Abstract
Human cytomegalovirus (HCMV) infection regulates a number of genes involved in the host antiviral response. We have previously reported that HCMV attenuates the expression of beta interferon (IFN-β) and a number of proinflammatory chemokines, and this attenuation is mediated by the HCMV immediate-early protein IE86. The present study seeks to identify the mechanism by which IE86 blocks IFN-β expression. We demonstrate that the induction of IFN-β during HCMV infection requires the activation of both the IRF-3 and the NFκB pathways. Therefore, IE86 may target either pathway to block IFN-β expression. Our results show that IE86 does not block IRF-3 phosphorylation, dimerization, nuclear translocation, or target gene expression. However, using gel shift analysis, we demonstrate that IE86 efficiently inhibits virus-induced binding of NFκB to the IFN-β promoter, resulting in attenuation of IFN-β and NFκB-dependent gene expression. Furthermore, IE86 expression inhibits tumor necrosis factor alpha-induced NFκB DNA binding and target gene expression. Together, these results identify IE86 as a NFκB antagonist, which results in the suppression of NFκB-dependent cytokine and chemokine gene expression.
Virus-infected cells respond to infection by inducing numerous signaling pathways that ultimately lead to the expression of cellular genes that limit viral replication and spread. This response is characterized by the induction of cytokines and proinflammatory chemokines. Cytokine and chemokine production are critical for the host to mount an effective antiviral response (27, 40, 41). For example, upon virus infection of the host cell, alpha/beta interferon (IFN-α/β) is expressed and secreted from the infected cell. This interferon then functions in an autocrine and paracrine fashion to induce a plethora of antiviral genes that can efficiently inhibit viral replication within the infected cell and the surrounding tissue. In addition, cells also produce chemokines (induced either directly upon infection or by interferon stimulation) which act to link the host innate immune response to the cell-mediated adaptive immune response. These small secretory proteins are critical for viral clearance.
Induction of IFN-β transcription involves critical signal transduction cascades which result in the recruitment and binding of cellular transcription factors to form an enhanceosome on the IFN-β promoter (58). Previous work has demonstrated that the cellular transcription factors nuclear factor kappa B (NFκB) and interferon regulatory factor 3 (IRF-3) are required for enhanceosome formation and IFN-β transcription (2, 5, 33, 50). Inhibition of either the NFκB or the IRF-3 pathway abrogates IFN-β transcription. A number of viruses have evolved mechanisms for inhibiting IFN-β expression by targeting the IRF-3 or NFκB pathways, which allows for viral persistence within the infected host. For example, the Ebola virus VP35, rotavirus NSP1, and human papillomavirus E6 proteins block specific steps required for the activation of IRF-3 (7, 25, 48). In addition, the NS3/4A protease cleaves the RIG-I-signaling adapter molecule IPS-1, which prevents the activation of both NFκB and IRF-3 during hepatitis C virus infection (10, 24, 38, 39).
Recently, our laboratory demonstrated that the human cytomegalovirus (HCMV) immediate-early 2 gene product IE86 can efficiently block expression of IFN-β and a number of proinflammatory chemokines (54, 55). However, the mechanism by which IE86 blocks the induction of IFN-β and these chemokines has not been elucidated. Given that activation of both NFκB and IRF-3 is required for IFN-β transcription and that IRF-3 and NFκB are regulated during HCMV infection (1, 13, 19-21, 32, 43, 46, 62), it is reasonable to suspect that IE86 may target one or both of these pathways to block IFN-β expression. In this report, we examine the effect of IE86 on the IRF-3 and NFκB pathways during HCMV infection. Our results demonstrate that IE86 does not inhibit the phosphorylation, homodimerization, nuclear translocation, or target gene expression of IRF-3. However, expression of IE86 can efficiently block the binding of NFκB to the IFN-β promoter and inhibit NFκB-dependent gene expression. In addition, IE86 can also block NFκB binding activity and NFκB-dependent gene expression following tumor necrosis factor alpha (TNF-α) treatment. Together, these results identify IE86 as an NFκB antagonist, which results in the suppression of NFκB-dependent cytokine and chemokine gene expression.
MATERIALS AND METHODS
Cell culture and virus infections.
Telomerase 12 human foreskin fibroblast (HFF) cells (12), 293, and Phoenix A (provided by Gary Nolan) cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal calf serum (Gemini), 100 units/ml penicillin, and 100 μg/ml streptomycin in an atmosphere of 5% CO2 at 37°C. HCMV stocks were generated and purified as previously described (54). For HCMV infection, cells were infected at a multiplicity of 5 PFU/cell with either purified wild-type HCMV (WT-HCMV; strain AD169), purified UV-irradiated HCMV (UV-HCMV; 360 mJ/cm2 in a Stratalinker), or purified recombinant HCMV. Sendai virus (Charles River laboratories) infections were performed as described previously (54). Cells were treated with TNF-α (50 ng/ml) (sc-4564; Santa Cruz) in serum-free Dulbecco's modified Eagle's medium.
Antibodies.
The following antibodies were obtained from commercial sources: α-pp65 (1205-S; Rumbaugh-Goodwin Institute), α-tubulin (TU-02; Santa Cruz), α-IE1/2 (MAb810; Chemicon), α-p50 (sc-7178; Santa Cruz), α-IκBαP (sc-8404; Santa Cruz), α-IκBα (sc-203; Santa Cruz), α-IRF-3 (sc-9082; Santa Cruz), α-adenovirus hexon (MAB8043; Chemicon), and α-GFP (sc-8334; Santa Cruz). pp71 antibodies were a generous gift from Thomas Shenk.
Generation of recombinant adenoviruses.
Adenovirus expressing the IκBα super repressor (IκBαSR) was generated by removing the IκBαSR cDNA from the pRep4-IκBαSR plasmid (provided by Aubrey Thompson) via a KpnI/HindIII double digestion and cloning the cDNA fragment into the pADTrack (29) vector that was also digested with KpnI and HindIII. The resulting plasmid was termed pADTrack-IκBαSR. Adenovirus was generated according to the AdEasy protocol (29). The generation of replication-defective adenoviruses expressing IE86, pp65, and green fluorescent protein (GFP) have previously been described (54). All adenoviruses express GFP in addition to the gene of interest, except AdGFP, which expresses only GFP. Stocks of the adenoviruses were generated and their titers determined on 293 cells as previously described (54). Importantly, expression from all adenoviruses was confirmed by Western blot analysis.
