Abstract
The structure of Red clover necrotic mosaic virus (RCNMV), an icosahedral plant virus, was resolved to 8.5 Å by cryoelectron microscopy. The virion capsid has prominent surface protrusions and subunits with a clearly defined shell and protruding domains. The structures of both the individual capsid protein (CP) subunits and the entire virion capsid are consistent with other species in the Tombusviridae family. Within the RCNMV capsid, there is a clearly defined inner cage formed by complexes of genomic RNA and the amino termini of CP subunits. An RCNMV virion has approximately 390 ± 30 Ca2+ ions bound to the capsid and 420 ± 25 Mg2+ ions thought to be in the interior of the capsid. Depletion of both Ca2+ and Mg2+ ions from RCNMV leads to significant structural changes, including (i) formation of 11- to 13-Å-diameter channels that extend through the capsid and (ii) significant reorganization within the interior of the capsid. Genomic RNA within native capsids containing both Ca2+ and Mg2+ ions is extremely resistant to nucleases, but depletion of both of these cations results in nuclease sensitivity, as measured by a significant reduction in RCNMV infectivity. These results indicate that divalent cations play a central role in capsid dynamics and suggest a mechanism for the release of viral RNA in low-divalent-cation environments such as those found within the cytoplasm of a cell.
The Tombusviridae family consists of small, icosahedral plant viruses that are transmitted through the soil and infect their hosts via the root system (34). Given that these viruses must survive harsh environments, it is not surprising that they possess unusually stable and robust capsids. This high degree of virion stability raises the question of how the intracellular environment triggers disassembly or minimally exposes the genome for translation. Structural analyses of several plant viruses have revealed that maintenance of a stable capsid conformation is dependent on the presence of divalent cations bound to the capsid (25, 33, 36). It has been a longstanding hypothesis that swelling and other conformational changes induced by ion extraction from virions are critical for a productive viral life cycle (13). For Tomato bushy stunt virus (TBSV), the type species of the genus Tombusvirus within the Tombusviridae family, it was hypothesized that Ca2+ ions would be released from viral capsids within infected cells, leading to virion swelling sufficient to expose the viral genome.
Crystallographic studies of TBSV (28) revealed a capsid formed by 180 chemically identical capsid protein (CP) subunits in three quasiequivalent conformations (A, B, and C). Each CP subunit is composed of three distinct structural domains, which include the RNA-interacting (R), shell (S), and protruding (P) domains. The conformational differences that distinguish the A, B, and C subunits are localized within the hinge regions between the respective S and P domains. These hinges point either down (in A-B dimers) or up (in C-C dimers). In addition, the loop that connects the R and S domains (the arm) is ordered in C subunits but disordered in A and B subunits. A similar structural organization was observed in Turnip crinkle virus (TCV) (50) and Carnation mottle virus (CarMV) (25), which are two other species in the Tombusviridae family and whose capsid structures have been solved to a resolution of ∼3 Å. All three viruses share a phylogenetically related CP, produce virions with capsids possessing T=3 icosahedral symmetry (9), and are ∼350 Å in diameter. Neither the packaged RNA nor the amino termini of the CP were sufficiently ordered to be resolved in these crystallographic studies.
Despite the lack of evidence of an organized genome within the Tombusviridae family members studied so far, a number of other RNA viruses, whose structures have been determined by X-ray crystallography and cryoelectron microscopy (cryoEM), have partially ordered genomes inside their capsids. The fraction of ordered RNA visualized in viruses varies significantly. For example, in Cowpea chlorotic mottle virus there is very little ordered RNA (40). Conversely, in the insect-infecting Nodaviridae family, Flock house virus and Pariacoto virus both package a substantial fraction of their RNA genomes within dodecahedral cages (45). Satellite Tobacco mosaic virus represents an extreme example, because essentially all of the genome within the virion is ordered and has been resolved in the X-ray structure (19, 20).
Red clover necrotic mosaic virus (RCNMV; genus Dianthovirus, family Tombusviridae) is morphologically similar to TBSV, CarMV, and TCV with virions ∼350 Å in diameter composed of 23% nucleic acid and 77% protein (26). However, RCNMV is significantly different by virtue of its RNA genome, which is split between two nonhomologous RNAs, a 3.9-kb polycistronic RNA-1 and a 1.5-kb monocistronic RNA-2 (21). Furthermore, the RCNMV genome is larger than that of any other monopartite member of the family Tombusviridae yet the virions are all similar in size. Recent virion characterization studies revealed that RCNMV virions exist as two distinct yet morphologically indistinguishable populations, with one population containing four copies of RNA-2 and a second, biologically active population containing one copy each of RNA-1 and RNA-2 (4). Such distinguishing features of the RCNMV virion have stimulated us to examine the RCNMV structure to better understand what makes it unique among members of the family Tombusviridae.
We have used cryoEM and three-dimensional (3D) image reconstruction to analyze the structure of RCNMV in its native form, as well as its form after removal of divalent cations. Though the native RCNMV capsid structure is similar to that of other viruses in the taxon, the genomic RNA of RCNMV is ordered and organized as an internal, dodecahedral cage. As demonstrated by spectroscopic analysis, RCNMV contains finite amounts of Ca2+ and Mg2+ ions. Selective extraction of these ions alters the capsid conformation and generates channels that expose the genomic RNA and likely provide a path for its exit into the cytosol of infected cells. This study suggests how divalent cations help stabilize RCNMV virions and how they may act as a switch to initiate disassembly.
MATERIALS AND METHODS
RCNMV propagation and purification.
Nicotiana clevelandii plants (4 to 6 weeks old) were rub inoculated with infectious RCNMV RNA transcripts (53) and maintained in a greenhouse at 20 to 24°C. Virions were purified from infected plants 7 to 10 days postinoculation (dpi) as previously described (53). Virion concentrations were determined by UV spectroscopy with an extinction coefficient (1 mg/ml, 1-cm light path at 260 nm) of 6.46 (21) and reaffirmed with the Coomassie Plus Protein Assay Reagent (Pierce Biotechnology, Rockford, IL). A typical RCNMV purification yielded 70 to 100 μg of virions per g of infected tissue.
Determination of virion Ca2+ and Mg2+ ion contents.
The Ca2+ and Mg2+ ion concentrations for purified RCNMV virions were determined by direct aspiration atomic absorption spectroscopy (14) with a Perkin-Elmer (Wellesley, MA) model 3110 spectrophotometer. Ca2+ and Mg2+ ion absorptions were monitored at 422.7 and 285.2 nm, respectively. Standards were prepared from dry CaCO3 and Mg(C2H3O2)2. The reported ion concentrations are the average of four determinations (duplicate measurements for two different viral preparations). The uncertainty is the standard deviation of the four determinations. A 2.5-μg sample of purified RCNMV virions was suspended directly in 200 μl of 0.2 M sodium acetate, pH 5.3. Ca2+ and Mg2+ ions were selectively removed through chelation by addition of an equal volume of EGTA or EDTA, respectively, to a final concentration of 10 mM and incubated overnight at 20°C. The EGTA or EDTA was removed by exchanging the solution three times with a 100-kDa limit Centricon filter unit (Millipore, Billerica, MA). To determine if divalent cations leach out of RCNMV virions under low-cation conditions (pseudophysiological conditions), virions were dialyzed in a Pierce Slide-A-Lyzer 10K MWCO dialysis cassette against two 1-liter exchanges of 18-mΩ deionized water over a 24-h incubation period at 20°C.
Infectivity bioassay.
The infectivities of treated virion preparations were assayed by inoculation onto 6-week-old Chenopodium quinoa plants, a local-lesion host for RCNMV. Each treatment was prepared in a 200-μl reaction volume. As appropriate, 4 μg of purified virions, 10 U of RNase (RNase A, T1, or V1; Ambion, Austin, TX) and either EGTA or EDTA (10 mM final concentration) were added to the inoculation buffer (10 mM sodium phosphate, pH 7.2). All samples were incubated for 4 h at room temperature prior to inoculation. For each treatment, 20 μl of the reaction mixture was applied to each of four carborundum-dusted leaves per plant by rub inoculation. Plants were monitored for local lesion development, which generally first appeared at 4 dpi, but lesions were counted at 7 dpi. The percent infectivity for each treatment was calculated by comparing the average number of lesions obtained per leaf versus that obtained for untreated virions.
