TNE buffer (50 mM Tris-HCl, 400 mM NaCl, 5 mM EDTA, pH 8.5) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 1 mM Na2S2O5) and then centrifuged at 3,500 × g for 5 min using a JA-14 rotor (Avanti JE; Beckman). The supernatant was saved, and the pellet was rehomogenized in 1,200 ml TNE buffer. The pooled supernatant was filtered through a nylon net (400 mesh) and centrifuged at 30,000 × g for 30 min. After the supernatant was discarded, the upper loose layer (pink) of pellet was rinsed out carefully using a Pasteur pipette, and the lower compact layer (gray) was resuspended in 10 ml TM buffer (50 mM Tris-HCl, 10 mM MgCl2, pH 7.5). The crude virus suspensions were pooled and centrifuged at 3,000 × g for 5 min using a JA-20 rotor, and the supernatant was centrifuged again at 30,000 × g for 20 min. After the supernatant and pink loose layer were removed, the white pellet was resuspended in 1.2 ml TM buffer containing 0.1% NaN3 and transferred to a 1.5-ml Eppendorf tube. The suspension was centrifuged three to five times at 650 × g for 5 min each time to remove pink impurities. Finally, the milk-like pure virus suspension was stored at 4°C until use. The purity of the virus preparation was evaluated by negative-staining transmission electron microscopy (TEM) (JEM 100 cxΠ).
Preparation of the envelope and nucleocapsid fractions.
The purified virus suspension was divided into two equal portions and centrifuged at 20,000 × g for 30 min at 4°C. The pellets were resuspended in 0.4 ml of salt-free buffer (20 mM Tris-HCl, 2 mM MgCl2, pH 7.5) and salt-containing buffer TMN (20 mM Tris-HCl, 150 mM NaCl, 2 mM MgCl2, pH 7.5). For each group, the virus suspension was divided into eight equal portions and subjected to treatment with four different detergents at two concentrations (0.1% and 1%), the nonionic detergent Triton X-100 (TX), octyl glucopyranoside (OG), the zwitterionic detergent 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and the ionic detergent sodium deoxycholate (NaDC), for 30 min at room temperature with gentle shaking. Subsequently, the samples were separated into two fractions, supernatant and pellet, by centrifugation at 20,000 × g for 20 min at 4°C. The pellet was rinsed with water to eliminate any residual supernatant solution and then resuspended in TMN buffer. Finally, all samples were mixed with an equal volume of 2× Laemmli sample buffer, resolved by SDS-PAGE, and stained with Coomassie brilliant blue (15).
Identification of the proteins by MALDI MS. (i) In-gel enzymatic digestion of protein.
Stained protein bands were excised from gels, cut into pieces, and transferred to a 1.5-ml microcentrifuge tube. The gel pieces were washed twice with 50% acetonitrile in 50 mM ammonium bicarbonate for 20 min each time, dehydrated with 100% acetonitrile for 10 min, and vacuum dried. For enzymatic digestion, 125 ng of modified trysin (sequencing grade; Promega) was dissolved in 10 μl of 50 mM ammonium bicarbonate and gradually added to the dry gel pieces. After being reswollen at 4°C for 30 min, samples were incubated overnight at 37°C. Following digestion, tryptic peptides were extracted twice with 60 μl of 50% acetonitrile and 0.1% trifluoroacetic acid for 30 min each time. The extracted solutions were pooled and vacuum dried for subsequent MS analysis.
(ii) MALDI-TOF/TOF MS.
The matrix used was a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile and 0.1% trifluoroacetic acid. The dried peptides were dissolved in 0.7 μl of the matrix, deposited on the target plate, and allowed to dry at room temperature. MALDI mass spectra were obtained using a 4700 Proteomics Analyzer with time-of-flight (TOF)/TOF optics (Applied Biosystems) fitted with a 355-nm Nd:YAG laser. Spectra were acquired in positive-ion reflector mode with an acceleration voltage of 20 kV. Instrumental control was performed with 4700 Explorer software, and GPS Explorer software was used to process data as well as to search the nonredundant database at the National Center for Biotechnology Information (NCBI), using the Mascot platform (Matrix Science, London, United Kingdom) with the following parameters: peptide mass fingerprint, 0.3 Da; MS/MS sequence tag, 0.4 Da; allowing one uncleaved tryptic site; and allowing oxidation of methionine and carbamidomethylation of cysteines.
Antibodies and Western blotting.
Polyclonal antibodies against specific WSSV structural proteins were generated in mouse by using the expressed recombinant proteins in Escherichia coli as described previously (40). For Western blotting, proteins dissolved in Laemmli sample buffer were separated by SDS-PAGE and then transferred onto a polyvinylidene fluoride membrane (Amersham) by semidry blotting at a constant current of 0.5 mA cm−2 for 1.5 h at room temperature. The blot was immersed in blocking buffer (20 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20, 3% nonfat milk, pH 7.5) at 4°C overnight, followed by incubation with diluted primary antibody for 1 h at room temperature. Subsequently, the blot was washed three times and incubated with secondary antibody (1:7,500), an alkaline phosphatase (AP)-conjugated goat anti-mouse immunoglobulin G (Promega), for 1 h at room temperature. After the blot was washed three times, the detection was performed with BCIP (5-bromo-4-chloro-3-indolylphosphate)/Nitro Blue Tetrazolium (Roche).