Generation of recombinant retroviruses.
pLXSN-IRF3ΔN was generated by using primers IRF3ΔN F (5′-AAGCTTATGGGAACCCCAAAGCCACGG-3′) and IRF3ΔN R (5′-TCTAGATCAGCTCTCCCCAGGGCCCTG-3′) to PCR amplify the IRF3ΔN open reading frame, using pCMVBL-IRF3ΔN (37) as a template. The PCR product was TA cloned into the pGEMT-Easy vector (Promega) and was subsequently sequenced. The IRF3ΔN cDNA was then removed by EcoRI digestion and cloned into the EcoRI-digested pLXSN vector (Clontech) to create pLXSN-IRF3ΔN. Retrovirus stocks were prepared as described previously (34). Briefly, 20 μg of the pLXSN or pLXSN-IRF3ΔN plasmids was transfected into Phoenix A cells by using Lipofectamine reagent (Invitrogen). Forty-eight hours after transfection, supernatant containing retrovirus was collected and cell debris removed via centrifugation (3,000 × g for 10 min). Polybrene (4 μg/ml) was added to the retrovirus-containing inoculum during infections. Following transduction, IRF3ΔN expression was confirmed by Western blot analysis.
Western blot analysis.
Western blot analyses were conducted as previously described (11). Briefly, cells were washed in phosphate-buffered saline (PBS) and harvested with a cell scraper, collected by centrifugation, and lysed in RIPA buffer (50 mM Tris-HCl, 1% NP-40, 0.25% Na-deoxycholate) with protease inhibitor cocktail (Roche). Cellular debris was removed by centrifugation, and the supernatant fluids were reserved. The protein concentration was determined by the Bradford assay (9). Equal amounts (40 μg) of protein were resolved by electrophoresis in the presence of sodium dodecyl sulfate (SDS) on 8.5 to 10% polyacrylamide gels. Proteins were transferred to nitrocellulose membrane (Optitran; Schleicher & Schuell) and probed with primary and secondary antibodies. Immunoreactive proteins were detected by the ECL chemiluminescent system (Amersham).
IRF-3 dimerization assay.
Dimerization assays were conducted as previously described (31). Briefly, cells were washed in PBS and harvested with a cell scraper, collected by centrifugation, and lysed in RIPA buffer without Na-deoxycholate (50 mM Tris-HCl, 1% NP-40). Cellular debris was removed by centrifugation, and the supernatant fluids were reserved. The protein concentration was determined by the Bradford assay (9). Equal amounts (10 μg) of protein were resolved in the absence of SDS by electrophoresis in 10% polyacrylamide gels. Proteins were transferred to nitrocellulose membrane (Optitran; Schleicher & Schuell) and probed with primary and secondary antibodies. Immunoreactive proteins were detected by the ECL chemiluminescent system (Amersham).
Immunofluorescence assay.
Cells were seeded onto sterilized coverslips in six-well culture dishes and infected the following day with virus. Cells were washed twice with PBS and subsequently fixed with 4% paraformaldehyde for 20 min. Cells were permeabilized with PBST (PBS, 0.1% Triton X-100, 0.05% Tween 20) for 25 min at room temperature and incubated with blocking solution (PBST, 0.5% bovine serum albumin, 1% goat serum) for an additional 30 min. Cells were then incubated with primary antibody for 1 h at room temperature, washed three times in PBST, and incubated with secondary antibodies conjugated to Alexa-488 or Alexa-546 for 1 h. Slides were washed in double-distilled H2O and nuclei stained with Hoechst (0.5 μg/ml) for 5 min. Coverslips were sealed on slides and cells visualized using a Zeiss Atto Arc HBO 110W upright microscope.
Northern blot analysis.
RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's protocol and quantitated on a Nanodrop spectrophotometer. Northern blot analysis was performed as previously described (55). Briefly, total RNA (6 to 10 μg) was separated by electrophoresis on a 1% formaldehyde gel and transferred to Nytran Supercharge membranes by using a Turboblotter (Schleicher & Schuell) according to the manufacturer's instructions. Membranes were cross linked using a Stratalinker and probed overnight with 32P-labeled probes generated by random priming in ULTRAhyb (Ambion) hybridization buffer at 48°C. Membranes were then washed twice in low-stringency wash buffer (0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS) and twice in high-stringency wash buffer (2× SSC, 0.1% SDS) at 45°C and exposed to film for autoradiography.
EMSA.
NF-κB-specific electrophoretic mobility shift assays (EMSA) were performed as previously described (5, 61). Briefly, nuclear extracts were prepared by lysing cells in cytosolic isolation buffer (10 mM HEPES [pH 7.6], 60 mM KCl, 1 mM EDTA, 0.1% NP-40, 1 mM dithiothreitol, proteinase inhibitor cocktail) and sedimenting nuclei by centrifugation (3,000 × g for 10 min). Nuclei were then washed in lysis buffer lacking NP-40 and subsequently lysed in nuclear lysis buffer (20 mM Tris-HCl [pH 8], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, proteinase inhibitor cocktail) and quantitated by the Bradford assay (9). Nuclear extracts (10 μg) were then incubated for 10 min in 19 μl of extract buffer [10 mM Tris-HCl (pH 7.9), 50 mM NaCl, 0.5 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 7.5 mM MgCl2, 1 μg poly(dI-dC)]. Unlabeled specific-competitor (PRDII) or nonspecific-competitor (mPRDII) (5′-GGCAAATTGCGGCAAATTGC-3′) double-stranded oligonucleotides were added during this incubation step when indicated. Double-stranded oligonucleotides (5′-GGGAAATTCCGGGAAATTCC-3′) containing two NFκB binding sites from the positive regulatory domain II (PRDII) region of the IFN-β promoter (5) were end labeled using [γ-32P]ATP plus T4 polynucleotide kinase and added to the reaction mixture (250,000 counts per minute). Binding mixtures were incubated at room temperature for 30 min. Samples were separated on a prerun 6% polyacrylamide gel (60:1 polyacrylamide/bis ratio). Gels were then dried and exposed to film for autoradiography.
RESULTS
Inhibition of IRF-3 or NFκB blocks IFN-β induction.
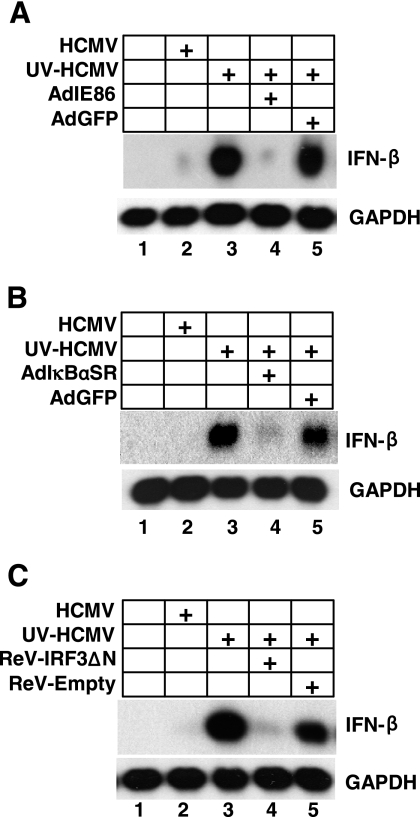
We and others have previously reported that IFN-β expression is attenuated during WT-HCMV infection, compared to that during infection with transcriptionally inactive UV-HCMV (Fig. 1A, compare lanes 2 and 3) (14, 51, 54). We have also demonstrated that the HCMV immediate-early 2 gene product IE86 can efficiently block the induction of IFN-β following infection with UV-HCMV (Fig. 1A) (54). However, the mechanism by which IE86 blocks IFN-β expression has not been elucidated.