DLS.
Dynamic light-scattering (DLS) data on virion size were collected on a Malvern 1000ES Zetasizer (Malvern Instruments, Worcestershire, United Kingdom). Forty micrograms of purified virions was suspended in 1 ml of 10 mM Tris-HCl, pH 7.0, unless otherwise noted. DLS measurements were conducted at 20°C, and the data were analyzed with a nonnegative least-squares algorithm (15).
cryoEM.
For cryoEM, an ∼1-mg/ml RCNMV virus suspension in 10 mM Tris-HCl, pH 6.5 to 6.8, and, when appropriate, a 10 mM final concentration of EGTA or EDTA, were plunge-frozen over holes in a holey carbon film (R2/2 Quantifoil; Micro Tools GmbH, Jena, Germany). The cryogrids were held at −174°C in a Gatan 626 Cryo-Holder (Pleasanton, CA). RCNMV micrographs were recorded on Kodak SO-163 film (Kodak, Rochester, NY) at 300 keV with a dose of ∼20 electrons/Å2 in an FEI CM300 FEG microscope as described elsewhere (30) at a nominal magnification of ×47,000. Focusing was performed off axis at a magnification of ×105,000 in an area adjacent to the one selected for imaging.
Image processing.
Images were digitized with a PhotoScan microdensitometer (Z/I Imaging, Huntsville, AL) with a 7-μm step size. The interactive graphics program ROBEM (http://cryoem.ucsd.edu/programs.shtm) was used to box images of individual RCNMV virions and to estimate the defocus and other characteristics of the contrast transfer function (CTF) for each micrograph. The CTF was determined by analyzing an incoherent average of the Fourier transforms of all of the particle images from each micrograph (55). CTF correction in images with Wiener-type filtering (5, 51) was performed before orientation or origin searching and refinement with the stand-alone program CTFCOR.
An ab initio model of the virus was obtained with the IMAGIC-5 package (49). The particle images were aligned and grouped into classes by multivariate statistical analysis and classification (47). Images making up a class were averaged to generate a characteristic view with an enhanced signal-to-noise ratio. The angular-reconstitution technique (46) was then used to determine the relative orientations of the views; these orientations were then used to calculate 3D maps (39). After several iterations of alignment, classification, and orientation determination, the 3D reconstruction was deemed reliable to a resolution of 18Å.
Subsequent refinement to a higher resolution made use of a modified version of the PFT protocol (54). This eventually led to a 3D reconstruction of native RCNMV at a resolution of 8.5Å. The resolution at each stage of both IMAGIC-5 and PFT refinement was estimated by Fourier shell correlation (37, 48).
For native, untreated RCNMV virions (RCNMVNAT), micrographs ranging from 0.6 to 2.9 μm underfocus were chosen for processing. A total of 5,069 RCNMV virion images were selected from 56 micrographs. The final 3D reconstruction included data from 2,546 virion images. The resolution of the 3D reconstruction steadily improved during the refinement process and reached 8.5 Å after the final cycle by a conservative 0.5 FSC criterion (35).
For Ca2+ ion-depleted virions (RCNMV−Ca), 4,447 virion images were selected from 19 micrographs (defocus range, 0.6 to 2.1 μm underfocus). The processing was performed analogous to RCNMVNAT. The final RCNMV−Ca 3D map was computed from 1,652 virion images and achieved an estimated resolution of about 9 Å.
For divalent-cation-depleted virions (RCNMV−Ca/−Mg), 1,878 virion images were selected from 23 micrographs, where the underfocus ranged between 0.77 and 4.2 μm. The 18-Å model of RCNMVNAT obtained in IMAGIC-5 was used as an initial template for orientation and origin searching of the RCNMV−Ca/−Mg images by the PFT protocol. Refinement eventually led to a final 3D map (resolution of 16.5 Å) computed from 1,001 virion images.
Difference maps of the RCNMVNAT, RCNMV−Ca, and RCNMV−Ca/−Mg reconstructions were computed in ROBEM or IMAGIC-5. For each difference map, the higher-resolution map was low-pass filtered to the lower resolution limit of the other map and the magnification and contrast of one map were scaled to best match those of the other map so the largest differences would most likely reflect genuine changes in the structures. The density and difference maps were surface rendered and displayed with a threshold of 1 sigma in IRIS Explorer (Numerical Algorithms Group, Oxford, United Kingdom). Atomic and pseudoatomic models were displayed in PyMOL (DeLano Scientific, San Francisco, CA).
Sequence similarity search, alignment, and fitting of pseudoatomic model into cryoEM maps.
Sequences similar to that of RCNMV CP (Swiss-Prot primary accession number P22955) were searched for with BLAST on the ExPASy website (http://www.expasy.org). Sequence alignments of the S and P domains separately relative to TBSV (P11795) were performed with T-COFFEE (http://www.ch.embnet.org/software/TCoffee.html) (32). Homology modeling with the crystal structure of TBSV CP (PDB entry 2TBV) as the template was performed with SWISS-MODEL (http://swissmodel.expasy.org) (38). Separate homology models of the S and P domains were fitted interactively into the cryoEM maps with the O program (18). Situs (52) was then used to refine the fit of these models into the cryoEM maps.
For RCNMV−Ca/−Mg virions, the conformations of the CP models needed to be adjusted to produce models that better fit the reconstructed density map. To accomplish this, we used the normal-mode flexible-fitting (NMFF) procedure (42), which relies on elastic-network normal-mode modeling (43) to fit each capsid subunit independently. In the elastic-network description, the cutoff was set to 8 Å, the rotations-translations of blocks method was used for the normal-mode analysis (41), and a total of 26 normal modes with the lowest frequencies were explored by the NMFF procedure. Only the Cα atom coordinates were used for the flexible fitting, and all atom models were constructed from the original homology models by energy minimization with the Cα trace obtained from NMFF as a constraint. Energy minimization was carried out with the CHARMM package (7).
RESULTS
RCNMV divalent-cation content.
Crystallographic studies have demonstrated that Ca2+ ions influence the conformation of plant RNA viruses. Within the resolved crystal structures of several species of the family Tombusviridae, the number of coordinated Ca2+ ions varies from 60 to 360 per capsid (24, 27, 28). Atomic absorption spectroscopy was used to determine that 390 ± 30 Ca2+ ions are present per RCNMV virion (Table 1). This corresponds to the presence of slightly more than two Ca2+ ions per CP subunit, which is comparable to the number observed in TBSV (28). In addition, there are 420 ± 25 Mg2+ ions per RCNMV virion (Table 1). Although the role and location of Mg2+ ions in RCNMV are unknown, we hypothesize that they are associated with the viral RNA inside the capsid.
TABLE 1.
Divalent-cation content of RCNMV virions as determined by atomic absorption spectroscopy
| Virion treatment | Avg no. of divalent cations/viriona ± SD
|
|
|---|---|---|
| Ca2+ | Mg2+ | |
| None | 390 ± 30 | 420 ± 25 |
| EGTAb | <30 | 125 ± 28 |
| EDTAc | <30 | 40 ± 2 |
| Dialysis against H2Od | <30 | 50 ± 4 |
Divalent-cation concentrations reported for each treatment are the averages of four measurements (duplicate determinations for two different viral preparations).
Virions were incubated in 10 mM EGTA at 20°C overnight prior to concentration and desalting with Centricon filter units.
Virions were incubated in 10 mM EDTA at 20°C overnight prior to concentration and desalting with Centricon filter units.
Virions were dialyzed against two 1-liter exchanges of 18-mΩ deionized H2O over a 24-h incubation period at 20°C.