Glycosylation and phosphorylation analysis of WSSV structural proteins.
The glycosylation detection of WSSV structural proteins in gels was performed according the manual of the Pro-Q Emeral 300 glycoprotein stain kit (Invitrogen). The glycoprotein stain reacts with periodate-oxidized carbohydrate groups and creates a bright green fluorescent signal easily visualized using 300 nm UV illumination on glycoproteins, providing much more sensitive glycoprotein detection than the standard periodic acid-Schiff base method using acidic fuchsin dye. The phosphorylation status of WSSV structural proteins was investigated using Western blotting as described above. The primary antibody was a biotin-conjugated mouse monoclonal antibody (Sigma) directed against phosphothreonine (1:10,000), phosphotyrosine (1:5,000), or phosphoserine (1:40,000), and the secondary antibody was AP-conjugated streptavidin (1:5,000; Promega).
Expression and purification of recombinant VP28 (rVP28).
The entire VP28 gene was amplified from the genomic DNA of WSSV with the forward primer 5′-GAGAGCATGCTTGATCTTTCTTTCACTCT-3′ and the reverse primer 5′-GAGAGGATCCCTCGGTCTCAGTGCCAG-3′ (SphI and BamHI restriction sites underlined). The PCR product was digested with SphI and BamHI and cloned into pQE-70 vector, and the recombinant plasmid was transformed into E. coli XL-Blue. The bacterial cultures were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 6 h at 37°C and then harvested. The recombinant protein was purified using a column of Ni-nitriloacetic acid resins (QIAGEN) under denatured conditions according to the instructions for QIAexpressionist and renatured as described previously (20).
Biotinylation of proteins and far-Western experiments.
The protein was dialyzed against phosphate-buffered saline (PBS) with a 10-fold molar excess of sulfo-NHS-LC-biotin (Pierce). The mixture was incubated at room temperature for 30 min and dialyzed against PBS to remove the unreacted biotin reagent.
The viral envelope proteins were separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane. Proteins adsorbed to the membrane were renatured in turn by incubation with renaturation buffer (20 mM Tris-HCl, 150 mM NaCl, 2% nonfat milk, 1 mM EDTA, 25 mM dithiothreitol, 0.1% Tween 20, 10% glycerol, pH 7.5) containing 6 M, 3 M, 1 M, or 0.1 M guanidine-HCl at room temperature for 1 h each time and then put in renaturation buffer overnight at 4°C. The blot was washed and incubated with 0.5 μg of biotinylated rVP28 in 5 ml renaturation buffer for 3 h at room temperature. The blot was subsequently washed three times and incubated with AP-conjugated streptavidin for 1 h at room temperature. After the blot was washed three times, the detection was performed as described above. Biotinylated glutathione S-transferase (GST) was used to incubate with viral envelope proteins as a control.
Coimmunoprecipitation experiments.
The viral envelope proteins were labeled with fluorescein isothiocyanate (FITC) as described previously (40). The FITC-labeled envelope proteins were incubated with anti-VP28 serum or anti-GST serum (1:250) as a negative control overnight at 4°C. Subsequently, protein A-Sepharose beads were added to the mixture and incubated for 1 h at 4°C. The Sepharose beads were collected by centrifugation and washed five times with PBS. The bound proteins were dissociated from the antibody by boiling in Laemmli sample buffer for 10 min and separated by SDS-PAGE. After electrophoresis, the gel was scanned by a Typhoon 9210 scanner (GE Healthcare).
RESULTS
Preparation of pure WSSV samples and separation of the envelope and nucleocapsid proteins.
Due to the lack of a continuous cell culture system for large-scale production of WSSV virions, it is rather difficult to purify a large amount of WSSV virions with high quality. To overcome the disadvantage, we established and optimized a simple and efficient protocol for WSSV purification. After multiple rounds of conventional differential centrifugations, the milk-like virus suspension was obtained from infected crayfish tissues and TEM observation revealed that most of the purified virions were intact (Fig. 1A).
FIG. 1.
(A) TEM of purified WSSV virions. Bar = 200 nm. (B) TEM of WSSV nucleocapsids after treatment with 1% TX and 0.15 M NaCl. Bar = 100 nm. (C) Coomassie brilliant blue-stained 12% SDS-PAGE gel of the supernatant (S) and pellet (P) fractions of WSSV virions after treatment with four detergents at different detergent (0.1% and 1%) and salt (0 M and 0.15 M NaCl) concentrations. M, protein molecular mass marker (kDa). VP95, VP28, VP26, VP24, VP19, and VP15 are indicated on the right.