FIG. 1.
IRF-3 and NFκB are both required for IFN-β expression. HFF cells were transduced with adenoviruses expressing GFP, IE86 (A), and IκBαSR (B) or retroviruses expressing IRF-3ΔN and empty vector (C). Transduced cells were then mock infected or infected with WT-HCMV or UV-inactivated HCMV. RNA was isolated 8 h postinfection and analyzed by Northern blot analysis for IFN-β and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcripts.
The IRF-3 and NFκB transcription factors are required for IFN-β transcription following infection with a number of viruses (2, 5, 20, 30, 44, 50, 58, 60). We therefore hypothesized that IE86 may target the IRF-3 and/or NFκB pathway to inhibit IFN-β expression. However, we first needed to confirm that both IRF-3 and NFκB are required for IFN-β expression following infection with UV-HCMV. To test this, HFFs were transduced with replication-defective viruses expressing either a nonphosphorylatable form of the IκBα repressor, IκBαSR (which blocks NFκB activation) (5), or a dominant-negative IRF-3 protein, IRF3ΔN (which blocks IRF-3 activation) (37). Transduced cells were then mock infected or infected with WT-HCMV or UV-inactivated HCMV. RNA was isolated 8 h postinfection and assayed for IFN-β expression by Northern blot analysis. As shown in Fig. 1B, expression of IκBαSR prior to infection with UV-HCMV efficiently inhibited the induction of IFN-β expression (Fig. 1B, compare lanes 3 and 4). IFN-β expression was also inhibited when IRF-3ΔN was expressed prior to UV-HCMV infection (Fig. 1C, compare lanes 3 and 4). However, transduction with a control virus did not block the induction of IFN-β following UV-HCMV infection. Together, these initial experiments establish that induction of IFN-β during UV-HCMV infection requires IRF-3 and NFκB, and thus, IE86 may target one or both of these pathways to block IFN-β expression.
IRF-3 activation and target gene expression are not attenuated by IE86.
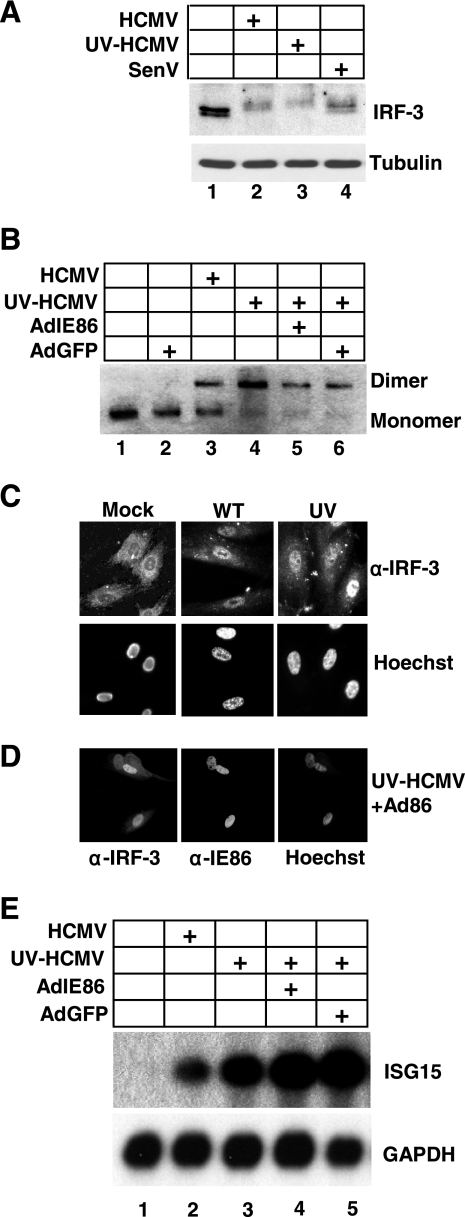
We next determined whether IRF-3 is activated during HCMV infection and whether IE86 is capable of blocking IRF-3 activation. IRF-3 is constitutively expressed and retained in the cytoplasm of uninfected cells. The C terminus of IRF-3 structurally obscures a nuclear import signal. Phosphorylation on key serine residues results in a conformation change which reveals the nuclear import signal and facilitates IRF-3 homodimerization and nuclear translocation (37). To assess IRF-3 activation during HCMV infection, we examined IRF-3 phosphorylation, the ability of IRF-3 to homodimerize, the ability of IRF-3 to translocate to the nucleus, and the ability of IRF-3 to activate gene expression. To monitor the phosphorylation state of IRF-3, HFF cells were either mock infected or infected with WT-HCMV, UV-HCMV, or Sendai virus. Cell lysates were harvested 6 h postinfection and assayed for IRF-3 expression by Western blot analysis. As shown in Fig. 2A, a slower-migrating form of IRF-3, which is consistent with hyperphosphorylation (37), is observed following infection with WT-HCMV, UV-HCMV, and Sendai virus, suggesting that IE86 does not block IRF-3 phosphorylation. To investigate this more directly, we assayed for IRF-3 homodimerization, which requires IRF-3 phosphorylation (37). HFF cells were transduced with replication-defective adenoviruses that express either IE86 or GFP and then mock infected or infected with WT-HCMV or UV-HCMV. As shown in Fig. 2B, only monomeric IRF-3 was present in mock-infected or GFP-transduced cells (Fig. 2B, lanes 1 and 2). However, IRF-3 was present as a dimer in cells that were infected with WT-HCMV or UV-HCMV (Fig. 2B, lanes 3 and 4). Importantly, expression of IE86 prior to UV-HCMV infection did not block IRF-3 dimerization (Fig. 2B, compare lanes 4 and 5). We next examined whether HCMV infection and/or IE86 expression could block IRF-3 nuclear translocation. HFF cells were mock infected or infected with WT-HCMV or UV-HCMV and fixed for immunofluorescent staining 3 h postinfection. As shown in Fig. 2C, IRF-3 is localized in the cytoplasm of mock-infected cells. However, upon infection with either WT-HCMV or UV-HCMV, IRF-3 translocates to the nucleus. Additionally, prior expression of IE86 had no effect on the ability of IRF-3 to translocate to the nucleus following UV-HCMV infection (Fig. 2D). Lastly, we determined whether IE86 could attenuate the expression of IRF-3-dependent genes. To test this, HFF cells were transduced with replication-defective adenoviruses that express either IE86 or GFP. Transduced cells were then mock infected or infected with WT-HCMV or UV-HCMV and assayed for expression of the IRF-3-dependent gene interferon-stimulated gene 15 (ISG15) (20, 26, 37). As shown in Fig. 2E, we observed a dramatic increase in ISG15 expression following both WT-HCMV and UV-HCMV infection. Importantly, prior expression of IE86 was unable to block the induction of ISG15 following UV-HCMV infection. However, prior expression of IRF-3ΔN efficiently blocked the expression of ISG15 (data not shown) (20). Similar results were obtained for the IRF-3-dependent genes ISG54, ISG60, and GBP1 (data not shown). Collectively, these results demonstrate that IE86 does not block IRF-3 phosphorylation, homodimerization, nuclear translocation, or gene expression.