An experiment was performed to mimic the host cell environment with respect to divalent-cation concentrations and to verify the hypothesis that loss of these ions from virions would lead to exposure of the RCNMV genome. Purified virus was exhaustively dialyzed against deionized water. Dialysis removed virtually all of the Ca2+ ions and substantially reduced the number of Mg2+ ions (only 50 ± 4 remained) (Table 1). This suggests that, while high concentrations of divalent cations are present within native virions, they can diffuse out in a medium with low cation concentrations.
To further investigate the role of Ca2+ ions in maintaining capsid integrity, virions were treated with EGTA, which preferentially chelates calcium ions (8). EGTA treatment removed all Ca2+ ions to the limit of detection and reduced the number of Mg2+ ions per virion from 420 ± 25 to 125 ± 28 (Table 1). On the basis of the known selectivity of EGTA, the loss of Mg2+ ions was interpreted to be the result of diffusion rather than nonspecific chelation (8). When virions were treated with 10 mM EDTA, which is a less selective divalent-cation chelator, virtually all of the Ca2+ ions were removed as well and the number of Mg2+ ions was reduced to the even lower number of 40 ± 2 per virion (Table 1).
Integrity of cation-depleted virions.
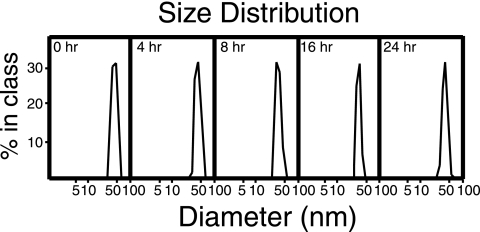
To test if the long-standing hypothesis that TBSV disassembly is initiated by Ca2+ ion removal (13) also holds for RCNMV, we determined if removal of Ca2+ and Mg2+ ions induces RCNMV disassembly. The integrity of cation-depleted RCNMV capsids was indirectly determined by DLS, which measures the changes in virion diameter before and after cation depletion. DLS measurements indicated that the cation-depleted capsids remained largely intact for at least 24 h (Fig. 1). Subsequent cryoEM analysis confirmed that virions remain intact after partial or complete removal of cations, even for periods as long as 7 days.
FIG. 1.
DLS determination of RCNMV virion size populations after depletion of divalent cations. DLS profiles of RCNMV virions were recorded after the noted treatment periods. CryoEM results confirmed that virion size did not change for a much longer period of time after the treatment (up to a week).
3D reconstruction of RCNMVNAT.
The RCNMVNAT vitrified sample consisted of monodisperse spherical particles (Fig. 2). After several iterations of alignment, classification, and orientation determination of particle images, followed by refinement, a de novo 3D RCNMV model at a resolution of 18 Å was obtained with IMAGIC. Further refinement of orientations and origins of individual virions with the 18-Å model as a template resulted in a 3D map of RCNMV at a resolution of 8.5 Å (conservative 0.5 Fourier shell correlation criterion) or a resolution of 7.5 Å with the 0.14 correlation criterion (35). The 3D reconstruction of RCNMVNAT (Fig. 3) clearly resolved the T=3 icosahedral symmetry (9) and showed that it is similar to that known for other species in the family Tombusviridae, i.e., TBSV (28), TCV (50), and CarMV (25). The close resemblance between RCNMVNAT and TBSV is supported by the similarity of their respective CP sequences (Fig. 4). The diameter of the RCNMVNAT virion is 366 Å, and the capsid shell, which comprises the entangled S domains of the three CP subunit conformations (A, B, and C), is ∼30 Å thick. Ninety distinct protrusions, each extending ∼37 Å above the capsid shell, arise from tight associations between pairs of neighboring P domains contributed by 30 C-C subunit homodimers and 60 A-B subunit heterodimers. The C-C protrusions are located at the icosahedral twofold axes, and the A and B subunits are arranged around the fivefold and threefold axes of symmetry, respectively (Fig. 3A). Each subunit has an overall L shape, with the bottom bar of the L corresponding to the S domain that forms the floor of the capsid shell and the vertical bar of the L corresponding to the P domain protrusions. The protrusions are approximately elliptical in cross section with dimensions of ∼50 Å (C-C) and 47 Å (A-B) along the long axis and 31 Å along the short axis. Each protrusion has a pair of dimples on either side (Fig. 3A) with an ∼13-Å-wide S-shaped ridge at the top (highest particle radius).
FIG. 2.
Cryoelectron micrograph of RCNMV virions embedded in vitreous ice. The characteristic bumpy appearance of virions is a consequence of the radially extended P domains of the CP subunits.
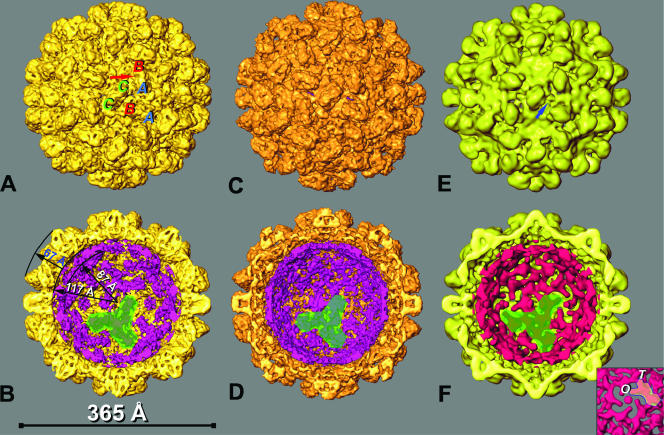
FIG. 3.
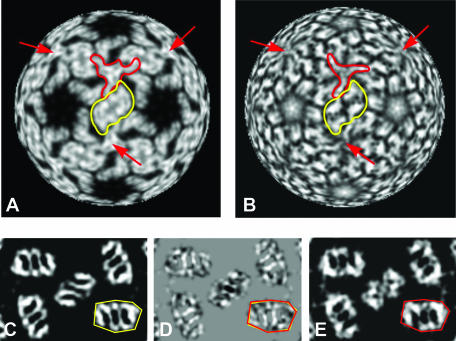
Shaded-surface representations of the 3D reconstructions of RCNMVNAT, RCNMV−Ca, and RCNMV−Ca/−Mg. (A) RCNMVNAT map at a resolution of 8.5 Å showing characteristic protrusions formed by P domains of the CP. The positions of the A, B, and C CP subunits within one asymmetric unit of the capsid are shown by corresponding letters. The red arrow points to the characteristic dimple in a P domain. (B) Cutaway view of RCNMVNAT. The RNA-protein inner cage, essentially disconnected from the capsid shell by a 7-Å gap, is shown in magenta with a single trimer of RNA-protein slabs highlighted in green. Twenty such trimers constitute the cage. (C) RCNMV−Ca map at 9 Å. (D) Cutaway view of RCNMV−Ca. The cage is essentially the same as in RCNMVNAT. (E) RCNMV−Ca/−Mg reconstruction at a resolution of 16.5 Å. (F) Cutaway view of RCNMV−Ca/−Mg with the cage colored red. Additional open channels through the capsid are visible (E, blue arrow). The inset is a view along the fivefold axis of panel F. The O and T labels identify two subdomains of RNA cage slabs splitting at the distal end of a single slab in RCNMVNAT or RCNMV−Ca.
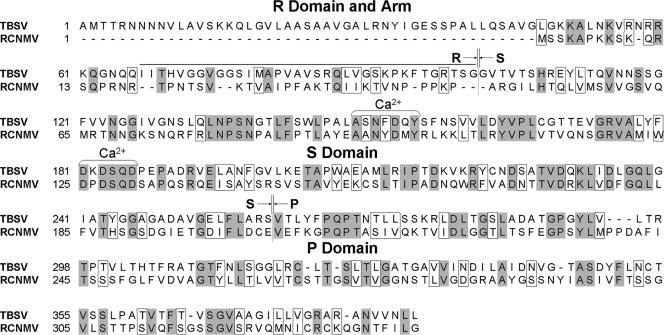
FIG. 4.
