Abstract
A rotavirus sample collection from 19 consecutive years was used to investigate the heterogeneity and the dynamics of evolution of G1 rotavirus strains in a geographically defined population. Phylogenetic analysis of the VP7 gene sequences of G1P[8] human rotavirus strains showed the circulation of a heterogeneous population comprising three lineages and seven sublineages. Increases in the circulation of G1 rotaviruses were apparently associated with the introduction of novel G1 strains that exhibited multiple amino acid changes in antigenic regions involved in rotavirus neutralization compared to the strains circulating in the previous years. The emergence and/or introduction of G1 antigenic variants might be responsible for the continuous circulation of G1 rotaviruses in the local population, with the various lineages and sublineages appearing, disappearing, or cocirculating in an alternate fashion under the influence of immune-pressure mechanisms. Sequence analysis of VP4-encoding genes of the G1 strains revealed that the older strains were associated with a unique VP4 lineage, while a novel VP4 lineage emerged after 1995. The introduction of human rotavirus vaccines might alter the forces and balances that drive rotavirus evolution and determine the spread of novel strains that are antigenically different from those included in the vaccine formulations. The continuous emergence of VP7-VP4 gene combinations in human rotavirus strains should be taken into consideration when devising vaccination strategies.
Rotavirus infections are the most important cause of severe gastroenteritis in infants and young children throughout the world and are responsible in developed countries for approximately one-third of diarrhea-associated hospitalizations and in developing countries for 400,000 to 500,000 deaths annually (55). Accordingly, the development of rotavirus vaccines is considered a global priority (48).
On the basis of the outer layer viral proteins VP7 and VP4, both of which elicit the production of neutralizing antibodies, human rotaviruses (HRVs) are classified into 11 G (VP7-specific) and 10 P (VP4-specific) types (23, 56, 59). Rotavirus strains of the G1 to G4 and G9 types account for more than 90% of the diarrhea episodes in children (38, 41, 59). The G1, G3, G4, and G9 types are usually associated with P[8] specificity, while G2 is associated with P[4] (23). The role of type-specific (homotypic) versus cross-protective (heterotypic) immunity for protection against rotavirus infection and disease is still debated. Taking into account the antigenic heterogeneity of rotaviruses, polyvalent vaccines have been developed. A live, attenuated rhesus-human reassortant tetravalent (G1 to G4) vaccine, RotaShield (12, 52, 61), was licensed in the United States in 1998 and withdrawn 1 year after its release as it was associated with an increased risk of intussusception (13, 49). Since then, a variety of approaches to the development of effective rotavirus vaccines have been undertaken. Recently, a promising monovalent G1P1A[8] human attenuated vaccine, Rotarix, which is reported to induce good heterotypic protection, has been licensed (20, 57). This formulation is derived from the fact that both P1A[8] and G1 specificities are unanimously acknowledged as the most prevalent and ubiquitous (27, 59). Documented, uninterrupted rotavirus surveillance has been conducted since 1985 in Palermo, Italy (4, 5). In 15 of the 20 years examined, G1P[8] HRVs have represented the first most common rotavirus serotype circulating. Fluctuations of the other rotavirus serotypes were observed over time, characterized by peaks of G4P[8] strains in 1990 to 1993, 1999 to 2001 and 2003; of G2P[4] strains in 1996 and 1997; and of G9P[8] strains in 1999 and 2000 (1, 2). Cross-sectional analyses showed that the epidemics of G2P[4] and G4P[8] were due to the introduction into the infant population of Palermo of new rotavirus strains rather than to the reemergence of old epidemic strains (1, 2).
Genetic and antigenic drift of viruses are regarded as important mechanisms driven by specific immunologic pressures. Positive selection of single amino acid mutations in defined virus epitopes steadily generates virus diversification. Sequence analysis of rotavirus VP7 has mapped highly divergent regions (VR1 to VR9) across the different rotavirus serotypes, and four such divergent regions are regarded as major antigen sites, namely, region A (amino acids [aa] 87 to 101), B (aa 143 to 152), C (aa 208 to 223), and F (aa 235 to 242) (14, 22, 39, 51). In addition, sequence analyses of VP7 of G1 HRVs has revealed the existence of at least four VP7 genetic lineages (7, 37), and VP7 sequence analysis of escape mutants selected with G1 serotype-specific monoclonal antibodies (MAbs) has given information on the amino acids involved in virus neutralization (18, 64).
In view of a possible introduction of the monovalent G1 vaccine in Europe, it is important to investigate the pattern of antigenic and genetic evolution of G1 human rotavirus isolates obtained from clinical samples over time to acquire information on the mechanisms of distribution and selection of rotavirus strains and lineages. The purpose of the present study was to analyze the VP7 and VP4 coding genes to evaluate the genetic variability of G1P[8] rotavirus strains infecting the infant population of Palermo, Sicily, over a 19-year period.
MATERIALS AND METHODS
Samples.
Two to four strains were selected on the basis of the electropherotypes predominant in each year, of the availability of fecal samples, and of double-stranded RNA conservation. Forty-one G1 rotavirus strains were selected from a total of 780 G1 strains obtained from children less than 5 years old hospitalized with acute gastroenteritis at the G. Di Cristina Children's Hospital of Palermo from 1986 to 2004. The VP7 serotype and the VP6 subgroup specificities of all of these strains were initially determined with MAbs, and all of the strains exhibited G1 serotype and subgroup II specificities.
Determination of serotype and subgroup.
G typing was performed according to previously published methods (4), with MAbs RV4:2, RV5:3, RV3:1, and ST3-3:1, respectively, which are reactive with G1-to-G4-specific viral protein VP7 (17). Subgrouping was performed with subgroup I- and subgroup II-specific MAbs (255/60 and 631/9) reactive with viral protein VP6, as described elsewhere (4).
RNA extraction and RT-PCR.