FIG. 2.
IE86 does not block IRF-3 activation or target gene expression. (A) HFF cells were mock infected or infected with WT-HCMV, UV-HCMV, or Sendai virus. Cell lysates were prepared 6 h postinfection and assayed for IRF-3 and tubulin expression by Western blot analysis. (B) HFF cells were transduced with adenoviruses expressing IE86 or GFP. Twenty-four hours posttransduction, cells were mock infected or infected with WT-HCMV or UV-HCMV at a multiplicity of 5 PFU/cell. Cell extracts were prepared 6 h postinfection and assayed for IRF-3 dimerization by native gel electrophoresis and Western blot analysis using an IRF-3 antibody. HFF cells (C) or HFF cells transduced with adenovirus expressing IE86 (D) were seeded onto coverslips and either mock infected, infected with WT-HCMV, or infected with UV-HCMV. Cells were fixed 3 h postinfection and assayed for IRF-3 and IE86 localization by immunofluorescence assay. Nuclei were stained with Hoechst. (E) HFF cells were transduced with adenoviruses expressing IE86 or GFP for 24 h. Cells were then infected with WT-HCMV or UV-HCMV at a multiplicity of 5 PFU/cell. RNA was isolated 8 h postinfection and assayed for ISG15 and GAPDH expression by Northern blot analysis.
NFκB DNA binding is attenuated by HCMV gene expression.
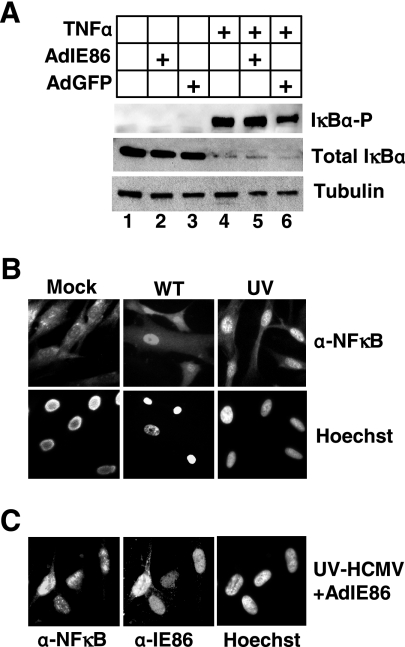
We next assayed the ability of IE86 to prevent NFκB activation during HCMV infection. Normally retained in the cytoplasm bound to the inhibitor complex IκB, NFκB is activated by the phosphorylation and degradation of the IκB inhibitor, which unmasks the nuclear localization signal of NFκB and facilitates rapid nuclear translocation (3). To assay the activation of NFκB, we first assayed for the phosphorylation and degradation of the inhibitor IκBα following treatment with TNF-α, a potent inducer of NFκB activation in the presence and absence of IE86. HFF cells were transduced with adenoviruses expressing either IE86 or GFP and treated with 50 ng/ml TNF-α for 30 min, and cell lysates were prepared for Western blot analysis. As shown in Fig. 3A, treatment with TNF-α resulted in the phosphorylation and degradation of IκBα (Fig. 3A, lane 4). In addition, prior expression of IE86 did not prevent TNF-α-induced IκBα phosphorylation or degradation (Fig. 3A, compare lanes 4 and 5).
FIG. 3.
NFκB is activated during HCMV infection. (A) HFF cells were mock transduced or transduced with adenoviruses expressing IE86 or GFP and treated with 50 ng/ml TNF-α. Cell lysates were prepared 30 min posttreatment and assayed for phosphorylated IκBα, total IκBα, and tubulin by Western blot analysis. HFF cells (B) or HFF cells transduced with adenovirus expressing IE86 (C) were seeded onto coverslips and either mock infected or infected with WT-HCMV or UV-HCMV. Cells were fixed 3 h postinfection and assayed for NFκB (p50) and IE86 localization by an immunofluorescence assay. Nuclei were stained with Hoechst.
We next determined whether HCMV infection or IE86 expression could block the nuclear translocation of NFκB by examining the subcellular localization of the p50 subunit of NFκB. As shown in Fig. 3B, NFκB is predominantly localized in the cytoplasm of mock-infected cells. However, upon infection with WT-HCMV or UV-HCMV, NFκB (p50) is rapidly translocated to the nucleus. Again, prior expression of IE86 was unable to block the nuclear translocation of p50 following infection with UV-HCMV (Fig. 3C) or TNF-α treatment (data not shown). Similar results were obtained when we examined the nuclear translocation of the p65 subunit of NFκB (data not shown).
IE86 inhibits NFκB DNA binding.