Amino acid sequence alignment of the RCNMV and TBSV CPs. The overall identity and similarity between the two proteins are 26 and 39%, respectively. The S domains are the most closely related (35% identical, 48% similar), and the P domains are more diverse (27% identical, 44% similar). There is no significant identity or similarity between the R domains and arm regions. Arrows delimit the structural domains. The sequence numbering and domain assignments are based on the TBSV X-ray structure (28). Identical residues are shaded gray, whereas similar residues are boxed. Brackets mark the conserved residues that provide the Ca2+ ligands in TBSV.
A prominent internal feature of the reconstruction is a cage-like density distribution lying between radii of 87 and 117 Å (Fig. 3B). This inner cage is composed of elongated slabs of density that appear to associate as trimers (one trimer is shaded green in Fig. 3B). The cage and the capsid have different symmetries; the capsid is composed of 180 CP subunits arranged with T=3 quasisymmetry, whereas the cage is composed of 60 slabs arranged with T=1 icosahedral symmetry. At the twofold axes, slabs are oriented in an antiparallel fashion and are separated by ∼50 Å. Similar cage-like structures have been observed in several other RNA viruses and virus-like particles (12, 20, 23, 29, 40, 44, 45).
Homology model of RCNMVNAT CP and fitting into density map.
An RCNMVNAT CP homology model was constructed to help better understand the structural organization of the virus. This model was created with SWISS-MODEL and the TBSV crystal structure because its CP had the highest sequence similarity to RCNMV among the solved Tombusviridae structures (Fig. 4). In creating S and P domain homology models for each of the RCNMV A, B, and C CP subunits, the C TBSV CP subunit was first subdivided into separate S, P, and extended-arm domains and the sequence for each of these pieces was separately aligned to the RCNMV sequence, followed by homology modeling for individual domains.
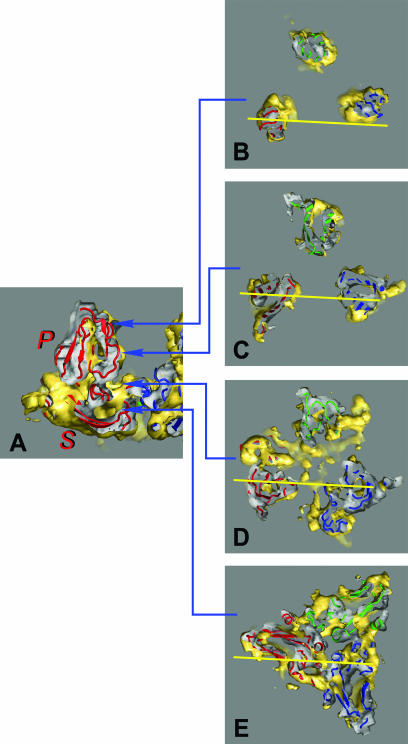
Each domain model was fitted as a separate rigid body (with Situs) to the A, B, and C subunit densities in the 8.5 Å RCNMVNAT cryoEM map. The separated S and P domains for each CP subunit were reunited postfitting to obtain final models for the complete A, B, and C subunits. These pseudoatomic models clearly demonstrated conformational differences between different CP subunits. A cross-correlation coefficient of 0.8 calculated between the EM density map and the homology model with an NMFF routine indicated good overall correspondence. In particular, the β-sheets comprising the S and P domains docked remarkably well into the EM density map and accounted for most of the observed capsid density. The central section of the B subunit (Fig. 5A) and cross-sections of the ABC trimer (Fig. 5B to E) are highlighted in Fig. 5. Slices through the P domains (Fig. 5B and C) exhibit clear, end-on views of β-sheets that fit well into the cryoEM map. Slices through the S domains (Fig. 5D and E) exhibit a more complex distribution of density that is consistent with the models, where the floor of the viral capsid is composed of a series of closely associated β-sheets. The structural transition from the RCNMVNAT to the RCNMV−Ca/−Mg state is shown in a simulated animation (see the supplemental material), where significant movement of the P and S domains is depicted.
FIG.5.
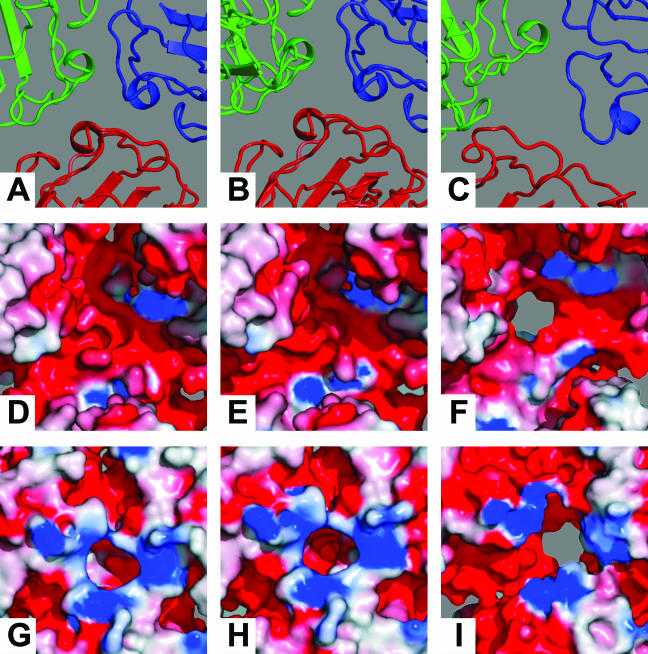
Homology models of the RCNMV CP subunits fitted into the RCNMVNAT density map. The yellow and gray shading highlights the outer and inner surfaces of the molecular envelope of the RCNMV cryoEM density map. The A, B, and C subunits are depicted in a colored cartoon representation in blue, red, and green, respectively. Panel A shows the B subunit viewed perpendicular to the central section through the capsid with a portion closest to the viewer removed. The locations of the 7 and P domains are denoted by labels S and P. Panels B to E show perpendicular sections through the RCNMVNAT ABC trimer at different levels. Panels B and C and panels D and E represent sections through the P and S domains, respectively. The yellow lines in each panel indicate the positions of the vertical central section shown in panel A.
To complete the fit and construct the entire RCNMV homology model, the extended arm of the TBSV C subunit was manually adjusted with the program O to fit into the tube-like density in the RCNMV reconstruction that lies between the capsid shell and the cage. This density easily accommodates the putative RCNMV CP arm sequence. The extended arm does not account for the density observed in the cage. Most of the CP sequence is accommodated within the capsid shell of the homology model; therefore, the slabs likely represent ordered segments of viral RNA or viral RNA-CP complexes (e.g., RNA-R domain complexes). Though the primary sequences of the TBSV and RCNMV extended arms exhibit little sequence identity, we compared the density distributions within the RCNMV and TBSV density maps (radial sections in Fig. 6A and B), where arm loops are most likely localized. These radial sections suggest very similar locations of the arm loops. The N terminus of the C subunit extended arm in the homology model lies ∼4.5 Å away from the threefold axes where three cage slabs associate to form trimers (Fig. 6B). They are coincident with density peaks that possibly originate from a β-annulus-like structure similar to that found in TBSV (Fig. 6A) (28).
FIG. 6.
Radial projections of virion density maps. Maps of TBSV (A) and RCNMVNAT (B) virions at a 124-Å radius corresponding to the S domains are shown. Protein density is shown in white. One C-C dimer is outlined in yellow for each virion. Trimers of extended arms, centered at one of the icosahedral threefold axes in each structure, are outlined in red. The N-terminal ends of the extended arms approach each other near the threefold axes and appear as triangular (TBSV) or globular (RCNMVNAT) peaks of density. Red arrows point to icosahedral threefold axes in the two structures. (C, D, and E) RCNMV density maps at a 162-Å radius corresponding to the P domains. RCNMVNAT (C), RCNMV−Ca (E), and RCNMVNAT-minus-RCNMV−Ca difference (D) maps are shown. Black and white striped features in the difference maps (D) highlight the small shifts and rotations of the P domains that accompany extraction of Ca2+ ions from RCNMVNAT. The yellow and red outlines in panels C and E, respectively, identify equivalent areas in RCNMVNAT and RCNMV−Ca. In panel D, the outlines shown in panels C and E are superimposed to indicate the small rotation (∼3°).