To separate optimally the envelope and nucleocapsid fractions from purified WSSV virion preparations, we first tested the effects of four different detergents (TX, OG, CHAPS, and NaDC) and their concentrations on the extraction of envelope proteins from the virions. After detergent treatment and centrifugation, the supernatant and pellet fractions were subsequently analyzed by SDS-PAGE. The efficiency of each detergent solution was determined by the intensities of known envelope proteins (e.g., VP28 and VP19) in the supernatant fraction. As can be seen in Fig. 1C, TX was more efficient in solubilizing envelope proteins than the other three detergents under low-detergent (0.1%) conditions. About 90% of VP28 and VP19 was soluble in the presence of TX (Fig. 1C, lane 1), whereas an estimated 90, 70, and 80% remained in the pellet fraction in the presence of OG (lane 10), CHAPS (lane 18), and NaDC (lane 26), respectively. In addition, for each of the four detergents, better protein solubilization was achieved when the detergent concentration was increased from 0.1% to 1% (Fig. 1C, lanes 5, 13, 21, and 29). We also tested the effect of the salt (NaCl) concentration on the extraction of envelope proteins. The results revealed that increasing the concentration of NaCl (up to 0.15 M) could enhance envelope protein solubility (Fig. 1C, lanes 3, 7, 11, 15, 19, 23, 27, and 31), especially in the case of VP26 and another protein with an apparent molecular mass of 95 kDa (termed VP95). Finally, we tested the effect of reducing agents, such as dithiothreitol, on the extraction of envelope proteins. The result showed that the addition of a reducing agent had no effect on the partitioning of proteins, suggesting that there are no disulfide bond interactions between envelope and nucleocapsid proteins (data not shown). Based on these data, we conclude that to obtain the optimal and most-reproducible extraction results, the use of 1% Triton X-100, as well as 0.15 M NaCl, was required. Under this condition, VP28 and VP19 were present completely in the supernatant fraction (Fig. 1C, lanes 7 and 8). Figure 1B showed the viral nucleocapsids from the resulting pellet of TX treatment. Another commonly used detergent, Nonidet P-40 (NP-40), was also utilized to solubilize envelope proteins, and the protein profile extracted was identical to that of TX (data not shown).
Identification of WSSV proteins.
Under optimal treatment conditions, WSSV envelope and nucleocapsid fractions were separated by SDS-PAGE; the resulting protein bands are shown clearly in Fig. 2. Twenty-five distinct protein bands ranging from 10 to 200 kDa could be observed in the viral envelope fraction. In the nucleocapsid fraction, only eight protein bands were readily seen. A total of 33 prominent bands (Fig. 2) were excised individually from the gels and further analyzed by MS. MALDI mass spectra of tryptic peptides of each protein were generated and sequenced against the NCBI nonredundant database using the Mascot platform. In total, 29 genes from the WSSV genome were identified with 6 to 64% coverage of amino acid sequences. Proteins encoded by wsv011 and wsv442 were found in more than one band, while both band 12 and band 13 contained two proteins (Table 1).
FIG. 2.
Coomassie brilliant blue-stained 12% SDS-PAGE gel of WSSV envelope proteins (EP) and nucleocapsid proteins (NP). Numbers indicate the excised bands. M, protein molecular mass marker (kDa).
TABLE 1.
WSSV envelope and nucleocapsid proteins identified in this study
| Band no. | Protein namea | WSSV ORF forb:
|
ORF sizec
|
Method(s)d | Sequence coverage (%) | Predicted structuree | |||
|---|---|---|---|---|---|---|---|---|---|
| CN | TW | TH | No. of amino acids | Mass (kDa) | |||||
| 1 | VP187 | wsv209 | WSSV264 | ORF108 | 1,606 | 174 | MS | 23 | TM |
| 2, 7, 8, 9, 11 | VP150 | wsv011 | WSSV067 | ORF036 | 1,301 | 144 | MS, Ab | 40 | TM, SP |
| 3 | VP124 | wsv216 | WSSV271 | ORF111 | 1,194 | 132 | MS | 23 | TM |
| 4 | VP110 | wsv035 | WSSV092 | ORF041 | 972 | 108 | MS | 39 | TM, SP, RGD |
| 5, 29 | VP95 | wsv442 | WSSV502 | ORF006 | 800 | 89 | MS, Ab | 35 | TM |
| 6 | VP90 | wsv327 | WSSV383 | ORF159 | 856 | 96 | MS | 24 | TM, RGD |
| 10 | VP56 | wsv325 | WSSV381 | ORF158 | 465 | 51 | MS | 34 | TM, SP |
| 12 | VP52A | wsv238 | WSSV294 | ORF119 | 486 | 51 | MS, Ab | 52 | TM, SP |
| 12 | VP52B | wsv256 | WSSV311 | ORF128 | 384 | 43 | MS, Ab | 31 | TM, SP |
| 13 | VP41A | wsv237 | WSSV293 | ORF118 | 292 | 33 | MS, Ab | 10 | |
| 13 | VP41B | wsv242 | WSSV298 | ORF120 | 300 | 34 | MS, Ab | 34 | TM |