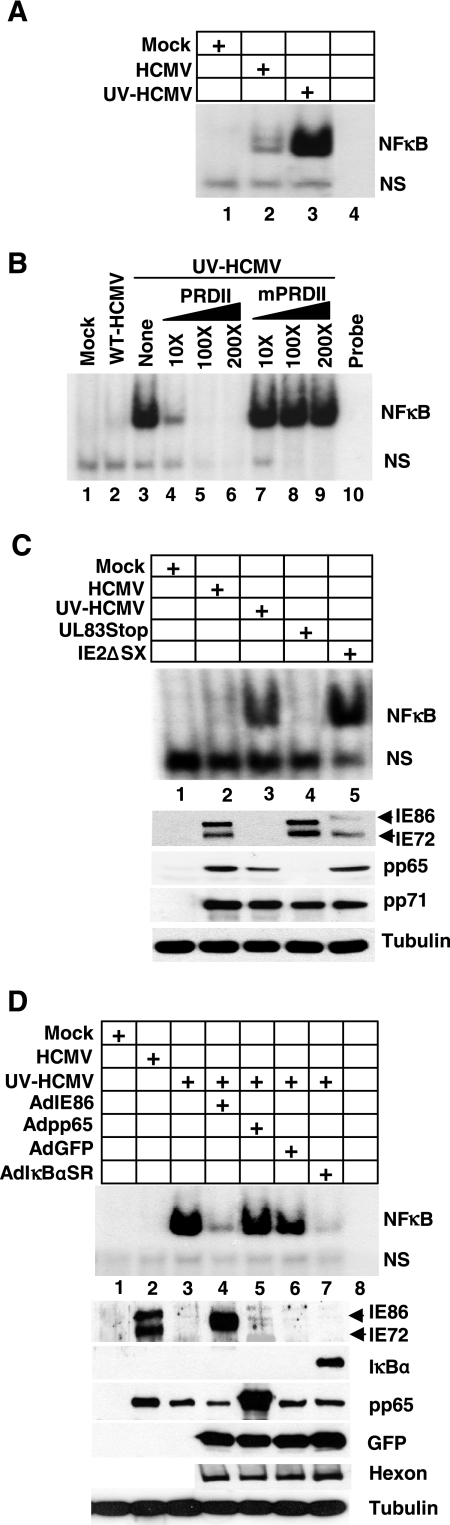
Since NFκB is activated and translocated to the nucleus in the presence of IE86, we determined whether HCMV infection or IE86 expression could attenuate the binding of NFκB to the IFN-β promoter. Therefore, EMSA were performed, using the NFκB binding site within the IFN-β promoter as probe (5). Cells were mock infected or infected with WT-HCMV or UV-HCMV. Nuclear extracts were prepared, incubated with labeled probe, and assayed for NFκB binding. As shown in Fig. 4A, NFκB binding was observed following both WT-HCMV and UV-HCMV infection. However, NFκB binding was significantly enhanced following infection with UV-HCMV compared to WT-HCMV infection (Fig. 4, compare lanes 2 and 3). To confirm that the binding observed following infection with UV-HCMV was specific for the NFκB binding site within the IFN-β promoter, we performed a competition assay with unlabeled probe. As shown in Fig. 4B, the addition of unlabeled probe (PRDII) to extracts from UV-HCMV-infected cells efficiently blocked NFκB binding in a dose-dependent manner. However, NFκB binding was not inhibited when unlabeled probe containing a 2-base-pair mismatch within the NFκB binding site (mPRDII) was added to the reaction mixture. These data suggest that HCMV gene expression and potentially IE86 may attenuate the binding of NFκB to its target sequence within the IFN-β promoter. To test whether IE86 blocks NFκB DNA binding during HCMV infection, we utilized an IE2 mutant virus termed IE2ΔSX, in which IE86 expression is both attenuated and severely delayed compared to wild-type infection (49, 55). Cells were infected with WT-HCMV, UV-HCMV, IE2ΔSX, or a pp65 mutant virus termed UL83Stop (55). Nuclear extracts were prepared 6 h postinfection and assayed for NFκB binding. As shown in Fig. 4C, infection with the IE2ΔSX virus results in a dramatic increase in NFκB DNA binding compared to WT-HCMV infection (Fig. 4C, compare lanes 2 and 5). In addition, NFκB DNA binding following infection with the IE2ΔSX virus was similar to that observed following infection with UV-HCMV (Fig. 4C, compare lanes 3 and 5), whereas the level of NFκB binding following infection with the UL83Stop virus was only slightly above that observed in mock-infected cells. A Western blot is included in Fig. 4C to show the expression of IE86, pp65, and pp71 following infection with these viruses.
FIG. 4.
IE86 attenuates NFκB DNA binding activity. (A) HFF cells were mock infected or infected with WT-HCMV or UV-HCMV at a multiplicity of 5 PFU/cell. Nuclear extracts were prepared 6 h postinfection and assayed by EMSA for binding of NFκB to the PRDII region of the IFN-β promoter. NS indicates a nonspecific shift. (B) HFF cells were infected with WT-HCMV or UV-HCMV. Six hours postinfection, nuclear lysates were isolated and assayed for NFκB binding. A competition analysis was performed on UV-HCMV extracts by adding unlabeled specific-competitor oligonucleotide probe (PRDII) or a mutated probe sequence (mPRDII) in increasing concentrations to the binding mixture to confirm the specificity of the NFκB DNA binding. (C) HFF cells were infected with HCMV, UV-HCMV, UL83Stop virus, or IE2ΔSX virus at a multiplicity of 5 PFU/cell. Nuclear extracts were prepared 6 h postinfection and assayed for NFκB binding by EMSA. A Western blot is also included to confirm the expression of the various viral proteins. (D) HFF cells were transduced with adenoviruses expressing IE86, pp65, GFP, or IκBαSR and then infected at a multiplicity of 5 PFU/cell with UV-HCMV. Nuclear lysates were prepared 6 h postinfection and assayed for NFκB binding by EMSA. A Western blot is included to confirm the expression of IE86, pp65, IκBα, GFP, adenovirus hexon, and tubulin.
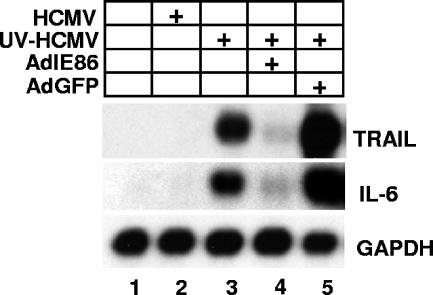
We next determined whether IE86 expression in the absence of virus infection could block UV-HCMV-induced NFκB DNA binding. Cells were transduced with adenovirus expressing IE86, pp65, IκBαSR, or GFP. Transduced cells were infected with UV-HCMV and nuclear extracts prepared for EMSA 6 h postinfection. As shown in Fig. 4D, prior expression of IE86 or IκBαSR efficiently inhibited NFκB DNA binding following UV-HCMV infection (Fig. 4D, compare lanes 4 and 7 to lane 3). However, prior expression of pp65 or GFP had no effect on NFκB DNA binding activity. A Western blot is included in Fig. 4D to confirm expression of IE86, pp65, IκBα, and GFP. Blots were also probed for the structural adenovirus hexon protein to confirm that an approximately equal number of virus particles was used for each transduction. Finally, we assayed for the ability of IE86 to attenuate NFκB-dependent gene expression. As shown in Fig. 5, expression of the NFκB-dependent genes tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) (6) and interleukin-6 (IL-6) (36) was induced following infection with UV-HCMV. However, expression of IE86 prior to infection with UV-HCMV effectively inhibited the expression of TRAIL and IL-6, whereas expression of GFP had no effect on their expression (Fig. 5).
FIG. 5.
IE86 attenuates NFκB target gene expression. Cells were transduced with adenovirus expressing either IE86 or GFP and then infected at a multiplicity of 5 PFU/cell with UV-HCMV. RNA was isolated 8 h postinfection and assayed for TRAIL, IL-6, and GAPDH expression by Northern blot analysis.
IE86 blocks virus- and TNF-α-induced NFκB DNA binding and target gene expression.