3D reconstruction of RCNMV−Ca virions.
The reconstruction of Ca2+-depleted virions (RCNMV−Ca) was determined ab initio in the same manner used to reconstruct the RCNMVNAT structure. The final 3D map was obtained at a resolution of 9 Å (Fig. 3C and D). The overall RCNMV−Ca structure appears quite similar to that of RCNMVNAT at a resolution of 9 Å, but detailed comparisons did reveal important differences. The CP subunits in RCNMV−Ca virions were rotated ∼2 to 4° clockwise relative to their orientations in RCNMVNAT and were translated in tangential directions by ∼4 to 8 Å (Fig. 6C, D, and E). Prominent densities in the difference map (Fig. 6D; black = positive differences; white = negative differences) highlight regions of the P domains that change upon Ca2+ chelation. All RCNMV−Ca subunits are synchronously shifted and rotated (Fig. 3), which leaves a slightly thinner capsid shell at the quasithreefold axes.
3D reconstruction of RCNMV−Ca/−Mg virions.
Preliminary analysis of cryoEM images of divalent-cation-depleted RCNMV (RCNMV−Ca/−Mg) confirmed that treatment does not significantly alter the virion size. Hence, we used the 3D reconstruction of RCNMVNAT at a resolution of 18 Å as the initial model for the orientation and origin search with images of RCNMV−Ca/−Mg to produce a density map with a final resolution of 16.5 Å. Although the size and overall organization of RCNMV−Ca/−Mg were similar to those of RCNMVNAT, including the presence of a well-defined cage (Fig. 3E and F), significant differences appeared throughout the structure. The protrusions are unaltered in height but become shorter (42 Å versus 50 Å) and wider (38 Å versus 31 Å) in RCNMV−Ca/−Mg and lack the ridge at high radius present in the protrusions of RCNMVNAT (compare Fig. 3A, C, and E). A new and prominent feature of the RCNMV−Ca/−Mg reconstruction is the presence of ∼11- to 13-Å-diameter holes that traverse the capsid shell at the pseudothreefold axes (Fig. 3E).
Even though the RCNMV−Ca/−Mg virions lack Mg2+ ions, the cage is still present and maintains a thickness and general organization similar to those found in RCNMVNAT. However, the slabs are less well defined in RCNMV−Ca/−Mg (Fig. 3F). The distal end of each slab in RCNMVNAT subdivides into two subdomains in RCNMV−Ca/−Mg. One of these subdomains forms a spoke-like structure around the fivefold axis (O); the other (T) lies perpendicular to O (Fig. 3, inset).
Fitting the homology model into EM maps of RCNMV−Ca and RCNMV−Ca/−Mg virions.
As described above for the RCNMVNAT reconstruction, the same A, B, and C subunit homology models were fitted into the RCNMV−Ca 9-Å density map. An overall cross-correlation coefficient of 0.75 signified general correspondence. Comparison of the RCNMVNAT and RCNMV−Ca models showed that the CP subunits are shifted and rotated 2 to 4° more clockwise around the quasithreefold axes after Ca2+ ion depletion (Fig. 7). This rearrangement of CP subunits leads to slight modifications in the shape of the protrusions, as well as the floor. The P domains in the CP subunits of a trimer tilt slightly away from the quasithreefold axis. At the same time, the S domains also tilt slightly, leading to a smoother, more rounded capsid floor.
FIG. 7.
Close-up views of interfaces of the A, B, and C CP subunits in pseudoatomic homology models. The loops and α-helices that form the lining of the quasithreefold axis move slightly away from the axis when Ca2+ ions are depleted and significantly farther when both Ca2+ and Mg2+ are depleted, resulting in the opening of an 11- to 13-Å channel through the capsid shell in RCNMV−Ca/−Mg virions. Electrostatic-potential diagrams show the charge distributions of the inner (middle row) and outer (lower row) surfaces. The concentration of positive charge just outside the cavities in RCNMVNAT and RCNMV−Ca or the channel in RCNMV−Ca/−Mg may serve to attract and position the RNA for exit through the charged channel. The outer surface of the capsid in the region surrounding the local threefold axis is predominantly negatively charged, which might help repel RNA as it emerges from the surface. (A to C) Close-up views of a portion of the A, B, and C subunit interfaces in the pseudoatomic homology models for the native RCNMVNAT (A), RCNMV−Ca (B), and RCNMV−Ca/−Mg (C) structures. The loops and α-helices that form the lining of the local threefold axis move slightly from the axis when Ca2+ ions are chelated (B) and significantly farther when both Ca2+ and Mg2+ ions are chelated (C), opening an 11- to 13-Å channel through the capsid shell. (D to I) Electrostatic-potential representations of CP trimers showing charge distributions on the outer (D to F) and inner (G to I) surfaces for RCNMVNAT (D and G), RCNMV−Ca (E and H), and RCNMV−Ca/−Mg (F and I). Red represents negative charges, whereas positively charged residues are shown in blue. On the inner capsid surface, the concentration of positive charge just outside the cavities in RCNMVNAT (G) and RCNMV−Ca (H) or the channel in RCNMV−Ca/−Mg (I) may serve to attract and position the RNA for exit through the negatively charged channel (not shown). The outer surface of the capsid in the region surrounding the local threefold axis is predominantly negatively charged, which might help repel RNA as it emerges from the surface and therefore may facilitate the binding of ribosomes to the protruding ends.
The homology models were initially fitted manually into the 16.5-Å resolution RCNMV−Ca/−Mg map. Here, the rigid-body fitting protocol failed to yield satisfactory results, especially for the S domains, where portions of the model ended up well outside the density for the virion shell, suggesting that the S domains had adopted a different conformation. NMFF procedures (42) were then used to dock the homology model into the RCNMV−Ca/−Mg map. The resulting model was much improved compared to that obtained by rigid-body fitting (Table 2), as most of the S domain residues now lay within the EM density envelope. Correlations between the model and the map improved, on average, from 0.73 to 0.82. To avoid problems potentially caused by overfitting, only the 20 lowest-frequency normal modes were permitted in the NMFF procedure (42). These modes corresponded to large-scale conformational changes and were sufficient for fitting maps at relatively low resolution.
TABLE 2.
Correlations between pseudoatomic models and RCNMV−Ca/−Mg map before and after application of NMFF procedure
| CP subunit | Correlation coefficient
|
|
|---|---|---|
| Before NMFF | After NMFF | |
| A | 0.76 | 0.85 |
| B | 0.67 | 0.82 |
| C | 0.76 | 0.80 |
Close inspection of the RCNMVNAT and RCNMV−Ca/−Mg capsid models demonstrated significant internal rearrangements of the CP subunits (Fig. 3 and 7). The S and P domains tilt in much the same way as in the RCNMV−Ca structure, but the tilts are more severe. On average, the P domains tilt away from the quasithreefold axis by ∼10° and rotate clockwise, thereby opening up this region of the virion surface even more than in the RCNMV−Ca map (Fig. 7). The S domains also tilt away from the axis and rotate around their long axes, making the capsid floor significantly smoother than in either the RCNMVNAT or the RCNMV−Ca structure (Fig. 3 and 7). At the same time, the subunits within the trimer move apart and this generates an 11- to 13-Å diameter channel in the capsid shell (Fig. 7). No such channel is visible in the RCNMVNAT or the RCNMV−Ca map (Fig. 7).
On the basis of the known TBSV crystal structure and our homology-modeling experiments, we speculate that Ca2+ ions are bound at the RCNMV S-S domain interfaces. Although Ca2+ ions cannot be directly resolved in the present reconstructed maps, the fitting of homology models into the maps, coupled with analysis of sequence data, allows us to define the putative Ca2+-binding sites. The CP sequence alignment demonstrates that there are conserved Ca2+-binding residues at these interfaces (RCNMV residues 93 to 99 and 125 to 130) (Fig. 4).
RCNMV RNA nuclease sensitivity is dependent on the divalent-cation content of capsids.