Rotavirus RNA was extracted from 10% fecal suspensions as described by Boom et al. (10). The extracted RNA was resuspended in RNase-free sterile water and used for reverse transcription (RT)-PCR with random primers (34). To determine the G and P genotypes, specimens were analyzed with type-specific primers by a heminested RT-PCR strategy. In a first PCR round, a 1,060-nucleotide-long fragment of the gene encoding VP7 was amplified with a generic oligonucleotide primer pair (Beg9-End9) (28, 36), and the G types were subsequently predicted in a second PCR round with a pool of internal primers specific for the G1, G2, G3, G4, and G9 genotypes in combination with the reverse consensus primer (28, 36). Similarly, an 876-bp fragment of the fourth genome segment, encompassing the VP8* portion of VP4, was amplified with generic primers Con3 and Con2, and P genotyping was carried out with internal primers specific for the P[4], P[6], P[8], and P[9] genotypes (26, 36). The PCR mixtures were prepared and thermal cycling was performed as previously described (36). Amplicons were analyzed by electrophoresis with a 2% SeaKem LE (Cambrex Bio Science Rockland Inc., Rockland, ME) agarose gel in Tris-acetate-EDTA at 6 V/cm for 60 min.
Sequence and phylogenetic analyses.
Amplicons of the genes encoding VP7 and VP4 were directly sequenced with primers Beg9-End9 and Con2-Con3, respectively. Amplicons were purified with GeneClean purification spin columns (Q-biogene, Cambridge, United Kingdom) prior to sequencing with the CEQ2000 Dye Terminator Cycle Sequencing Quick Start Kit (Beckman-Coulter). All methods were carried out by following the manufacturers' instructions. Sequences were resolved with an automated sequencer (CEQ; Beckman-Coulter). Sequence alignment was performed with CLUSTAL W (60). Phylogenetic analysis was carried out with the MEGA software, version 3.0 (40), with the Kimura two-parameter model as a method of substitution and the neighbor-joining method to construct the phylogenetic tree. The statistical significance of the inferred phylogenies was estimated by bootstrap analysis with 1,000 pseudoreplicate data sets. Phylogenetic analysis was performed on partial VP7 (962-bp) and VP4 (500-bp) nucleotide sequences and on the deduced amino acid sequences (aa 25 to 295 for VP7 and aa 10 to 154 for VP4). The sequences were aligned with and compared to additional sequences of the G1 and P[8] rotavirus genotypes, respectively, obtained from online databases.
Nucleotide sequence accession numbers.
All of the sequences obtained in the present study have been deposited in the GenBank sequence database. Accession numbers of the VP7 gene sequences are DQ377566 to DQ377601. Accession numbers of the VP4 gene segments are DQ377602 to DQ377630.
RESULTS
Phylogenetic and sequence analyses of VP7 and VP8*.
G1 strains, representative of the predominant double-stranded RNA electropherotypes, were selected for each year. Some older strains were not amplified by RT-PCR, most likely because of degradation or alterations of the stored samples. Of the 19 years considered, amplicons were not obtained for 1988 and 1991. After the initial determination of the VP7 serotype by enzyme-linked immunosorbent assay (data not shown), genotyping by RT-PCR of the VP7 and VP4 genes of the 41 Italian strains confirmed their characterization as G1P[8]. Hence, partial sequences of segments 9 and 4 of the Italian strains were used in parallel to construct phylogenetic trees. The VP7 and VP4 lineage designations were used as previously described (1, 7, 19, 35, 37, 47).
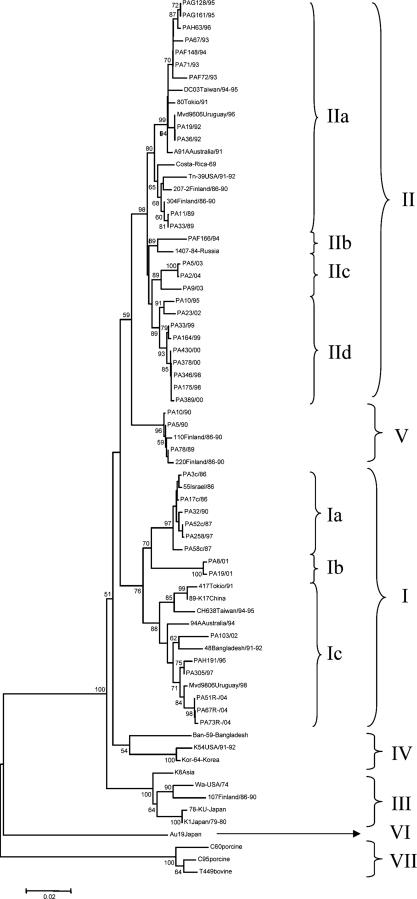
As shown in Fig. 1, the G1 HRVs segregated into six lineages (I to VI) and the Italian G1 HRVs clustered into three such genetic lineages, I, II, and V. VP7 lineage I included 14 Italian G1 strains within three sublineages (Ia to Ic) pertaining to eight different rotavirus years (1986 to 2004). Sublineage Ia included six strains circulating in 1986 to 1997, sublineage Ib included two strains circulating in 2001, while sublineage Ic included six strains circulating in the years 1996 to 2004. VP7 lineage II included 24 G1 strains recovered in 11 different years (1989 to 2004) and further distinguishable into four sublineages (IIa to IId). Sublineage IIa included 11 strains from the years 1989 to 1996, sublineage IIb included 1 strain from 1994, sublineage IIc included three strains from 2003 and 2004, and sublineage IId included 9 strains from the years 1995 to 2002. Three strains detected in the years 1989 and 1990 were related to Finnish strains collected during the same period and formed a separate lineage tentatively designated V. It is of note that in our phylogenetic reconstruction, two additional separate branches were resolved. One branch (designated lineage VI) was constituted exclusively by the atypical human Japanese strain Au19, which was detected in 1997 and is characterized by the rare supershort electropherotype and the P[6]-III VP4 genotype (50). The other branch included animal G1 strains, porcine isolates C60 and C95 (15), and bovine strain T449 (8), and it was designated lineage VII. No Italian HRV strains were found to cluster with G1 strains in lineage IV or, notably, in G1 lineage III, which includes reference G1 strains Wa and KU, as well as RotaShield vaccine strain D, all dating back to the 1970s (37).
FIG. 1.
Phylogenetic analysis of partial VP7 nucleotide sequences (47 to 1008) of genotype G1 strains. The phylogenetic tree was constructed by the neighbor-joining method and Kimura's two-parameter model, and statistical support was provided by bootstrapping of 1,000 pseudoreplicates. Bootstrap values above 49% are given at branch nodes.