| 14 | VP39 | wsv339 | WSSV395 | ORF162 | 283 | 32 | MS | 29 | |
| 15 | VP38 | wsv259 | WSSV314 | ORF129 | 309 | 35 | MS | 37 | |
| 16 | VP33 | wsv254 | WSSV309 | ORF127 | 281 | 32 | MS | 31 | RGD |
| 17 | VP32 | wsv198 | WSSV253 | ORF102 | 278 | 31 | MS | 28 | |
| 18 | VP31 | wsv340 | WSSV396 | ORF163 | 261 | 30 | MS | 12 | RGD |
| 19 | VP28 | wsv421 | WSSV480 | ORF001 | 204 | 22 | MS | 45 | TM, SP |
| 20 | VP26 | wsv311 | WSSV367 | ORF153 | 204 | 22 | MS | 56 | TM, SP |
| 21 | VP24 | wsv002 | WSSV058 | ORF031 | 208 | 23 | MS | 38 | TM, SP |
| 22 | VP19 | wsv414 | WSSV473 | ORF182 | 121 | 13 | MS | 6 | TM, SP |
| 23 | VP16 | wsv321 | WSSV377 | ORF155 | 117 | 13 | MS | 12 | TM, SP |
| 24 | VP14 | WSSV349 | ORF144 | 97 | 11 | MS | 25 | TM, SP | |
| 25 | VP12 | wsv009 | WSSV065 | ORF034 | 95 | 11 | MS | 9 | |
| 26 | VP664 | wsv360 | WSSV419 | ORF167 | 6,077 | 664 | MS | 64 | TM, RGD |
| 27 | VP190 | wsv289 | WSSV344 | ORF143 | 1,565 | 174 | MS | 68 | TM, SP |
| 28 | VP136 | wsv271 | WSSV326 | ORF134 | 1,218 | 135 | MS | 41 | TM, RGD |
| 30 | VP76 | wsv220 | WSSV275 | ORF112 | 674 | 76 | MS, Ab | 28 | |
| 31 | VP60 | wsv415 | WSSV474 | ORF183 | 544 | 62 | MS | 32 | TM |
| 32 | VP51 | wsv308 | WSSV364 | ORF151 | 466 | 52 | MS, Ab | 12 | |
| 33 | VP15 | wsv214 | WSSV269 | ORF109 | 80 | 9 | Ab | ||
Based on the apparent molecular mass of the protein on 12% SDS-PAGE.
Based on three WSSV isolates. CN, China; TW, Taiwan; TH, Thailand. The accession numbers are AF332093, AF440570, and AF369029, respectively.
Size of ORFs in amino acids and predicted molecular mass (kDa).
Method used for protein identification. MS, MALDI MS; Ab, antibody (Western blotting).
The presence of putative structures. SP, signal peptides; RGD, cell attachment domain.
Band 33 of the nucleocapsid fraction could not be identified by MALDI MS. Based on its apparent molecular mass, it should be the major nucleocapsid protein VP15 (31). Sequence analysis shows that VP15 contains 14 arginines and 13 lysines in a total of 61 amino acids (45), so it seemed that the tryptic fragments of VP15 were too small to identify by MALDI MS. Western blotting using specific antibody against VP15 confirmed that this band was indeed VP15 (data not shown).
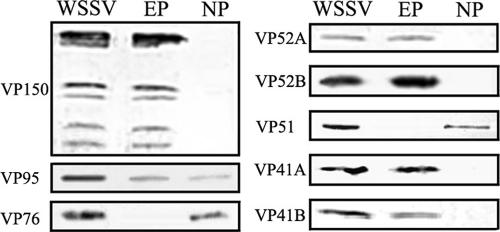
Of 30 proteins identified in this study, 22 were detected in the envelope fraction, 7 in the nucleocapsid fraction, and 1 in both the envelope and the nucleocapsid fractions (Fig. 2; Table 1). Inconsistent with our data, the nucleocapsid proteins VP76 and VP51 were thought to be WSSV envelope proteins in previous reports (11, 12). We therefore prepared nine antibodies against specific structural proteins, including VP150, VP95, VP76, VP52A, VP52B, VP51, VP41A, VP41B, and VP12. Immunoblotting using these antibodies was performed for not only further confirming the MS data but also making an unambiguous characterization of proteins with controversy. Except for VP12, eight other proteins were detected by Western blotting using specific antibodies as expected (Fig. 3). VP150, VP52A, VP52B, VP41A, and VP41B were found completely in the viral envelope fraction, confirming that they are all vial envelope proteins. In addition, five bands (Fig. 2, bands 2, 7, 8, 9, and 11) in the envelope fraction were detected by Western blotting with antibody against VP150. We speculate that four proteins with smaller molecular masses are likely truncated forms of VP150 (see Discussion). VP76 and VP51 were present in the nucleocapsid fraction exclusively after normal detergent extraction. We further attempted to reextract the pellet with a high-salt (0.8 M NaCl and 0.8 KCl) buffer and a low pH (0.1 M glycine, pH 2.5). VP76 and VP51 remained in the pellet fraction after two stringent treatments (data not shown). Based on these data, we can conclude that VP76 and VP51 should be components of the nucleocapsid and not the envelope. VP95 was confirmed to partition consistently in both the envelope and the nucleocapsid fractions (see Discussion). The results of Western blotting indicated that our MS data were credible to some extent.
FIG. 3.
Western blotting of eight structural proteins of WSSV. The intact virions (WSSV) were divided into the envelope (EP) and nucleocapsid (NP) fractions after treatment with 1% TX and 0.15 M NaCl.
Characteristics of WSSV structural proteins.
Among 30 structural proteins identified, 20 have potential transmembrane (TM) domains, 12 have a predicted signal peptide, and 6 have an RGD motif (Table 1). Other than these predicated structures, there is no obvious sequence homology with any known proteins (43).