The loss of stabilizing Ca2+ ions from plant virus capsids was hypothesized to be an early event in the virus life cycle following entry of virions into cells (13). On the basis of the results obtained in this study (RCNMV dialysis against deionized water), it is likely that the RCNMV loses both Ca2+ and Mg2+ ions upon exposure to the cytosol. The DLS and cryoEM studies described here have shown that loss of divalent cations does not disassemble the RCNMV capsid; rather, it opens channels through the capsid, coupled with rearrangement of the viral RNA (Fig. 3 and 7).
To determine if depletion of capsid divalent cations increases the accessibility of viral RNA to the external environment, the infectivity of divalent-cation-depleted RCNMV was determined after nuclease treatment. There was no observed difference in infectivity between treated and untreated RCNMVNAT virions (Table 3), suggesting that the RCNMV genome is fully protected within virion capsids containing the full complement of divalent cations. Depletion of Ca2+ ions resulted in a 33 to 81% reduction in infectivity after nuclease treatment, with the amount of reduction dependent on the nuclease used (Table 3). Infectivity was further reduced by nuclease treatment after both Ca2+ and Mg2+ ions were removed. Only a slight decrease (∼11%) in the infectivity of RCNMV−Ca versus RCNMV−Ca/−Mg was observed after RNase V1 treatment compared to a more-than-50% decrease observed with RNases A and T1 (Table 3). Interestingly, both single-strand-specific RNases (A and T1) and a double-strand-specific RNase (V1) also cause a decrease in the infectivity of cation-depleted virions. To confirm that nuclease digestion occurred in vitro, RNA was extracted from nuclease-treated, cation-depleted virions and shown by electrophoresis to be digested (data not shown). The loss of infectivity after nuclease treatment suggests that removal of divalent cations from the capsid causes both single- and double-stranded portions of the RCNMV RNA genome to become exposed to the environment.
TABLE 3.
Local-lesion infectivity assay of chelator-treated RCNMV virions on C. quinoa after exposure to nucleases
| Chelator treatmenta | % Infectivity after treatmentb with:
|
|||
|---|---|---|---|---|
| No RNase | RNase A | RNase T1 | RNase V1 | |
| Mock treatmentc | 0 | 0 | 0 | 0 |
| None | 100 (53 ± 14) | 100 (53 ± 2) | 100 (53 ± 11) | 100 (45 ± 13) |
| EGTA | 106 (56 ± 6) | 19 (10 ± 3) | 49 (26 ± 4) | 67 (30 ± 6) |
| EDTA | 91 (48 ± 4) | 9 (5 ± 1) | 23 (12 ± 5) | 56 (25 ± 6) |
Four micrograms of purified RCNMV virions was treated with or without a chelator, followed by nuclease digestion prior to inoculation onto C. quinoa leaves.
Infectivity is expressed as a percentage of the number of lesions formed per leaf by treated virions versus untreated virions. The average number of lesions observed and standard deviation from four leaves are shown in parentheses.
Mock treatment was done with buffer but no RCNMV virions.
DISCUSSION
Structural analysis of RCNMV provides additional evidence that viruses in the Tombusviridae family have a conserved morphology despite differences in their amino acid sequences. This suggests that the assembly and disassembly mechanisms of species in the family Tombusviridae are, in general, similar, with possible individual variations on a common theme. The unusually robust virions of TBSV, RCNMV, and CarMV hint at a trigger factor that causes at least weakening, if not complete disassembly, of the capsid, followed by genome leakage in the cytosol and production of new virions. The RCNMV structures with different divalent-cation contents obtained in the present study revealed significant conformational rearrangements upon cation depletion with channels opening in the capsid. These channels could provide conduits for the viral RNA to exit the capsid and become available for replication.
CP sequence comparisons among the diverse members of the Tombusviridae family revealed significant variation in the number of residues that comprise the arms and R domains, with the longest sequences belonging to TBSV and Tobacco necrosis virus. The putative R domain and arm regions of the RCNMV CP are 52 residues shorter than the corresponding regions in TBSV (Fig. 4) and may correlate with the necessity to package a larger genome (∼5.3 to 5.8 kb in RCNMV versus ∼4.9 kb in TBSV). This hypothesis for structure conservation has been recently confirmed by the ability of a series of TBSV CP N-terminal deletion mutants to form virus-like particles (16). Coincidentally, a 52-residue TBSV deletion mutant (CP-NΔ52) produces particles which are morphologically very similar to wild-type TBSV virions, and this resembles the situation with the shorter RCNMV CP. The major structural differences between RCNMV and TBSV can be partially attributed to the shorter CP amino terminus in RCNMV.
One of the most striking features of the RCNMV virion 3D reconstruction is the cage, a structure that is composed of 60 slabs organized as 20 trimers, each with a prominent knob at its center (Fig. 3). The cage and capsid shell are separated by a 7- to 9-Å gap. The pseudoatomic model of the RCNMV CP subunit accounts for most of the densities in the capsid shell and also the connecting densities corresponding to the extended arms. Altogether, with the exception of 14 to 16 N-terminal residues, the model accounts for the entire CP sequence. The N-terminal residues may be part of the cage slabs. Each slab is ∼16 Å thick and ∼25 Å wide in cross-section. A feature this large can easily accommodate a single-stranded RNA (ssRNA) fragment complexed with the N-terminal part corresponding to the R domain of CP. Excluded-volume estimates suggest that the cage accounts for as much as 35 to 40% of the total RCNMV RNA genome; the remaining genomic RNA lies inside the cage and appears to be more loosely packed and is most likely disordered or does not conform to icosahedral symmetry.
The presence of an ordered genome has been described for a number of viruses. Within the family Tombusviridae, cryoEM reconstructions of TBSV (1) and Hibiscus chlorotic ringspot virus (12) revealed the presence of a layer inside the virion capsid tentatively formed by the CP R domains complexed with genomic RNA. However, these regions in TBSV and Hibiscus chlorotic ringspot virus appeared diffuse with no characteristic discernible details like the discrete slabs observed in the RCNMV cage. This lack of an ordered inner structure in TBSV does not correlate with infectivity since a TBSV CP deletion mutant lacking the 30 N-proximal residues of the RNA-binding domain forms virus-like particles which vary in size yet are just as infectious as wild-type virions (11).
In TBSV, a β-annulus formed by three interlocking extended arms (28) is located at the threefold axis inside the capsid shell. Each arm extends away from the annulus and connects to a corresponding S domain. Similar density features at the same threefold location are observed in the RCNMVNAT (Fig. 6A, red arrows and red outlines) and RCNMV−Ca maps. The β-annuli in the RCNMV density map appear as knobs at the center of the slab trimer, and the arms appear as well-defined, curved tubes of density that run from the knob toward the capsid shell (data not shown). On the basis of the sequence alignment of TBSV and RCNMV (Fig. 4), RCNMV appears to lack a globular R domain like that known to be present in TBSV. In RCNMV, the N-terminal sequence is predominantly basic and we believe that up to 16 N-terminal residues interact with ordered RNA and are involved in the formation of the slabs.
In addition to determining the structure of the RCNMVNAT capsid, this study has focused on the role of Ca2+ and Mg2+ ions in RCNMV 3D organization and assembly. Atomic absorption spectroscopy showed that each virion contains significant amounts of divalent cations. When native virions were exposed to low Ca2+ or Mg2+ ion conditions, such as exhaustive dialysis against deionized water, these cations were leached from the virions. To probe their role in RCNMV capsid stability, structural and functional studies were performed with divalent cations selectively removed from virions by the chelating agents EGTA (Ca2+) and EDTA (Ca2+ and Mg2+). Comparative analysis of the structures obtained has shown that removal of Ca2+ ions induces movements of the S and P domains (Fig. 3 and 7). The removal of both Ca2+ and Mg2+ ions triggers more global rearrangement in the RCNMV structure (Fig. 3 and 7). These data indicate that Ca2+ ions are not solely responsible for structural dynamics of RCNMV virions; Mg2+ ions also significantly contribute to the rearrangements.