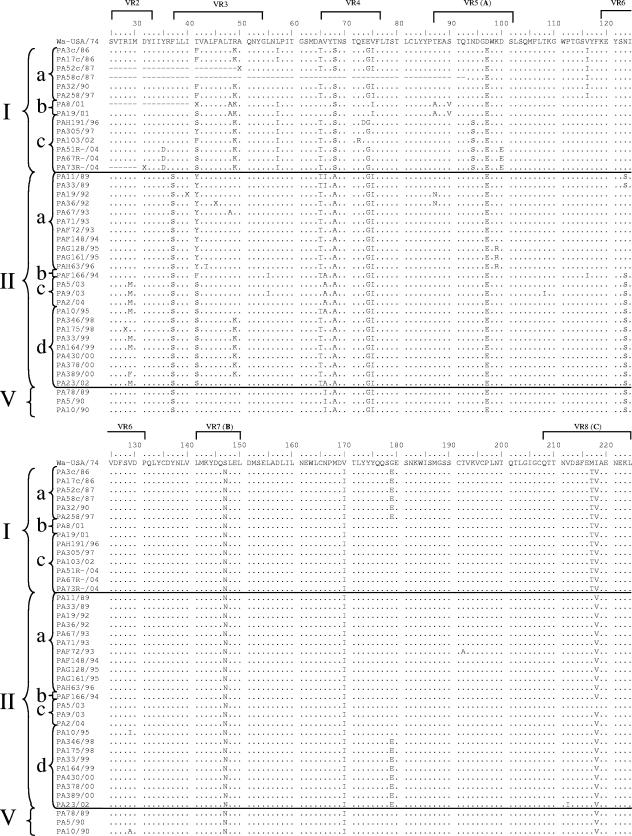
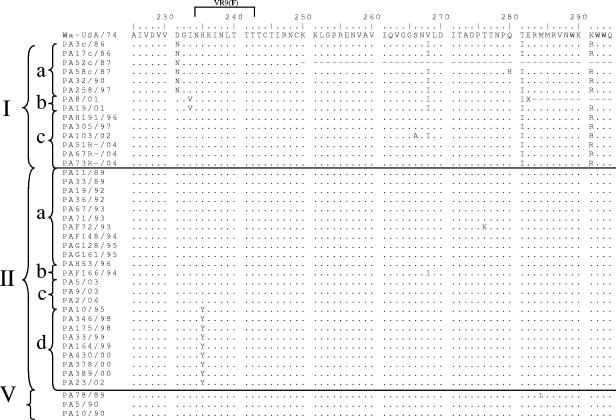
The degree of nucleotide identity among the Italian G1 strains in each lineage was >94.5% within lineage I and >96.6% within lineage II. Interlineage nucleotide identities between Italian G1 HRV strains of the two main lineages, I and II, ranged from 92.7 to 95.4%. An alignment of the VP7 deduced amino acid sequences was performed. The VP7 amino acid sequences of the Italian lineage I and II strains showed amino acid identity values of >95.3% and 96.5% compared to G1 HRVs of lineages I and II, respectively, and amino acid identity values ranging from 93.1 to 93.7% compared to the reference Wa strain (lineage III). Most of the amino acid changes between strain Wa and the Italian G1 strains of lineages I and II accumulated within the VP7 hypervariable regions (Fig. 2). Outside the variable regions, two peculiar amino acid substitutions were found at positions 281 and 291. There were clear synapomorphies (shared-derived substitutions) within the lineage G1-I HRVs, i.e., Ser68, Thr217, Ile281, and Arg291, while Ile66 was typical of lineage G1-V HRVs and yet it was also present in two G1-IIa strains. Amino acids Ser37 and Ala68 were highly conserved across lineage II and V strains. Synapomorphies, mostly scattered outside the variable regions, were detected in the various sublineages of lineage I. Ile116, Glu179, and Asn231 were conserved within sublineage Ia; Ala48, Ala87, Val90, and Val233 were conserved within sublineage Ib, while Ser94 was conserved in sublineage Ic. Ile57 was present in all lineage I strains except for three sublineage Ia HRVs. Also in lineage II there were amino acid substitutions characteristic for the various sublineages. At position 41, there were Tyr, Phe, and Ser in sublineage IIa, IIb, and IIc-d strains, respectively; Ser123 was present in all lineage II strains, except for six sublineage IIa strains; Tyr235 was found in sublineage IId strains. The substitutions Glu97, Asn147, and Ile170 were present in all Italian strains compared to reference strain Wa, regardless of their lineage and year of isolation.
FIG. 2.
Alignment of the VP7 deduced amino acid sequences of Italian G1 strains compared to reference strain Wa. Conserved amino acid residues are indicated by dots.
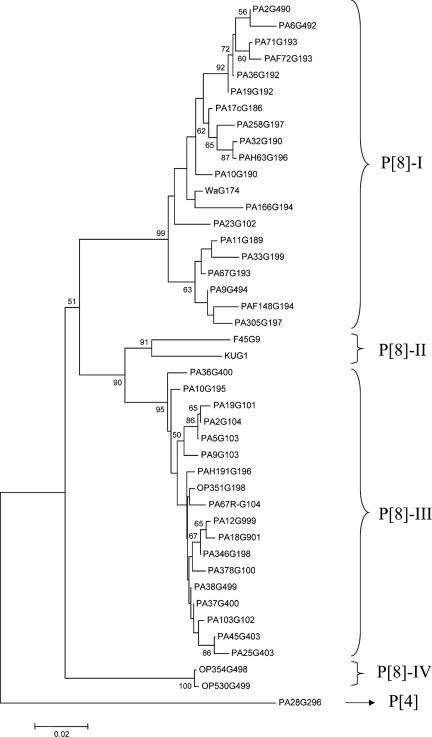
In the VP8* tree (Fig. 3), P[8] HRVs clustered in four distinct lineages (I to IV). The VP8* sequences of 27 of the 41 Italian G1 HRVs clustered into two defined lineages. Lineage P[8]-I included 16 G1 strains collected in the years 1986 to 2002, and lineage P[8]-III included 11 G1 strains isolated in the years 1995 to 2004. In addition, the VP8* sequences of the Italian G1P[8] strains were compared with the VP8* sequences of G4P[8] and G9P[8] Italian HRVs circulating in the same period (1, 3). The nucleotide sequence variation in VP8* was <6.3% among the Italian strains of lineage P[8]-I and <3.7% among strains of lineage P[8]-III. Amino acid sequence variation was <6.9% within each of the two lineages P[8]-I and P[8]-III, while interlineage variation ranged from 5.2% to 16%. The amino acids at positions 120 and 135 were conserved within the P[8]-I lineage but were distinct from those in the P[8]-III lineage (data not shown).