In this study, the glycosylation and phosphorylation statuses of the WSSV structural proteins were further analyzed. The structural proteins were separated by SDS-PAGE followed by glycoprotein detection. The result showed that all of the WSSV structural proteins were not glycosylated (data not shown), consistent with a previous report (31). The phosphorylation experiment depended on the specificity of antibodies for phosphothreonine, phosphotyrosine, and phosphoserine. As shown in Fig. 4, both VP28 and VP19 were phosphorylated on threonine residues, whereas no band was detected using antibody against phosphotyrosine or phosphoserine. In addition, no positive signal was detected in the nucleocapsid samples (data not shown), suggesting that the nucleocapsid proteins are not phosphorylated.
FIG. 4.
Phosphorylation analysis of WSSV envelope proteins. The left panel shows the SDS-PAGE gel of WSSV envelope proteins, the middle three panels show Western blotting of an equivalent gel probed with a biotin-conjugated mouse monoclonal antibody directed against phosphothreonine (P-Thr), phosphotyrosine (P-Tyr), and phosphoserine (P-Ser), and the right two panels show Western blotting with anti-VP28 (1:3,000) and anti-VP19 (1:3,000) antibodies.
Interaction of VP28 with VP26 and VP24.
Our previous attempt using native PAGE to separate WSSV envelope proteins indicated that interactions between viral envelope proteins are likely to exist (unpublished data). A recent study showed that VP24 interacts with the most abundant envelope-associated protein, VP28 (41). Since VP28 is such an important protein that it might play a key role in the virus life cycle, we are interested in determining whether VP28 interacts with other structural proteins to facilitate functional exploration. Far-Western experiments using biotinylated rVP28 were performed. The viral structural proteins were separated by SDS-PAGE, transferred to a membrane, renatured, and subsequently incubated with biotinylated rVP28 or biotinylated GST as a negative control. After detection, we observed two prominent bands corresponding to VP26 and VP24 (Fig. 5A). No signal was detected in the GST control.
FIG. 5.
(A) Far-Western analysis using biotinylated rVP28. The left panel shows the SDS-PAGE gel of WSSV envelope proteins, the middle two panels show far-Western analysis of an equivalent gel with biotinylated rVP28 (Bio-VP28) and biotinylated GST (Bio-GST), and the right two panels show Western blotting with anti-VP26 (1:3,000) and anti-VP24 (1:3,000) antibodies. (B) Coimmunoprecipitation of VP26 and VP24 with anti-VP28 antibody. The FITC-labeled envelope proteins (FITC-EP) were prepared and incubated with antiserum directed to VP28 (Anti-VP28) or GST (Anti-GST). The immunoprecipitated products were resolved by SDS-PAGE and visualized by fluorescent scanning. The numbers on the left correspond to the molecular masses of the marker proteins. VP28, VP26, and VP24 are indicated on the right.
To further confirm the far-Western results, coimmunoprecipitation analysis was performed. As shown in Fig. 5B, both VP26 and VP24 were immunoprecipitated with anti-VP28 antibody but not with anti-GST antibody. Since anti-VP28 antibody did not cross-react with VP26 and VP24 (Fig. 4), these results indicated that VP28 interacted with VP26 and VP24. It is worth noting that VP28 precipitated with antibody migrated as three bands with similar molecular masses. This observation is in agreement with previous studies showing that VP28 has three forms (11, 29).
DISCUSSION
To date, the entry pathway or assembly process of WSSV has not yet been defined due to the lack of adequate understanding of the biological properties of the viral envelope and nucleocapsid proteins. In order to obtain information about the full set of viral proteins that compose the virion, the purity and quantity of virus preparations are very critical. In previous reports, WSSV virions were isolated directly from the hemolymph of the infected crayfish by density gradient centrifugation and the resulting preparations contained generally crayfish hemocyanin, a major hemolymph protein (∼72 kDa) (29, 32). In addition to hemocyanin, some other cellular proteins, such as actin and vitellogenin, were also found in the virion protein profiles (29). We consider that these cellular proteins are not constituents of WSSV virions, but nonspecifically copurified with the virions, and that the presence of these contaminants in virion preparations may interfere with the identification of the viral structural proteins, especially some quite low-abundant proteins. On the other hand, we cannot exclude the possibility that some viral nonstructural proteins adhere to the cellular proteins, especially hemocyanin, which is considered a multifunctional molecule possessing osmoregulation, protein storage, or enzymatic activities (38) as well as antiviral or antibacterial properties (8, 17). In this study, we purified a large amount of intact WSSV virions with higher purity by using a more efficient method and made a more clear protein profile of WSSV virions, in which those cellular proteins were not observed. In addition, our previous study revealed that the purified WSSV virions by this approach still keep strong infectivity (42). Subsequently, a total of 30 structural proteins were identified from purified WSSV virions. We could be reasonably sure that this protein profile is more accurate and comprehensive than those that have previously been reported, because nearly all of the proteins that appear in the SDS-PAGE gel were identified as WSSV-encoded proteins by MALDI MS and/or Western blotting.