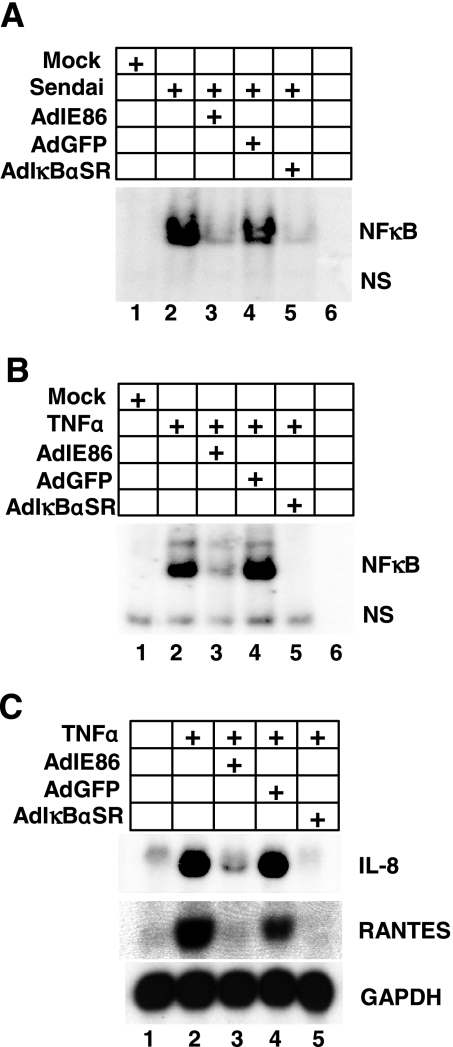
We next determined whether IE86 was capable of blocking NFκB-dependent DNA binding and/or gene expression following exposure to stimuli other than HCMV infection. To test this, cells were transduced with adenoviruses that express IE86 or GFP and then infected with Sendai virus or treated with TNF-α and assayed for NFκB DNA binding activity. Both Sendai virus and TNF-α are potent inducers of NFκB (5). As shown in Fig. 6A, infection with Sendai virus results in a robust induction of binding of NFκB to the IFN-β promoter (Fig. 6A, compare lanes 1 and 2). Interestingly, prior expression of IE86 inhibited NFκB DNA binding induced by Sendai virus infection (Fig. 6A, compare lanes 2 and 3). In addition, expression of IE86 inhibited the NFκB DNA binding activity observed following TNF-α treatment (Fig. 6B, compare lanes 2 and 3). Finally, we examined the effect of IE86 expression on TNF-α-induced gene expression. Cells were transduced with adenovirus expressing IE86, GFP, or IκBαSR and then treated with TNF-α. RNA was isolated 6 h posttreatment and assayed by Northern blot analysis for expression of the TNF-α-induced, NFκB-dependent genes IL-8 (35) and regulated upon activation normal T-cell expressed and secreted (RANTES) (42). As shown in Fig. 6C, treatment of cells with TNF-α induced the expression of IL-8 and RANTES (Fig. 6C, compare lanes 1 and 2). Expression of IL-8 and RANTES following TNF-α treatment is also dependent on NFκB signaling since their expression is effectively blocked in the presence of the IκBαSR (Fig. 6C, compare lanes 2 and 5). Interestingly, expression of IE86 prior to TNF-α treatment effectively blocked the expression of IL-8 and RANTES (Fig. 6C, compare lanes 2 and 3). Taken together, our results demonstrate that IE86 effectively inhibits both virus-induced and TNF-α-induced NFκB DNA binding and gene expression.
FIG. 6.
IE86 attenuates NFκB DNA binding following TNF-α treatment. HFF cells were transduced with adenovirus expressing IE86, GFP, or IκBαSR and then infected with Sendai virus at 100 HAU/ml (A) or treated with 50 ng/ml TNF-α (B). Nuclear extracts were prepared 6 h postinfection or -treatment and assayed for NFκB binding by EMSA. NS indicates a nonspecific shift. (C) Cells were transduced with adenovirus expressing IE86, GFP, or IκBαSR for 24 h. Transduced cells were then treated with TNF-α. RNA was isolated 6 h posttreatment and assayed for IL-8 and RANTES expression by Northern blot analysis.
DISCUSSION
Previous reports have demonstrated that HCMV can attenuate the expression of IFN-β and proinflammatory chemokines and that this attenuation is dependent on a newly synthesized viral protein expressed early during infection (14, 51). We recently demonstrated that the HCMV immediate-early 2 gene product IE86 can efficiently block the induction of IFN-β and a number of chemokines following HCMV infection (54, 55). However, the mechanism by which IE86 inhibits IFN-β and chemokine expression has remained elusive. Therefore, we set out to investigate how IE86 attenuates IFN-β and chemokine production during HCMV infection.
IRF-3 and NFκB are required for UV-HCMV-induced IFN-β expression.
The IRF-3 and NFκB pathways are required for IFN-β induction following certain viral infections (2, 5, 33, 50). In addition, a number of viruses express proteins that specifically target the IRF-3 and/or NFκB pathway in order to inhibit the expression of IFN-β (7, 10, 22, 25, 39, 48, 52, 57). Therefore, we examined whether either pathway is required for the induction of IFN-β observed following infection with UV-HCMV. Using dominant negative repressors that block activation of IRF-3 or NFκB, we were able to demonstrate that inhibition of either pathway will block IFN-β induction following infection with UV-inactivated HCMV (Fig. 1B and C). Therefore, IE86 may target either the IRF-3 or the NFκB pathway to block IFN-β expression.
IE86 does not target IRF-3 activation or gene expression.
Others have demonstrated that IRF-3 is activated at early times after HCMV infection (8, 13, 19, 20, 28, 43, 46, 59). Our results confirm these previous observations and demonstrate that IRF-3 is phosphorylated, homodimerizes, translocates to the nucleus, and activates target gene expression following both wild-type and UV-HCMV infection (Fig. 2A to C and E), suggesting that IE86 does not inhibit IRF-3 activation. Using a replication-defective adenovirus that expresses IE86, we were able to directly examine whether IE86 is capable of blocking IRF-3 activation. As shown in Fig. 2, expression of IE86 prior to infection with UV-HCMV did not inhibit IRF-3 homodimerization, nuclear translocation, or target gene expression. These results are in agreement with a recent report by DeFilippis et al. which used small interfering RNA directed against IRF-3 to demonstrate that a subset of genes induced during WT-HCMV infection, including IFN-β and ISG15, requires IRF-3 activation (20). Together, these results demonstrate that IRF-3 is activated and that IE86 does not target the IRF-3 pathway during HCMV infection.
IE86 attenuates NFκB activation by inhibiting DNA binding activity.