FIG. 3.
Phylogenetic analysis of partial VP4 nucleotide sequences (25 to 500) of type P[8] strains. The phylogenetic tree was constructed by the neighbor-joining method and Kimura's two-parameter model, and statistical support was provided by bootstrapping of 1,000 pseudoreplicates. The sequence of P[4] strain PA28 was used as the outgroup. Bootstrap values above 49% are given at branch nodes.
Dynamics of the evolution of G1 HRVs in Italy.
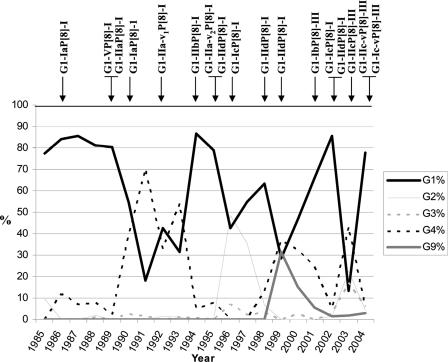
Figure 4 shows the prevalence of rotavirus isolates with G1 to G4 or G9 serotype specificity in the infant population of Palermo in 1985 to 2004 and the correlation observed between the peaks of G1 infections and the various VP7/VP4 lineages and sublineages. All of the G1 strains detected before 1994 belonged to lineage P[8]-I, while in 1995 G1 strains of lineage P[8]-III were detected, reaching a predominant diffusion in the subsequent years. Strains of the two different VP4 lineages were cocirculating in 1996 and 2002. As shown in Table 1, circulation of strains belonging to different G1 lineages or sublineages showed a pattern of consistent fluctuation since 1986. The various peaks of infections by G1 HRVs were apparently associated with the introduction of novel G1 HRV strains. G1 strains of the same sublineages were detected across a broad time span (sublineage Ia in the years 1986, 1987, 1990, and 1997; sublineage IId in the period 1995 to 2002), suggesting that some strains persisted over the years despite the periodic introduction of novel G1 strains. Conversely, a temporally limited circulation was described for G1 HRVs of lineage V (1989 and 1990) and for G1 HRVs of sublineage Ib (2001). A cocirculation of HRVs belonging to different VP7 lineages or sublineages was documented in 1989, 1990, 1994 to 1997, 2002, and 2004. Table 1 shows the amino acid substitutions in the main VP7 antigenic regions A, C, and F over the years. A 217-Thr→Met change was observed in region C between the G1-IIa and G1-V strains detected in 1989 and 1990 and the G1-Ia strains detected in 1986 and 1987. Sequences of G1 strains from 1988 and 1991 were not available. G1-IIa strains persisted through 1995 and 1996, although the mutation 87-Thr→Asn was present in the strains of 1992 (variant G1-IIa-v1) and the mutation 99-Lys→Arg was present in the strains detected after 1994 (variant G1-IIa-v2). Novel G1 strains G1-IIb and G1-IId were detected in the G1 epidemics of 1994 and 1995, respectively. Strain G1-IId displayed a peculiar 235-Lys→Tyr (region F) change and reemerged in 1998, persisting through 1999, 2000, and 2002. The strains detected in 2002 displayed the mutation 212-Val→Ile in region C (variant G1-IId-v). Again, in the 1996 and 1997 G1 epidemics, a novel strain, G1-Ic, was prominent and differed from the G1 strains of the former year in the amino acid changes 94-Asn→Ser, 99-Arg→Lys, and 217-Met→Thr (G1-IIa-v2 strains) or 94-Asn→Ser, 217-Met→Thr, and 235-Tyr→Lys (G1-IId strains). The 2001 and 2002 epidemic peaks of G1 rotaviruses were characterized by the broadest occurrence of G1 strains, with 70 to 85% of the infant rotavirus infections in Palermo due to G1 strains. In the 2001 rotavirus epidemic, a new strain, G1-Ib, appeared. This strain displayed multiple changes compared to preexisting G1-IId strains (87-Thr→Ala, 90-Ser→Val, 217-Met→Thr, and 235-Tyr→Lys). In 2002, the G1-Ib strain was replaced by G1-Ic and G1-IId-v strains exhibiting three and five changes, respectively, in the antigenic regions. Finally, during 2003 and 2004, a novel G1 strain, G1-IIc (and its variant 218-Val→Ile), appeared with two, three, or five changes compared to the strains circulating in the previous years. In addition, in 2004 a variant of the G1-Ic strains emerged with a single change, 100-Asp→Glu (antigenic region A).
FIG. 4.
Percent temporal distribution of G1 to G4 and G9 types of rotavirus strains circulating in Palermo, Italy, from 1985 to 2004. Arrows indicate the appearance of novel G1P[8] strains named after their VP7 and VP4 sublineages.
TABLE 1.
Pattern of variability in VP7 antigenic regions A, C, and F of Italian G1 strains of the different lineages in different years
| G1 straina | Year(s) | VR5 (region A, aa 87-101) | VR8 (region C, aa 208-221) | VR9 (region F, aa 233-242) |
|---|---|---|---|---|
| Ia | 1986, 1987, 1990, 1997 | TEASTGINDGEWKDS | QTTNVDSFETVAEN | NHKINLTTT |
| IIa | 1989, 1993, 1994 | --------------- | ---------M---- | --------- |
| V | 1989, 1990 | --------------- | ---------M---- | --------- |
| IIa-v1 | 1992 | N-------------- | ---------M---- | --------- |
| IIa-v2 | 1995, 1996 | ------------R-- | ---------M---- | --------- |
| IIb | 1994 | --------------- | ---------M---- | --------- |
| IId | 1995, 1998, 1999, 2000, 2002 | --------------- | ---------M---- | --Y------ |
| Ic | 1996, 1997, 2002 | -------S------- | -------------- | --------- |
| Ib | 2001 | A--V----------- | -------------- | V-------- |
| IId-v | 2002 | --------------- | ----I----M---- | --Y------ |
| IIc | 2003, 2004 | --------------- | ---------M---- | --------- |
| IIc-v | 2003 | --------------- | ---------MI--- | --------- |
| Ic-v | 2004 | -------S-----E- | -------------- | --------- |
| Wa | 1974 | ----------D---- | ---------MI--- | --------- |
The different G1 lineages are indicated by Roman numerals, sublineages are indicated by letters, and variants are indicated by the letter v with or without a subscript number.