Detergents are indispensable in the isolation of envelope proteins from the enveloped virion to study their intrinsic structural and functional properties. However, not all types of detergents are suitable for each envelope protein (18). In this study, the effects of different types of detergents clearly indicated that the envelope of WSSV is most susceptible to the nonionic detergent TX or NP-40. We also noted that most of the envelope-associated proteins can be released from WSSV virions and maintain solubility with TX, with two exceptions. Both VP26 and VP95 partitioned in both the envelope and nucleocapsid fractions at zero or very low NaCl concentrations (Fig. 1C, lanes 5 and 6). In our previous study, we have provided evidence that VP26 is associated with both the envelope and the nucleocapsid of WSSV, and we have speculated that it might function as a matrix-like linker protein between the envelope and the nucleocapsid (40). In this study, we undertook a more detailed biochemical characterization of VP26. The results showed that the extraction of VP26 was variable at different salt concentrations and that VP26 was present exclusively in the envelope fraction when virions were subjected to treatment with 1% Triton X-100 and 0.5 M NaCl (data not shown). On the basis of our present data, we suggest that VP26 is only loosely bound to the nucleocapsid so that it can be released completely from the nucleocapsid with detergent under high-salt conditions. Similar results regarding dual association of VP26 were independently obtained by Tsai et al. (28). As for VP95, however, we consider this possibility unlikely, since about 40% of VP95 still remained in the nucleocapsid fraction even after treatment with 1% Triton X-100 under high-salt conditions (0.8 M NaCl and 0.8 M KCl) (unpublished data).
Bands 2, 7, 8, 9, and 11 of the envelope fraction were all identified by MS as the gene product of ORF wsv011, which encodes a 144-kDa polypeptide that shows a partial homology to flagellins of E. coli (43). Except band 2, the apparent molecular masses of the other four proteins determined from the 1D gel were smaller than expected, and the tryptic mass spectra of four bands obtained from MS were found to map exclusively to the C-terminal domain of the protein, suggesting that the N-terminal domains of four proteins were all removed. In a previous study, a protein with an apparent molecular mass of 53 kDa was also identified as the gene product of ORF wsv011 (referred to as VP53A) (29), which is in agreement with our observation (the molecular mass of band 11 is 53 kDa). Here, we offer two possible explanations for the presence of multiple forms of VP150. First, they might represent a deliberate proteolytic processing of the protein which appears to be necessary for virus assembly, as is observed with many viruses (1, 25, 36). Another possibility could be that VP150 is highly sensitive to proteases and easily degraded during the process of purification. These truncated forms are derived from the different degraded products of VP150.
Glycosylated envelope proteins are very common in animal viruses and often play critical roles in receptor attachment and membrane fusion (9, 34). For WSSV, however, none of the structural proteins appear to be glycosylated, which is an unusual feature among enveloped animal viruses. Differing from those of many enveloped viruses, the formation of the WSSV envelope takes place in the nucleus, which might be responsible for the lack of glycosylation in WSSV. In addition to glycosylation, protein phosphorylation is a basic mechanism for the posttranslational modification of protein function in eukaryotic cells (14). Our studies revealed that both VP28 and VP19 are threonine phosphorylated. For WSSV structural proteins, this study represents their first characterization by phosphorylation analysis. In vaccinia virus, envelope protein phosphorylation is required to form the viral membranes associated with immature virions (1). Further research will be performed to test which kinase mediates phosphorylation of VP28 and VP19 and whether envelope protein phosphorylation plays a role in WSSV morphogenesis.
As previously described, the most abundant envelope protein, VP28, encoded by ORF wsv421, has three forms with similar molecular masses (11, 29). In this study, we also detected three proteins corresponding to VP28, thus confirming previous data. Moreover, in our initial attempt to separate envelope proteins by 2D gels, we noted that VP28 migrated as three spots with similar molecular masses but different pIs (data not shown). It is not known what might lead to these different pI forms of the protein. Attempts to show glycosylation modification added to the protein have been unsuccessful. Therefore, we speculate that these forms may correspond to different phosphorylation states (known to make the pIs of proteins more acidic) of VP28.
Four proteins identified in the nucleocapsid fraction are predicted to have TM domains (Table 1); however, these are perhaps just hydrophobic regions that might not function as TM domains, since the TM signals of these four proteins are much lower than those of typical envelope proteins, such as VP28 and VP19. Among 23 proteins in the envelope fraction, several proteins do not possess potential TM domains and behave mostly like soluble proteins, indicating that these proteins are not integral envelope proteins. Therefore, we postulate that these proteins attain their envelope association through interaction with other integral envelope proteins. In many enveloped viruses, such as vaccinia virus and herpesvirus, a number of interactions have been demonstrated between structural proteins (5, 7, 13, 35), and protein-protein interactions are suggested to be essential for virion morphogenesis (27). Further exploration of the biochemical interactions of WSSV structural proteins might help to elucidate the molecular mechanisms of virion morphogenesis. In this study, additional in vitro experiments provided evidence for the direct interaction of VP28 with VP26 and VP24. Since VP26 is believed to be a key element in the linkage between the viral nucleocapsid and the envelope, it is possible that the stability of a ternary complex composed of VP28, VP26, and VP24 is important for virion morphogenesis, especially when the naked nucleocapsid is wrapped inside the envelope. This possibility will be investigated in the future.
In conclusion, this study is the most comprehensive biochemical analysis for the structural proteins of WSSV so far. Efforts to identify additional associations between structural proteins are in progress. We anticipate that further exploration of the function of structural proteins will facilitate a better understanding of the molecular mechanism underlying WSSV infection and assembly and may be helpful for the diagnosis and control of virus infection.