Upon loss of divalent cations, each of the 180 P domains exhibited increased flexibility and they reoriented by 30 or more degrees (Fig. 3). As a result, the P-P interfaces are changed significantly. Increased mobility of the P domains appears to explain, at least in part, why the resolution of the RCNMV−Ca/−Mg map was limited to 16.5 Å. Nevertheless, use of the NMFF procedure provided a means to quantitatively model the changes observed in the cryoEM map. The flexible fitting procedure showed dramatic changes in S domains upon divalent-cation depletion that cannot be described in terms of simple rotations and translations. Instead, the S domains appear to adopt a conformation different from that seen in RCNMVNAT or RCNMV−Ca virions. These conformational changes in the P and S domains are responsible for forming the 11- to 13-Å-diameter channels at the quasithreefold axes (Fig. 3 and 7). The differences were especially prominent for both α-helices and for the loop (R165 to D174) lining the axis. In its native conformation, loop I158 to G181 lies completely outside of the density envelope in the RCNMV−Ca/−Mg map when fitted as a rigid body. The NMFF modeling experiments strongly suggest that refolding of this loop occurs (Table 2). It is noteworthy in this context that the swollen TBSV structure did not exhibit any significant CP subunit refolding (33). The conformational changes observed in RCNMV upon loss of divalent cations most likely results from the combined effect of Ca2+ and Mg2+ ions. Loss of Ca2+ alone does not significantly alter virion integrity. Removal of Ca2+ seems to initiate structural changes such as small-scale movements of the P and S domains and the slight increase in intersubunit distances at the quasithreefold axes of the capsid, where a channel appears after both cations are lost. This fact alone suggests that the RCNMV cage is tightly linked to the capsid and might exert a force when cations leave the virions, leading to opening of the channels. Additionally, our experiments with cation depletion demonstrated that pH changes used in earlier experiments with TBSV (13) are not necessary for the channels opening in RCNMV.
Depletion of both Ca2+ and Mg2+ ions also leads to more significant changes in the structure of RCNMV (Fig. 3). We assume that Mg2+ ions bind to the ssRNA genome, neutralizing its charge and aiding in its condensation in virions. Loss of Mg2+ ions could create a charge imbalance and lead to the observed splitting of the cage slabs (Fig. 3, inset).
The channels that arise at the quasithreefold axes in the RCNMV−Ca/−Mg virions are too constricted to permit nucleases in their native form (>55 Å in diameter) to penetrate the capsid and to cleave the packaged RNA genome. However, these channels are sufficient in size to permit ssRNA (11 to 13 Å in diameter) to leak from the capsid. Such leakage would correlate with increased nuclease sensitivity of RCNMV−Ca/−Mg to single-strand-specific RNases (Table 3). The inner part of the channel (the entrance) includes a few basic residues but is predominantly lined with negatively charged residues (Fig. 7). The termini of the genomic RNAs might be attracted to the entrance of the channel, but repulsive forces within the channel would facilitate transit of the RNA to the cytosol. The changes observed in both the cage and the outer shell of RCNMV−Ca/−Mg may point to the actual RNA release mechanism that occurs for RCNMVNAT in vivo. Interestingly, studies of Cowpea chlorotic mottle virus have similarly suggested that viral RNA release by free diffusion occurs through channels at the quasithreefold axes (17). Channels similar to those in RCNMV were observed in a recent cryoEM reconstruction of TBSV virions that were depleted of divalent cations, followed by a rise in pH to 7.5 (1). Free-Ca2+ and -Mg2+ ion concentrations are typically in the millimolar range in soil and groundwater (3, 6). In contrast, they range between submicromolar and micromolar levels in the cytosol. These low levels are maintained and regulated by several enzymes (22, 31). Such low cation concentrations could trigger leaching of Ca2+ and Mg2+ ions from RCNMV virions as they enter cells and in turn would produce the conformational changes that open channels for RNA to exit. In addition, it is also known that a cellular response to stress and infection is to increase cytosolic calcium (2). Further, once a cell begins apoptosis, regulation of cytosolic calcium is lost (10), ensuring an increased supply of calcium for virion formation late in the infection cycle.
Supplementary Material
Acknowledgments
We thank Y. Ji and C. Xiao, and R. Ashmore for use of their image-processing programs.
This research was supported in part by NIH grant GM-33050 to T.S.B., NSF grant MCB-0077964 to S.A.L. and T.L.S., a grant from the W. M. Keck Foundation to the Purdue Structural Biology Group for the purchase of a CM 300 FEG microscope, and a Purdue University reinvestment grant to the Structural Biology Group.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 18 August 2006.
REFERENCES
- 1.Aramayo, R., C. Merigoux, E. Larquet, P. Bron, J. Perez, C. Dumas, P. Vachette, and N. Boisset. 2005. Divalent ion-dependent swelling of Tomato Bushy Stunt Virus: a multi-approach study. Biochim. Biophys. Acta 1724:345-354. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson, M. M., L. D. Keppler, E. W. Orlandi, C. J. Baker, and C. F. Mischke. 1990. Involvement of plasma membrane calcium influx in bacterial induction of the K/H and hypersensitive responses in tobacco. Plant Physiol. 92:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber, S. A. 1995. Soil nutrient bioavailability, p. 271. Wiley-Interscience, New York, N.Y.
- 4.Basnayake, V. R., T. L. Sit, and S. A. Lommel. 2006. The genomic RNA packaging scheme of Red clover necrotic mosaic virus. Virology 345:532-539. [DOI] [PubMed] [Google Scholar]
- 5.Bowman, V. D., E. S. Chase, A. W. Franz, P. R. Chipman, X. Zhang, K. L. Perry, T. S. Baker, and T. J. Smith. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 76:12250-12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briggs, J. C., and J. F. Ficke. 1977. Quality of rivers of the United States, 1975 water year—based on the National Stream Quality Accounting Network (NASQAN). U.S. Geological Survey water-supply paper 1540-C.
- 7.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4:187-217. [Google Scholar]
- 8.Caldwell, P. C. 1970. Calcium chelation and buffers, p. 10-16. In A. W. Cuthbert (ed.), Calcium and cellular function. Macmillan, London, United Kingdom.
- 9.Caspar, D. L., and A. Klug. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27:1-24. [DOI] [PubMed] [Google Scholar]
- 10.Clough, S. J., K. A. Fengler, I. C. Yu, B. Lippok, R. K. Smith, Jr., and A. F. Bent. 2000. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. USA 97:9323-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desvoyes, B., and H. B. Scholthof. 2002. Host-dependent recombination of a Tomato bushy stunt virus coat protein mutant yields truncated capsid subunits that form virus-like complexes which benefit systemic spread. Virology 304:434-442. [DOI] [PubMed] [Google Scholar]
- 12.Doan, D. N., K. C. Lee, P. Laurinmaki, S. Butcher, S. M. Wong, and T. Dokland. 2003. Three-dimensional reconstruction of hibiscus chlorotic ringspot virus. J. Struct. Biol. 144:253-261. [DOI] [PubMed] [Google Scholar]
- 13.Durham, A. C., D. A. Hendry, and M. B. Von Wechmar. 1977. Does calcium ion binding control plant virus disassembly? Virology 77:524-533. [DOI] [PubMed] [Google Scholar]
- 14.Fishman, M. J., and S. C. Downs. 1966. Methods for analysis of selected metals in water by atomic absorption: U.S. Geological Survey water-supply paper 1540-C, p. C26-C28.
- 15.Hallett, F. R., J. Watton, and P. Krygsman. 1991. Vesicle sizing: number distributions by dynamic light scattering. Biophys. J. 59:357-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu, C., P. Singh, W. Ochoa, D. J. Manayani, M. Manchester, A. Schneemann, and V. S. Reddy. 2006. Characterization of polymorphism displayed by the coat protein mutants of tomato bushy stunt virus. Virology 349:222-229. [DOI] [PubMed] [Google Scholar]
- 17.Isea, R., C. Aponte, and R. Cipriani. 2004. Can the RNA of the cowpea chlorotic mottle virus be released through a channel by means of free diffusion? A test in silico. Biophys. Chem. 107:101-106. [DOI] [PubMed] [Google Scholar]
- 18.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47(Pt. 2):110-119. [DOI] [PubMed] [Google Scholar]
- 19.Larson, S. B., J. Day, A. Greenwood, and A. McPherson. 1998. Refined structure of satellite tobacco mosaic virus at 1.8 Å resolution. J. Mol. Biol. 277:37-59. [DOI] [PubMed] [Google Scholar]
- 20.Larson, S. B., and A. McPherson. 2001. Satellite tobacco mosaic virus RNA: structure and implications for assembly. Curr. Opin. Struct. Biol. 11:59-65. [DOI] [PubMed] [Google Scholar]
- 21.Lommel, S. A. 1983. Ph.D. dissertation. University California, Berkeley.