DISCUSSION
The use of molecular methods for rotavirus characterization has allowed accurate evaluation of the genetic evolution of rotavirus strains circulating throughout the world. A number of studies have investigated the genetic and antigenic variations in the VP7 gene of G1 HRVs (7, 21, 37, 42, 45, 47, 54, 64). These studies revealed a high genetic homogeneity of G1 strains isolated in a single outbreak and the concomitant occurrence of G1 strains belonging to the same lineage in different parts of the world, suggesting a lack of defined geographical or temporal patterns. An exception to this phenomenon is the substantial diversity observed between the G1 rotaviruses collected throughout the world in recent years and the G1 rotaviruses detected in the 1970s in the United States and Asia (G1-III [Wa-like]) that apparently are no longer circulating worldwide. Altogether, this may suggest continual circulation across the various geographical settings of rotavirus strains, with reemergence of old strains in new locations. However, in all those studies the collection of G1 strains from the various locations was limited temporally and the exact dynamics and mechanisms of diversification of G1 serotype strains could be conjectured but not clearly verified.
The availability of a rotavirus collection dating back to the mid 1980s in our laboratory allowed us to perform an in-depth investigation of the temporal pattern of variation of G1 HRVs. This is the first study to describe the temporal evolution of G1 strains in a well-defined geographical area over an extended time span. All of the Italian strains were included in three discrete VP7 lineages (I, II, and V), and no G1-III strains (Wa like) were detected. Italian G1 rotavirus strains were distinct from G1 reference strain Wa at both the nucleotide and amino acid levels, notably in antigenic regions A, B, C, and F, where neutralization epitopes have been mapped that are involved in serotype determination and protection. The changes 147-Ser→Asn and 97-Asp→Glu, which are considered to be critical in preserving neutralization epitopes (18, 22), were highly conserved in Italian strains, as in most G1 rotaviruses isolated after 1980. Also, the amino acid change 291-Lys→Arg, affecting a conserved region that triggers rotavirus binding to MA104 cells (24) and which is involved in rotavirus neutralization (18), was present in all Italian G1-I strains. By contrast, the amino acid substitution 94-Asn→Ser was present only in Italian strains belonging to sublineage Ic. Residue 94 is considered to be essential for discrimination of G1 monotypes (18) and for distinguishing between lineages I and II (21). Also, the presence in convalescent-phase sera of antibodies blocking the binding of a VP7 serotype-specific MAb mapping to aa 94 has been correlated with resistance to disease in adult volunteers challenged with virulent G1 strain D (29).
While between 1985 and 2004, G2P[4], G4P[8], and G9P[8] strains varied in their distribution or completely disappeared for short or long periods of time, G1 strains have been continuously circulating in the infant population of Palermo, a pattern that has been described in various other geographical settings as well (6, 9, 11, 25, 35, 46, 58, 59). However, unlike the G2P[4] and G4P[8] Italian rotavirus strains, a highly heterogeneous population of G1 strains has been circulating in Italy over the whole study period. A high overall variability was demonstrated in the VP7 sequences of the G1 rotaviruses analyzed in this study, with at least 23 amino acid substitutions being detected. By contrast, only 5 amino acid changes were observed in G2 strains and 12 were observed in G4 strains (1, 2). The high intraserotypic genetic and antigenic heterogeneity is a possible reason for the consistent predominance of G1 serotype rotaviruses throughout the world over the years. In this scenario, a number of lineages and sublineages may have circulated or cocirculated, with novel strains emerging, some strains disappearing, and other strains persisting. During a nearly 20-year period, repeated introductions of novel G1 strains were described in Palermo and peaks of G1 rotavirus infections in the infant population were temporally associated with the onset of novel G1 strains. Amino acid changes were constantly observed in VP7 antigenic regions A, C, and F of strains appearing in consecutive years. For instance, during a G1 epidemic in 2001, a novel strain, G1-Ib, appeared that displayed up to five amino acid changes in the antigenic regions from the G1 strains spreading in the former year. Some strains reemerged after long intervals, remaining apparently unaltered after 12 years (G1-Ia strains). Other strains persisted for several years but developed additional changes in antigenic regions A and C, such as 87-Thr→Asn (a G1-IIa-v1 strain), 99-Lys→Arg (a G1-IIa-v2 strain), 100-Asp→Glu (G1-Ic-v strains), and 212-Val→Ile (a G1-IId-v strain).
Along with rearrangement, reassortment, and interspecies transmission, positive accumulation of point mutations is regarded as a powerful mechanism that steadily generates rotavirus diversification (23). This mechanism of evolution is well documented for influenza viruses and plays an important role in the continuous requirement for vaccinal updates. Ten changes in vaccine composition have been recommended between 1986 and 1998 to account for new antigenic variants of H3N2 influenza virus A. H3 hemagglutinin exhibited a rate of accumulation of amino acid substitutions of ca. 5 × 10−3 per year (30), while analysis of Italian G1-I HRV strains detected from 1986 to 2004 showed a lower rate of accumulation of amino acid substitutions, no more than ca. 2 × 10−3 per year. However, the results obtained by analyzing VP7 of the G1 HRVs revealed a more complex scenario because several G1 lineages circulate among the human population at any time in any place, while a single lineage was found in H3 hemagglutinin. In this, rotaviruses appear to be more similar to influenza B viruses (44), where the alternate circulation of different strains and lineages is regarded as a mechanism that reduces the selection pressure by recycling of different lineages, thus resulting in lower evolution rates.