Acknowledgments
This investigation was supported financially by National Basic Research Program “973” of China (2006CB101801) and the National Natural Science Foundation of China (30330470).
Mass spectrometry analysis was performed by the Research Center for Proteome of Fudan University. We thank Chenglin Wu, Shengliang Xiong, Qi Ding, Zuliang Jie, and Fanyu Lin for their contributions to this study.
Footnotes
Published ahead of print on 23 August 2006.
REFERENCES
- 1.Betakova, T., E. J. Wolffe, and B. Moss. 1999. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J. Virol. 73:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, P. S., H. C. Chen, and Y. C. Wang. 1998. Detection of white spot syndrome associated baculovirus in experimentally infected wild shrimp, crab and lobsters by in situ hybridization. Aquaculture 164:233-242. [Google Scholar]
- 3.Chazal, N., and D. Gerlier. 2003. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67:226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, L., C. F. Lo, Y. L. Chiu, F. C. Chen, and G. H. Kou. 2000. Experimental infection of white spot syndrome virus (WSSV) in benthic larvae of mud crab Scylla serrata. Dis. Aquat. Org. 40:157-161. [DOI] [PubMed] [Google Scholar]
- 5.Chiu, W. L., and W. Chang. 2002. Vaccinia virus J1R protein: a viral membrane protein that is essential for virion morphogenesis. J. Virol. 76:9575-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbel, V., Zuprizal, Z. Shi, C. Huang, Sumartono, J. M. Arcier, and J. R. Bonami. 2001. Experimental infection of European crustaceans with white spot syndrome virus (WSSV). J. Fish Dis. 24:377-382. [Google Scholar]
- 7.Cudmore, S., R. Blasco, R. Vincentelli, M. Esteban, B. Sodeik, G. Griffiths, and J. Krijnse Locker. 1996. A vaccinia virus core protein, p39, is membrane associated. J. Virol. 70:6909-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Destoumieux-Garzon, D., D. Saulnier, J. Garnier, C. Jouffrey, P. Bulet, and E. Bachere. 2001. Crustacean immunity. Antifungal peptides are generated from the C terminus of shrimp hemocyanin in response to microbial challenge. J. Biol. Chem. 276:47070-47077. [DOI] [PubMed] [Google Scholar]
- 9.Granof, A., and R. G. Webster. 1999. Encyclopedia of virology, 2nd ed. Academic Press, San Diego, Calif.
- 10.Hameed, A. S. S., K. Yoganadhan, S. Sathish, M. Rasheed, V. Murugan, and K. Jayaraman. 2001. White spot syndrome virus (WSSV) in two species of freshwater crabs (Paratelphusa hydrodomous and P. pulvinata). Aquaculture 201:179-186. [Google Scholar]
- 11.Huang, C., X. Zhang, Q. Lin, X. Xu, Z. Hu, and C. L. Hew. 2002. Proteomic analysis of shrimp white spot syndrome viral proteins and characterization of a novel envelope protein VP466. Mol. Cell. Proteomics 1:223-231. [DOI] [PubMed] [Google Scholar]
- 12.Huang, R., Y. Xie, J. Zhang, and Z. Shi. 2005. A novel envelope protein involved in white spot syndrome virus infection. J. Gen. Virol. 86:1357-1361. [DOI] [PubMed] [Google Scholar]
- 13.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter, T., and J. A. Cooper. 1985. Protein-tyrosine kinases. Annu. Rev. Biochem. 54:897-930. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophase T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lan, Y., W. Lu, and X. Xu. 2002. Genomic instability of prawn white spot bacilliform virus (WSBV) and its association to virus virulence. Virus Res. 90:269-274. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S. Y., B. L. Lee, and K. Soderhall. 2003. Processing of an antibacterial peptide from hemocyanin of the freshwater crayfish Pacifastacus leniusculus. J. Biol. Chem. 278:7927-7933. [DOI] [PubMed] [Google Scholar]
- 18.le Maire, M., P. Champeil, and J. V. Moller. 2000. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 1508:86-111. [DOI] [PubMed] [Google Scholar]
- 19.Leu, J.-H., J.-M. Tsai, H.-C. Wang, A. H.-J. Wang, C.-H. Wang, G.-H. Kou, and C.-F. Lo. 2005. The unique stacked rings in the nucleocapsid of the white spot syndrome virus virion are formed by the major structural protein VP664, the largest viral structural protein ever found. J. Virol. 79:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, L., X. Xie, and F. Yang. 2005. Identification and characterization of a prawn white spot syndrome virus gene that encodes an envelope protein VP31. Virology 340:125-132. [DOI] [PubMed] [Google Scholar]
- 21.Li, Q., Y. Chen, and F. Yang. 2004. Identification of a collagen-like protein gene from white spot syndrome virus. Arch. Virol. 149:215-223. [DOI] [PubMed] [Google Scholar]