Analysis of the early events in NFκB activation revealed that IE86 does not prevent the phosphorylation or degradation of the α subunit of the IκB inhibitor (Fig. 3A), nor does IE86 prevent the nuclear translocation of the NFκB subunit p50 or p65 (Fig. 3B and C and data not shown). However, using electrophoretic mobility shift assays, we demonstrate that IE86 can attenuate NFκB DNA binding activity. First, infection with wild-type HCMV results in an increase in the binding of NFκB to the IFN-β promoter compared to what was found for mock-infected cells (Fig. 4A, compare lanes 1 to 2), confirming previous studies that demonstrate HCMV infection results for both NFκB activation and DNA binding activity (18, 21, 23, 61). However, NFκB DNA binding was significantly enhanced following infection with UV-HCMV (Fig. 4A), suggesting that viral gene expression and, more specifically, IE86 may be involved in attenuating NFκB DNA binding activity. We used two independent methods to determine that IE86 is capable of attenuating NFκB DNA binding. First, we utilized the IE2ΔSX virus, which has amino acids 136 to 290 deleted from exon 5 of IE2 (49). The IE2ΔSX virus is viable but expresses IE86 at dramatically reduced levels and with delayed kinetics compared to those for IE86 expression during wild-type or revertant virus infection (49). We have previously demonstrated that infection with the IE2ΔSX virus results in an increase in IFN-β and RANTES transcript accumulation similar to that observed following UV-HCMV infection (55). Therefore, if IE86 is involved in blocking NFκB DNA binding, we would predict that infection with the IE2ΔSX virus would result in an increase in NFκB DNA binding compared to wild-type infection. As demonstrated in Fig. 4B, NFκB DNA binding was dramatically enhanced following infection with the IE2ΔSX virus compared to that following wild-type infection (Fig. 4C, compare lanes 2 and 5). In addition, NFκB DNA binding following infection with the IE2ΔSX virus was comparable to that observed following infection with UV-HCMV (Fig. 4C, compare lanes 3 and 5). Infection with a control mutant virus, UL83Stop, had no effect on NFκB DNA binding activity and looked identical to wild-type HCMV infection (Fig. 4C, compare lanes 2 and 4). We also utilized an adenovirus expression system to demonstrate that expression of IE86 prior to infection with UV-HCMV inhibited the binding of NFκB to the IFN-β promoter, whereas expression of the HCMV tegument protein pp65 or GFP had no effect on NFκB DNA binding (Fig. 4D). In addition, we showed that IE86 can inhibit the expression of the NFκB-dependent genes TRAIL and IL-6 during HCMV infection. We also demonstrated that IE86 can attenuate NFκB DNA binding and gene expression in the absence of HCMV infection. Using Sendai virus or the proinflammatory cytokine TNF-α as an inducer of NFκB, we showed that IE86 can attenuate both Sendai virus- and TNF-α-induced NFκB DNA binding (Fig. 6A and B) and TNF-α-induced NFκB-dependent gene expression (Fig. 6C).
Our results show that IE86 can function as an NFκB antagonist and suppress both virus- and TNF-α-induced NFκB DNA binding activity and subsequent NFκB-dependent cytokine and chemokine gene expression. This suppression of cytokine and chemokine expression during HCMV infection likely provides for a cellular environment that is conducive to viral replication and persistence. This report identifies a novel strategy employed by HCMV to attenuate the host antiviral cytokine and chemokine response early in infection by suppressing NFκB DNA binding. This augments a growing list of mechanisms by which HCMV inhibits cytokine and/or chemokine function. These mechanisms include the expression of chemokine mimics (45), the expression of chemokine binding proteins (56), and the expression of G-protein-coupled chemokine receptors (4, 47, 53). Additionally, a recent report suggests that HCMV may express additional proteins involved in blocking inflammatory chemokine expression. Jarvis et al. demonstrated that HCMV expresses a late protein during infection that is capable of attenuating both TNF-α- and IL-1β-induced chemokine expression (32). Interestingly, the mechanism by which this late viral protein inhibits chemokine expression involves blocking the activation of NFκB by preventing IκBα phosphorylation. Therefore, it is likely that HCMV may express at least two different proteins (IE86 and a yet-unidentified late protein) that target NFκB activation at different steps and function at different stages during HCMV replication to block IFN-β and chemokine expression.
The mechanism by which IE86 attenuates NFκB DNA binding is currently unclear. To date, a direct interaction of IE86 with members of the NFκB family has not been reported. We have been unable to detect an interaction between IE86 and the NFκB subunit p50 or p65 during HCMV infection (data not shown). There is, however, significant overlap between the interacting partners of the NFκB family members and those of IE86, including TATA binding protein, CBP, and Jun (15).Therefore, the binding of IE86 to the NFκB subunits or a necessary interacting partner may attenuate sequence-specific DNA binding. Additionally, NFκB activation is dependent upon phosphorylation and acetylation of the p65 subunit for maximal DNA binding and transcriptional activation (16, 17). An IE86-mediated block to p65 phosphorylation or acetylation may account for the attenuated NFκB DNA binding. We are currently investigating these possibilities in an attempt to reveal the mechanism by which IE86 inhibits NFκB DNA binding.
Acknowledgments
This work was supported in part by University of Minnesota grant in aid no. 19990 and Minnesota Medical Foundation grant no. 3624-9221-06 to W.A.B.
We are grateful to Deborah Spector, Gary Nolan, Michael Gale, Jr., Thomas Shenk, and Aubrey Thompson for reagents. We also thank Michael Gale, Jr., Aubrey Thompson, and Stacy Cantrell for critically reading the manuscript.
REFERENCES
- 1.Abate, D. A., S. Watanabe, and E. S. Mocarski. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 78:10995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 3.Aggarwal, B. B., G. Sethi, A. Nair, and H. Ichikawa. 2006. Nuclear factor-{kappa}B: a holy grail in cancer prevention and therapy. Curr. Signal Transduct. Ther. 1:25-52. [Google Scholar]
- 4.Alcami, A. 2003. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3:36-50. [DOI] [PubMed] [Google Scholar]
- 5.Algarte, M., H. Nguyen, C. Heylbroeck, R. Lin, and J. Hiscott. 1999. IκB-mediated inhibition of virus-induced beta interferon transcription. J. Virol. 73:2694-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baetu, T. M., H. Kwon, S. Sharma, N. Grandvaux, and J. Hiscott. 2001. Disruption of NF-kappaB signaling reveals a novel role for NF-kappaB in the regulation of TNF-related apoptosis-inducing ligand expression. J. Immunol. 167:3164-3173. [DOI] [PubMed] [Google Scholar]
- 7.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehme, K. W., J. Singh, S. T. Perry, and T. Compton. 2004. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J. Virol. 78:1202-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Breiman, A., N. Grandvaux, R. Lin, C. Ottone, S. Akira, M. Yoneyama, T. Fujita, J. Hiscott, and E. F. Meurs. 2005. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKɛ. J. Virol. 79:3969-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bresnahan, W. A., I. Boldogh, T. Ma, T. Albrecht, and E. A. Thompson. 1996. Cyclin E/Cdk2 activity is controlled by different mechanisms in the G0 and G1 phases of the cell cycle. Cell Growth Differ. 7:1283-1290. [PubMed] [Google Scholar]
- 12.Bresnahan, W. A., G. E. Hultman, and T. Shenk. 2000. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J. Virol. 74:10816-10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Browne, E. P., and T. Shenk. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. USA 100:11439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castillo, J. P., and T. F. Kowalik. 2002. Human cytomegalovirus immediate early proteins and cell growth control. Gene 290:19-34. [DOI] [PubMed] [Google Scholar]