Although VP7 is the surface protein represented in most current vaccines, VP4 may also be important in inducing protective immunity (33, 53, 62, 63) and rotavirus vaccines including the most common P serotype, P1A[8], have undergone evaluation in field trials (16). The older Italian G1P[8] HRVs displayed a P[8]-I lineage, while a second lineage, P[8]-III, emerged after 1995 and was also detected in all of the G4 and G9 HRV strains circulating in the same years (1, 3). Accordingly, the P[8] lineages did not display repeated switches between consecutive years. The only exception to this was observed in the G1 epidemic of 2002, when G1 strains accounted for 85% of the rotavirus gastroenteritis in children. The 2002 G1 HRVs were both P[8]-I and thus differed in the outer capsid proteins from the G1 strains spreading in 2001, as well as from the G9 and G4 strains cocirculating in the same years. Antigenic differences have been observed among the various P[6] lineages, I, II, and III (31, 43, 50), while it is not clear whether the various P[8] lineages also differ antigenically. Monitoring the temporal and geographical patterns of distribution of the various P[8] lineages and assessing their antigenic relationships will provide useful information to understand the role of VP4 in the mechanisms and patterns of rotavirus evolution.
In our study, it was possible to portray the evolutionary dynamics of G1P[8] strains in a geographically defined population in the absence of vaccine-induced immune pressure after routine prophylaxis or vaccine trials. It is possible that critical amino acid substitutions in key VP7 epitopes among circulating strains may allow escape from the limited antibody repertoire of young infants. However, it is not possible to know whether a similar model may apply to the evolutionary dynamics of the various rotavirus strains in a vaccinated population, where children are protected by a strong active immune response elicited by repeated vaccine administrations. Whether, in vaccinated populations, escape mechanisms due to antigenic differences among the various G1 VP7 lineages may result in vaccine failures or in selection of some lineages over time is a question that needs to be addressed in the near future. The results obtained in recent vaccine trials suggest that the vaccine G1 strains confer good homotypic protection against G1 strains from the field (20, 57). However, in vaccinees receiving rhesus rotavirus-based quadrivalent or monovalent (G1) vaccine, significant differences have been observed in neutralizing antibodies titers against G1 vaccine strain D (lineage III [Wa-like]) and G1 strains of other lineages (37). In a similar fashion, by cross-neutralization among G9 strains of various lineages (I to III), Hoshino et al. (32) demonstrated that antisera to lineage III may neutralize viruses of lineage II up to 64-fold less efficiently and viruses of lineage I up to 1,064-fold less efficiently. In order to ensure the continued effectiveness of vaccines, it is important to investigate the antigenic relationships between the various G1 lineages and extend G1 rotavirus surveillance studies by monitoring the circulation dynamics of the various strains.
Acknowledgments
This work was supported by grants from the Ministero della Sanità (Italian Ministry of Health) (Progetto Finalizzato 2003: diversità genetica ed antigenica dei rotavirus, studio dei meccanismi evolutivi ed implicazioni ai fini diagnostici e vaccinali) and from the Ministero dell'Istruzione, dell'Università e della Ricerca (Italian Ministry of Education, University and Research) (Fondi di Ateneo ex 60%).
Footnotes
Published ahead of print on 23 August 2006.
REFERENCES
- 1.Arista, S., G. M. Giammanco, S. De Grazia, C. Colomba, and V. Martella. 2005. Genetic variability among serotype G4 Italian human rotaviruses. J. Clin. Microbiol. 43:1420-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arista, S., G. M. Giammanco, S. De Grazia, C. Colomba, V. Martella, A. Cascio, and M. Iturriza-Gomara. 2005. G2 rotavirus infections in an infantile population of the South of Italy: variability of viral strains over time. J. Med. Virol. 77:587-594. [DOI] [PubMed] [Google Scholar]
- 3.Arista, S., G. M. Giammanco, S. De Grazia, M. C. Migliore, V. Martella, and A. Cascio. 2004. Molecular characterization of the genotype G9 human rotavirus strains recovered in Palermo, Italy, during the winter of 1999-2000. Epidemiol. Infect. 132:343-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arista, S., L. Giovannelli, D. Pistoia, A. Cascio, M. Parea, and G. Gerna. 1990. Electropherotypes, subgroups and serotypes of human rotavirus strains causing gastroenteritis in infants and young children in Palermo, Italy, from 1985 to 1989. Res. Virol. 141:435-448. [DOI] [PubMed] [Google Scholar]
- 5.Arista, S., E. Vizzi, D. Ferraro, A. Cascio, and R. Di Stefano. 1997. Distribution of VP7 serotypes and VP4 genotypes among rotavirus strains recovered from Italian children with diarrhea. Arch. Virol. 142:2065-2071. [DOI] [PubMed] [Google Scholar]
- 6.Banyai, K., J. R. Gentsch, R. Schipp, F. Jakab, E. Meleg, I. Mihaly, and G. Szucs. 2005. Dominating prevalence of P[8],G1 and P[8],G9 rotavirus strains among children admitted to hospital between 2000 and 2003 in Budapest, Hungary. J. Med. Virol. 76:414-423. [DOI] [PubMed] [Google Scholar]
- 7.Berois, M., S. Libersou, J. Russi, J. Arbiza, and J. Cohen. 2003. Genetic variation in the VP7 gene of human rotavirus isolated in Montevideo- Uruguay from 1996-1999. J. Med. Virol. 71:456-462. [DOI] [PubMed] [Google Scholar]
- 8.Blackhall, J., R. Bellinzoni, N. Mattion, M. K. Estes, J. L. La Torre, and G. Magnusson. 1992. A bovine rotavirus serotype 1: serologic characterization of the virus and nucleotide sequence determination of the structural glycoprotein VP7 gene. Virology 189:833-837. [DOI] [PubMed] [Google Scholar]