- 22.Luan, S., J. Kudla, M. Rodriguez-Concepcion, S. Yalovsky, and W. Gruissem. 2002. Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14(Suppl.):S389-S400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucas, R. W., S. B. Larson, M. A. Canady, and A. McPherson. 2002. The structure of tomato aspermy virus by X-ray crystallography. J. Struct. Biol. 139:90-102. [DOI] [PubMed] [Google Scholar]
- 24.Montelius, I., L. Liljas, and T. Unge. 1988. Structure of EDTA-treated satellite tobacco necrosis virus at pH 6.5. J. Mol. Biol. 201:353-363. [DOI] [PubMed] [Google Scholar]
- 25.Morgunova, E. Y., Z. Dauter, E. Fry, D. I. Stuart, V. Y. Stel'mashchuk, A. M. Mikhailov, K. S. Wilson, and B. K. Vainshtein. 1994. The atomic structure of Carnation Mottle Virus capsid protein. FEBS Lett. 338:267-271. [DOI] [PubMed] [Google Scholar]
- 26.Musil, M., and J. Gallo. 1982. Serotypes of red clover necrotic mosaic virus. I. Characterization of three serotypes. Acta Virol. 26:497-501. [PubMed] [Google Scholar]
- 27.Oda, Y., K. Saeki, Y. Takahashi, T. Maeda, H. Naitow, T. Tsukihara, and K. Fukuyama. 2000. Crystal structure of tobacco necrosis virus at 2.25 Å resolution. J. Mol. Biol. 300:153-169. [DOI] [PubMed] [Google Scholar]
- 28.Olson, A. J., G. Bricogne, and S. C. Harrison. 1983. Structure of tomato busy stunt virus. IV. The virus particle at 2.9 Å resolution. J. Mol. Biol. 171:61-93. [DOI] [PubMed] [Google Scholar]
- 29.Opalka, N., M. Tihova, C. Brugidou, A. Kumar, R. N. Beachy, C. M. Fauquet, and M. Yeager. 2000. Structure of native and expanded sobemoviruses by electron cryo-microscopy and image reconstruction. J. Mol. Biol. 303:197-211. [DOI] [PubMed] [Google Scholar]
- 30.Orlova, E. V., M. B. Sherman, W. Chiu, H. Mowri, L. C. Smith, and A. M. Gotto. 1999. Three-dimensional structure of low density lipoproteins by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA 96:8420-8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pittman, J. K., and K. D. Hirschi. 2003. Don't shoot the (second) messenger: endomembrane transporters and binding proteins modulate cytosolic Ca2+ levels. Curr. Opin. Plant Biol. 6:257-262. [DOI] [PubMed] [Google Scholar]
- 32.Poirot, O., K. Suhre, C. Abergel, E. O'Toole, and C. Notredame. 2004. 3DCoffee@igs: a web server for combining sequences and structures into a multiple sequence alignment. Nucleic Acids Res. 32:W37-W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson, I. K., and S. C. Harrison. 1982. Structure of the expanded state of tomato bushy stunt virus. Nature 297:563-568. [Google Scholar]
- 34.Rochon, D., K. Kakani, M. Robbins, and R. Reade. 2004. Molecular aspects of plant virus transmission by olpidium and plasmodiophorid vectors. Annu. Rev. Phytopathol. 42:211-241. [DOI] [PubMed] [Google Scholar]
- 35.Rosenthal, P. B., and R. Henderson. 2003. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333:721-745. [DOI] [PubMed] [Google Scholar]
- 36.Rossmann, M. G., C. Abad-Zapatero, J. W. Erickson, and H. S. Savithri. 1983. RNA-protein interactions in some small plant viruses. J. Biomol. Struct. Dyn. 1:565-579. [DOI] [PubMed] [Google Scholar]
- 37.Saxton, W. O., and W. Baumeister. 1982. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 127:127-138. [DOI] [PubMed] [Google Scholar]
- 38.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serysheva, I. I., E. V. Orlova, W. Chiu, M. B. Sherman, S. L. Hamilton, and M. van Heel. 1995. Electron cryomicroscopy and angular reconstitution used to visualize the skeletal muscle calcium release channel. Nat. Struct. Biol. 2:18-24. [DOI] [PubMed] [Google Scholar]
- 40.Speir, J. A., S. Munshi, G. Wang, T. S. Baker, and J. E. Johnson. 1995. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 3:63-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tama, F., F. X. Gadea, O. Marques, and Y. H. Sanejouand. 2000. Building-block approach for determining low-frequency normal modes of macromolecules. Proteins 41:1-7. [DOI] [PubMed] [Google Scholar]
- 42.Tama, F., O. Miyashita, and C. L. Brooks III. 2004. Flexible multi-scale fitting of atomic structures into low-resolution electron density maps with elastic network normal mode analysis. J. Mol. Biol. 337:985-999. [DOI] [PubMed] [Google Scholar]
- 43.Tama, F., and Y. H. Sanejouand. 2001. Conformational change of proteins arising from normal mode calculations. Protein Eng. 14:1-6. [DOI] [PubMed] [Google Scholar]
- 44.Tang, L., K. N. Johnson, L. A. Ball, T. Lin, M. Yeager, and J. E. Johnson. 2001. The structure of pariacoto virus reveals a dodecahedral cage of duplex RNA. Nat. Struct. Biol. 8:77-83. [DOI] [PubMed] [Google Scholar]
- 45.Tihova, M., K. A. Dryden, T. V. Le, S. C. Harvey, J. E. Johnson, M. Yeager, and A. Schneemann. 2004. Nodavirus coat protein imposes dodecahedral RNA structure independent of nucleotide sequence and length. J. Virol. 78:2897-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Heel, M. 1987. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy 21:111-124. [DOI] [PubMed] [Google Scholar]
- 47.van Heel, M., and J. Frank. 1981. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy 6:187-194. [DOI] [PubMed] [Google Scholar]
- 48.van Heel, M., and G. Harauz. 1986. Resolution criteria for three dimensional reconstructions. Optik 73:119-122. [Google Scholar]
- 49.van Heel, M., G. Harauz, and E. V. Orlova. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 50.Wei, N., L. A. Heaton, T. J. Morris, and S. C. Harrison. 1990. Structure and assembly of turnip crinkle virus. VI. Identification of coat protein binding sites on the RNA. J. Mol. Biol. 214:85-95. [DOI] [PubMed] [Google Scholar]
- 51.Wiener, N. 1949. Extrapolation, interpolation, and smoothing of stationary time series with engineering applications. The MIT Press, Cambridge, Mass.
- 52.Wriggers, W., and S. Birmanns. 2001. Using Situs for flexible and rigid-body fitting of multiresolution single-molecule data. J. Struct. Biol. 133:193-202. [DOI] [PubMed] [Google Scholar]
- 53.Xiong, Z. G., and S. A. Lommel. 1991. Red clover necrotic mosaic virus infectious transcripts synthesized in vitro. Virology 182:388-392. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, X., S. B. Walker, P. R. Chipman, M. L. Nibert, and T. S. Baker. 2003. Reovirus polymerase lambda 3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat. Struct. Biol. 10:1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, Z. H., S. Hardt, B. Wang, M. B. Sherman, J. Jakana, and W. Chiu. 1996. CTF determination of images of ice-embedded single particles using a graphics interface. J. Struct. Biol. 116:216-222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.