- 22.Lo, C. F., C. H. Ho, S. E. Peng, C. H. Chen, H. E. Hsu, Y. L. Chiu, C. F. Chang, K. F. Liu, M. S. Su, C. H. Wang, and G. H. Kou. 1996. White spot syndrome associated virus (WSBV) detected in cultured and captured shrimp, crabs and other arthropods. Dis. Aquat. Org. 27:215-225. [Google Scholar]
- 23.Mayo, M. A. 2002. Virus taxonomy-Houston 2002. Arch. Virol. 147:1071-1076. [DOI] [PubMed] [Google Scholar]
- 24.Mayo, M. A. 2002. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 147:1655-1656. [DOI] [PubMed] [Google Scholar]
- 25.Moss, B., and E. N. Rosenblum. 1973. Protein cleavage and poxvirus morphogenesis: tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J. Mol. Biol. 81:267-269. [DOI] [PubMed] [Google Scholar]
- 26.Pandey, A., and M. Mann. 2000. Proteomics to study genes and genomes. Nature 405:837-846. [DOI] [PubMed] [Google Scholar]
- 27.Szajner, P., H. Jaffe, A. S. Weisberg, and B. Moss. 2003. Vaccinia virus G7L protein interacts with the A30L protein and is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 77:3418-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai, J.-M., H.-C. Wang, J.-H. Leu, A. H.-J. Wang, Y. Zhuang, P. J. Walker, G.-H. Kou, and C.-F. Lo. 2006. Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion. J. Virol. 80:3021-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai, J.-M., H.-C. Wang, J.-H. Leu, H.-H. Hsiao, A. H.-J. Wang, G.-H. Kou, and C.-F. Lo. 2004. Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus. J. Virol. 78:11360-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Hulten, M. C. W., J. Witteveldt, S. Peters, N. Kloosterboer, R. Tarchini, M. Fiers, H. Sandbrink, R. K. Lankhorst, and J. M. Vlak. 2001. The white spot syndrome virus DNA genome sequence. Virology 286:7-22. [DOI] [PubMed] [Google Scholar]
- 31.van Hulten, M. C. W., M. Reijns, A. M. G. Vermeesch, F. Zandbergen, and J. M. Vlak. 2002. Identification of VP19 and VP15 of white spot syndrome virus (WSSV) and glycosylation status of the WSSV major structural proteins. J. Gen. Virol. 83:257-265. [DOI] [PubMed] [Google Scholar]
- 32.van Hulten, M. C. W., M. Westenberg, S. D. Goodall, and J. M. Vlak. 2000. Identification of two major virion protein genes of white spot syndrome virus of shrimp. Virology 266:227-236. [DOI] [PubMed] [Google Scholar]
- 33.van Hulten, M. C. W., R. W. Goldbach, and J. M. Vlak. 2000. Three functionally diverged major structural proteins of white spot syndrome virus evolved by gene duplication. J. Gen. Virol. 81:2525-2529. [DOI] [PubMed] [Google Scholar]
- 34.van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Moniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.). 2000. Virus taxonomy: seventh report of the International Committee on Taxonomy of Virus. Academic Press, San Diego, Calif.
- 35.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, C. H., C. F. Lo, J. H. Leu, C. M. Chou, P. Y. Yeh, M. C. Tung, C. F. Chang, M. S. Su, and G. H. Kou. 1995. Purification and genomic analysis of baculovirus associated with white spot syndrome (WSBV) of Penaeus monodon. Dis. Aquat. Org. 23:239-242. [Google Scholar]
- 38.Weber, R. E., and S. N. Vinogradov. 2001. Nonvertebrate hemoglobins: functions and molecular adaptations. Physiol. Rev. 81:569-628. [DOI] [PubMed] [Google Scholar]
- 39.Wongteerasupaya, C., J. E. Vickers, S. Sriurairatana, G. L. Nash, A. Akarajamorn, V. Boonsaeng, S. Panyim, A. Tassankajon, B. Withyanchumnarnkul, and T. W. Flegel. 1995. A non-occluded, systemic baculovirus that occurs in cells of ectodermal and mesodermal origin and causes high mortality in black tiger prawn Penaeus monodon. Dis. Aquat. Org. 21:69-77. [Google Scholar]
- 40.Xie, X., and F. Yang. 2005. Interaction of white spot syndrome virus VP26 protein with actin. Virology 336:93-99. [DOI] [PubMed] [Google Scholar]
- 41.Xie, X., and F. Yang. 2006. White spot syndrome virus VP24 interacts with VP28 and is involved in virus infection. J. Gen. Virol. 87:1903-1908. [DOI] [PubMed] [Google Scholar]
- 42.Xie, X., H. Li, L. Xu, and F. Yang. 2005. A simple and efficient method for purification of intact white spot syndrome virus (WSSV) viral particles. Virus Res. 108:63-67. [DOI] [PubMed] [Google Scholar]
- 43.Yang, F., J. He, X. Lin, Q. Li, D. Pan, X. Zhang, and X. Xu. 2001. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 75:11811-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, X., C. Huang, X. Tang, Y. Zhuang, and C. L. Hew. 2004. Identification of structural proteins from shrimp white spot syndrome (WSSV) by 2 DE-MS. Proteins 55:229-235. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, X., X. Xu, and C. L. Hew. 2001. The structure and function of a gene encoding a basic peptide from prawn white spot syndrome virus. Virus Res. 79:137-144. [DOI] [PubMed] [Google Scholar]