- 9.Bon, F., C. Fromantin, S. Aho, P. Pothier, E. Kohli and the AZAY Group. 2000. G and P genotyping of rotavirus strains circulating in France over a three-year period: detection of G9 and P[6] strains at low frequencies. J. Clin. Microbiol. 38:1681-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buesa, J., C. O. de Souza, M. Asensi, C. Martinez, J. Prat, and M. T. Gil. 2000. VP7 and VP4 genotypes among rotavirus strains recovered from children with gastroenteritis over a 3-year period in Valencia, Spain. Eur. J. Epidemiol. 16:501-506. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 1999. Rotavirus vaccine for the prevention of rotavirus gastroenteritis among children. Recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. 48:1-20. [PubMed] [Google Scholar]
- 13.Centers for Disease Control and Prevention. 1999. Withdrawal of rotavirus vaccine recommendation. Morb. Mortal. Wkly. Rep. 48:1007. [PubMed] [Google Scholar]
- 14.Ciarlet, M., Y. Hoshino, and F. Liprandi. 1997. Single point mutations may affect the serotype reactivity of serotype G11 porcine rotavirus strains: a widening spectrum? J. Virol. 71:8213-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciarlet, M., and F. Liprandi. 1994. Serological and genomic characterization of two porcine rotaviruses with serotype G1 specificity. J. Clin. Microbiol. 32:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark, H. F., P. A. Offit, R. W. Ellis, J. J. Eiden, D. Krah, A. R. Shaw, M. Pichichero, J. J. Treanor, F. E. Borian, L. M. Bell, and S. A. Plotkin. 1996. The development of multivalent bovine rotavirus (strain WC3) reassortant vaccine for infants. J. Infect. Dis. 174(Suppl. 1):S73-S80. [DOI] [PubMed] [Google Scholar]
- 17.Coulson, B. S., K. J. Fowler, J. R. White, and R. G. Cotton. 1987. Non-neutralizing monoclonal antibodies to a trypsin-sensitive site on the major glycoprotein of rotavirus which discriminate between virus serotypes. Arch. Virol. 93:199-211. [DOI] [PubMed] [Google Scholar]
- 18.Coulson, B. S., and C. Kirkwood. 1991. Relation of VP7 amino acid sequence to monoclonal antibody neutralization of rotavirus and rotavirus monotype. J. Virol. 65:5968-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Vos, B., T. Vesikari, A. C. Linhares, B. Salinas, I. Perez-Schael, G. M. Ruiz-Palacios, L. Guerrero Mde, K. B. Phua, A. Delem, and K. Hardt. 2004. A rotavirus vaccine for prophylaxis of infants against rotavirus gastroenteritis. Pediatr. Infect. Dis. J. 23:S179-S182. [DOI] [PubMed] [Google Scholar]
- 21.Diwakarla, C. S., and E. A. Palombo. 1999. Genetic and antigenic variation of capsid protein VP7 of serotype G1 human rotavirus isolates. J. Gen. Virol. 80:341-344. [DOI] [PubMed] [Google Scholar]
- 22.Dyall-Smith, M. L., I. Lazdins, G. W. Tregear, and I. H. Holmes. 1986. Location of the major antigenic sites involved in rotavirus serotype-specific neutralization. Proc. Natl. Acad. Sci. USA 83:3465-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 24.Frenchick, P., M. I. Sabara, M. K. Ijak, and L. A. Babiuk. 1988. Immune responses to synthetic peptide vaccines of veterinary importance, p. 141-151. In B. Kurstak, R. G. Marusyk, F. A. Murphy, and M. H. V. Van Regenmortel (ed.), Applied virology research, vol. 1. Plenum Publishing Corp., New York, N.Y. [Google Scholar]
- 25.Gault, E., R. Chikhi-Brachet, S. Delon, N. Schnepf, L. Albiges, E. Grimprel, J. P. Girardet, P. Begue, and A. Garbarg-Chenon. 1999. Distribution of human rotavirus G types circulating in Paris, France, during the 1997-1998 epidemic: high prevalence of type G4. J. Clin. Microbiol. 37:2373-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gentsch, J. R., P. A. Woods, M. Ramachandran, B. K. Das, J. P. Leite, A. Alfieri, R. Kumar, M. K. Bhan, and R. I. Glass. 1996. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J. Infect. Dis. 174(Suppl. 1):S30-S36. [DOI] [PubMed] [Google Scholar]
- 28.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green, K. Y., and A. Z. Kapikian. 1992. Identification of VP7 epitopes associated with protection against human rotavirus illness or shedding in volunteers. J. Virol. 66:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hay, A. J., V. Gregory, A. R. Douglas, and Y. P. Lin. 2001. The evolution of human influenza viruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshino, Y., R. W. Jones, and A. Z. Kapikian. 2002. Characterization of neutralization specificities of outer capsid spike protein VP4 of selected murine, lapine, and human rotavirus strains. Virology 299:64-71. [DOI] [PubMed] [Google Scholar]
- 32.Hoshino, Y., R. W. Jones, J. Ross, S. Honma, N. Santos, J. R. Gentsch, and A. Z. Kapikian. 2004. Rotavirus serotype G9 strains belonging to VP7 gene phylogenetic sequence lineage 1 may be more suitable for serotype G9 vaccine candidates than those belonging to lineage 2 or 3. J. Virol. 78:7795-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoshino, Y., L. J. Saif, M. M. Sereno, R. M. Chanock, and A. Z. Kapikian. 1988. Infection immunity of piglets to either VP3 or VP7 outer capsid protein confers resistance to challenge with a virulent rotavirus bearing the corresponding antigen. J. Virol. 62:744-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iturriza-Gomara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 1999. Comparison of specific and random priming in the reverse transcriptase polymerase chain reaction for genotyping group A rotaviruses. J. Virol. Methods 78:93-103. [DOI] [PubMed] [Google Scholar]
- 35.Iturriza-Gomara, M., J. Green, D. W. Brown, M. Ramsay, U. Desselberger, and J. J. Gray. 2000. Molecular epidemiology of human group A rotavirus infections in the United Kingdom between 1995 and 1998. J. Clin. Microbiol. 38:4394-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iturriza-Gomara, M., G. Kang, and J. Gray. 2004. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J. Clin. Virol. 31:259-265. [DOI] [PubMed] [Google Scholar]