- 16.Chen, L. F., and W. C. Greene. 2004. Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell Biol. 5:392-401. [DOI] [PubMed] [Google Scholar]
- 17.Chen, L. F., S. A. Williams, Y. Mu, H. Nakano, J. M. Duerr, L. Buckbinder, and W. C. Greene. 2005. NF-κB RelA phosphorylation regulates RelA acetylation. Mol. Cell. Biol. 25:7966-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compton, T., E. A. Kurt-Jones, K. W. Boehme, J. Belko, E. Latz, D. T. Golenbock, and R. W. Finberg. 2003. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J. Virol. 77:4588-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeFilippis, V., and K. Fruh. 2005. Rhesus cytomegalovirus particles prevent activation of interferon regulatory factor 3. J. Virol. 79:6419-6431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeFilippis, V. R., B. Robinson, T. M. Keck, S. G. Hansen, J. A. Nelson, and K. J. Fruh. 2006. Interferon regulatory factor 3 is necessary for induction of antiviral genes during human cytomegalovirus infection. J. Virol. 80:1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeMeritt, I. B., L. E. Milford, and A. D. Yurochko. 2004. Activation of the NF-κB pathway in human cytomegalovirus-infected cells is necessary for efficient transactivation of the major immediate-early promoter. J. Virol. 78:4498-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donelan, N. R., B. Dauber, X. Wang, C. F. Basler, T. Wolff, and A. Garcia-Sastre. 2004. The N- and C-terminal domains of the NS1 protein of influenza B virus can independently inhibit IRF-3 and beta interferon promoter activation. J. Virol. 78:11574-11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eickhoff, J. E., and M. Cotten. 2005. NF-kappaB activation can mediate inhibition of human cytomegalovirus replication. J. Gen. Virol. 86:285-295. [DOI] [PubMed] [Google Scholar]
- 24.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 102:2986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graff, J. W., D. N. Mitzel, C. M. Weisend, M. L. Flenniken, and M. E. Hardy. 2002. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J. Virol. 76:9545-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grandvaux, N., B. R. tenOever, M. J. Servant, and J. Hiscott. 2002. The interferon antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 15:259-267. [DOI] [PubMed] [Google Scholar]
- 28.Gravel, S. P., and M. J. Servant. 2005. Roles of an IkappaB kinase-related pathway in human cytomegalovirus-infected vascular smooth muscle cells: a molecular link in pathogen-induced proatherosclerotic conditions. J. Biol. Chem. 280:7477-7486. [DOI] [PubMed] [Google Scholar]
- 29.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiscott, J., P. Pitha, P. Genin, H. Nguyen, C. Heylbroeck, Y. Mamane, M. Algarte, and R. Lin. 1999. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 19:1-13. [DOI] [PubMed] [Google Scholar]
- 31.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375-388. [DOI] [PubMed] [Google Scholar]
- 32.Jarvis, M. A., J. A. Borton, A. M. Keech, J. Wong, W. J. Britt, B. E. Magun, and J. A. Nelson. 2006. Human cytomegalovirus attenuates interleukin-1β and tumor necrosis factor alpha proinflammatory signaling by inhibition of NF-κB activation. J. Virol. 80:5588-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juang, Y. T., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinsella, T. M., and G. P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405-1413. [DOI] [PubMed] [Google Scholar]
- 35.Kunsch, C., and C. A. Rosen. 1993. NF-kappa B subunit-specific regulation of the interleukin-8 promoter. Mol. Cell. Biol. 13:6137-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Libermann, T. A., and D. Baltimore. 1990. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol. Cell. Biol. 10:2327-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 103:6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 40.Mogensen, T. H., and S. R. Paludan. 2001. Molecular pathways in virus-induced cytokine production. Microbiol. Mol. Biol. Rev. 65:131-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mogensen, T. H., and S. R. Paludan. 2001. Virus-cell interactions: impact on cytokine production, immune evasion and tumor growth. Eur. Cytokine Netw. 12:382-390. [PubMed] [Google Scholar]
- 42.Moriuchi, H., M. Moriuchi, and A. S. Fauci. 1997. Nuclear factor-kappa B potently up-regulates the promoter activity of RANTES, a chemokine that blocks HIV infection. J. Immunol. 158:3483-3491. [PubMed] [Google Scholar]
- 43.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noah, D. L., M. A. Blum, and B. Sherry. 1999. Interferon regulatory factor 3 is required for viral induction of beta interferon in primary cardiac myocyte cultures. J. Virol. 73:10208-10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Penfold, M. E., D. J. Dairaghi, G. M. Duke, N. Saederup, E. S. Mocarski, G. W. Kemble, and T. J. Schall. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. USA 96:9839-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Randolph-Habecker, J. R., B. Rahill, B. Torok-Storb, J. Vieira, P. E. Kolattukudy, B. H. Rovin, and D. D. Sedmak. 2002. The expression of the cytomegalovirus chemokine receptor homolog US28 sequesters biologically active CC chemokines and alters IL-8 production. Cytokine 19:37-46. [DOI] [PubMed] [Google Scholar]
- 48.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanchez, V., C. L. Clark, J. Y. Yen, R. Dwarakanath, and D. H. Spector. 2002. Viable human cytomegalovirus recombinant virus with an internal deletion of the IE2 86 gene affects late stages of viral replication. J. Virol. 76:2973-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schafer, S. L., R. Lin, P. A. Moore, J. Hiscott, and P. M. Pitha. 1998. Regulation of type I interferon gene expression by interferon regulatory factor-3. J. Biol. Chem. 273:2714-2720. [DOI] [PubMed] [Google Scholar]
- 51.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spann, K. M., K. C. Tran, and P. L. Collins. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J. Virol. 79:5353-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stropes, M. P., and W. E. Miller. 2004. Signaling and regulation of G-protein coupled receptors encoded by cytomegaloviruses. Biochem. Cell Biol. 82:636-642. [DOI] [PubMed] [Google Scholar]
- 54.Taylor, R. T., and W. A. Bresnahan. 2005. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J. Virol. 79:3873-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor, R. T., and W. A. Bresnahan. 2006. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J. Virol. 80:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, D., W. Bresnahan, and T. Shenk. 2004. Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc. Natl. Acad. Sci. USA 101:16642-16647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 59.Yang, S., J. Netterwald, W. Wang, and H. Zhu. 2005. Characterization of the elements and proteins responsible for interferon-stimulated gene induction by human cytomegalovirus. J. Virol. 79:5027-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yie, J., K. Senger, and D. Thanos. 1999. Mechanism by which the IFN-beta enhanceosome activates transcription. Proc. Natl. Acad. Sci. USA 96:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yurochko, A. D., E. S. Hwang, L. Rasmussen, S. Keay, L. Pereira, and E. S. Huang. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J. Virol. 71:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yurochko, A. D., T. F. Kowalik, S. M. Huong, and E. S. Huang. 1995. Human cytomegalovirus upregulates NF-kappa B activity by transactivating the NF-kappa B p105/p50 and p65 promoters. J. Virol. 69:5391-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]