- 37.Jin, Q., R. L. Ward, D. R. Knowlton, Y. B. Gabbay, A. C. Linhares, R. Rappaport, P. A. Woods, R. I. Glass, and J. R. Gentsch. 1996. Divergence of VP7 genes of G1 rotaviruses isolated from infants vaccinated with reassortant rhesus rotaviruses. Arch. Virol. 141:2057-2076. [DOI] [PubMed] [Google Scholar]
- 38.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 39.Kobayashi, N., K. Taniguchi, T. Urasawa, and S. Urasawa. 1991. Analysis of the neutralization epitopes on human rotavirus VP7 recognized by monotype-specific monoclonal antibodies. J. Gen. Virol. 72:1855-1861. [DOI] [PubMed] [Google Scholar]
- 40.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 41.Laird, A. R., J. R. Gentsch, T. Nakagomi, O. Nakagomi, and R. I. Glass. 2003. Characterization of serotype G9 rotavirus strains isolated in the United States and India from 1993 to 2001. J. Clin. Microbiol. 41:3100-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, C. N., C. C. Lin, C. L. Kao, C. L. Zao, M. C. Shih, and H. N. Chen. 2001. Genetic characterization of the rotaviruses associated with a nursery outbreak. J. Med. Virol. 63:311-320. [DOI] [PubMed] [Google Scholar]
- 43.Li, B., and M. Gorziglia. 1993. VP4 serotype of the Gottfried strain of porcine rotavirus. J. Clin. Microbiol. 31:3075-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindstrom, S. E., Y. Hiromoto, H. Nishimura, T. Saito, R. Nerome, and K. Nerome. 1999. Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes. J. Virol. 73:4413-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lynch, M., B. Lee, P. Azimi, J. Gentsch, C. Glaser, S. Gilliam, H. G. Chang, R. Ward, and R. I. Glass. 2001. Rotavirus and central nervous system symptoms: cause or contaminant? Case reports and review. Clin. Infect. Dis. 33:932-938. [DOI] [PubMed] [Google Scholar]
- 46.Maunula, L., and C. H. van Bonsdorff. 1995. Rotavirus serotypes and electropherotypes in Finland from 1986 to 1990. Arch. Virol. 140:877-890. [DOI] [PubMed] [Google Scholar]
- 47.Maunula, L., and C. H. von Bonsdorff. 1998. Short sequences define genetic lineages: phylogenetic analysis of group A rotaviruses based on partial sequences of genome segments 4 and 9. J. Gen. Virol. 79:321-332. [DOI] [PubMed] [Google Scholar]
- 48.McCarthy, M. 2003. Project seeks to “fast track” rotavirus vaccine. Lancet 361:582. [DOI] [PubMed] [Google Scholar]
- 49.Murphy, T. V., P. J. Smith, P. M. Gargiullo, and B. Schwartz. 2003. The first rotavirus vaccine and intussusception: epidemiological studies and policy decisions. J. Infect. Dis. 187:1309-1313. [DOI] [PubMed] [Google Scholar]
- 50.Nakagomi, T., Y. Horie, Y. Koshimura, H. B. Greenberg, and O. Nakagomi. 1999. Isolation of a human rotavirus strain with a super-short RNA pattern and a new P2 subtype. J. Clin. Microbiol. 37:1213-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishikawa, K., Y. Hoshino, K. Taniguchi, K. Y. Green, H. B. Greenberg, A. Z. Kapikian, R. M. Chanock, and M. Gorziglia. 1989. Rotavirus VP7 neutralization epitopes of serotype 3 strains. Virology 171:503-515. [DOI] [PubMed] [Google Scholar]
- 52.Offit, P. A. 1996. Host factors associated with protection against rotavirus disease: the skies are clearing. J. Infect. Dis. 174(Suppl. 1):S59-S64. [DOI] [PubMed] [Google Scholar]
- 53.Offit, P. A., H. F. Clark, G. Blavat, and H. B. Greenberg. 1986. Reassortant rotaviruses containing structural proteins vp3 and vp7 from different parents induce antibodies protective against each parental serotype. J. Virol. 60:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Halloran, F., M. Lynch, B. Cryan, and S. Fanning. 2002. Application of restriction fragment length polymorphism analysis of VP7-encoding genes: fine comparison of Irish and global rotavirus isolates. J. Clin. Microbiol. 40:524-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parashar, U. D., E. G. Hummelman, J. S. Bresee, M. A. Miller, and R. I. Glass. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9:565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rahman, M., J. Matthijnssens, S. Nahar, G. Podder, D. A. Sack, T. Azim, and M. Van Ranst. 2005. Characterization of a novel P[25],G11 human group a rotavirus. J. Clin. Microbiol. 43:3208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salinas, B., I. Perez Schael, A. C. Linhares, G. M. Ruiz Palacios, M. L. Guerrero, J. P. Yarzabal, Y. Cervantes, S. Costa Clemens, S. Damaso, K. Hardt, and B. De Vos. 2005. Evaluation of safety, immunogenicity and efficacy of an attenuated rotavirus vaccine, RIX4414: a randomized, placebo-controlled trial in Latin American infants. Pediatr. Infect. Dis. J. 24:807-816. [DOI] [PubMed] [Google Scholar]
- 58.Sanchez-Fauquier, A., I. Wilhelmi, J. Colomina, E. Cubero, and E. Roman. 2004. Diversity of group A human rotavirus types circulating over a 4-year period in Madrid, Spain. J. Clin. Microbiol. 42:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 15:29-56. [DOI] [PubMed] [Google Scholar]
- 60.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Velazquez, F. R., D. O. Matson, J. J. Calva, L. Guerrero, A. L. Morrow, S. Carter-Campbell, R. I. Glass, M. K. Estes, L. K. Pickering, and G. M. Ruiz-Palacios. 1996. Rotavirus infections in infants as protection against subsequent infections. N. Engl. J. Med. 335:1022-1028. [DOI] [PubMed] [Google Scholar]
- 62.Ward, R. L., D. R. Knowlton, G. M. Schiff, Y. Hoshino, and H. B. Greenberg. 1988. Relative concentrations of serum neutralizing antibody to VP3 and VP7 proteins in adults infected with a human rotavirus. J. Virol. 62:1543-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward, R. L., M. M. McNeal, D. S. Sander, H. B. Greenberg, and D. I. Bernstein. 1993. Immunodominance of the VP4 neutralization protein of rotavirus in protective natural infections of young children. J. Virol. 67:464-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xin, K. Q., S. Morikawa, Z. Y. Fang, A. Mukoyama, K. Okuda, and H. Ushijima. 1993. Genetic variation in VP7 gene of human rotavirus serotype 1 (G1 type) isolated in Japan and China. Virology 197:813-816. [DOI] [PubMed] [Google Scholar]