Abstract
We report the first genome-wide comparison of in vivo promoter activities of a group of human-specific endogenous retroviruses in healthy and cancerous germ line tissues. To this end, we employed a recently developed technique termed genomic repeat expression monitoring. We found that at least 50% of human-specific long terminal repeats (LTRs) possessed promoter activity, and many of them were up- or downregulated in a seminoma. Individual LTRs were expressed at markedly different levels, ranging from ∼0.001 to ∼3% of the housekeeping beta-actin gene transcript level. We demonstrated that the main factors affecting the LTR promoter activity were the LTR type (5′-proviral, 3′ proviral, or solitary) and position with regard to genes. The averaged promoter strengths of solitary and 3′-proviral LTRs were almost identical in both tissues, whereas 5′-proviral LTRs displayed two- to fivefold higher promoter activities. The relative content of promoter-active LTRs in gene-rich regions was significantly higher than that in gene-poor loci. This content was maximal in those regions where LTRs “overlapped” readthrough transcripts. Although many promoter-active LTRs were mapped near known genes, no clear-cut correlation was observed between transcriptional activities of genes and neighboring LTRs. Our data also suggest a selective suppression of transcription for LTRs located in gene introns.
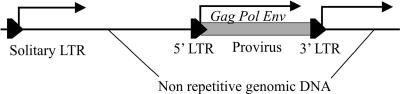
Retroelements (REs) occupy up to 30 to 40% of vertebrate genomes (21, 36, 48) and are suggested to be potent agents of genomic instability. They cause numerous host DNA rearrangements due to recombination events (19), by transduction of 5′ (22) or 3′ (37) RE flanking sequences into new genomic loci, by creating pseudogenes (14), or by causing RNA recombination (5, 10). Recently expanded gene classes in the human genome, such as those involved in immunity or responses to external stimuli, have transcripts enriched in REs, suggesting a significant role of REs in the diversification and evolution of mammalian genes (45). As mobile carriers of transcriptional regulatory modules, REs can affect host gene expression (27, 31, 42), thus probably taking part in speciation processes (25). In particular, REs might be at least partly responsible for phenotypic differences between Homo sapiens and its closest relatives, Pan paniscus and Pan troglodytes chimpanzees (11, 43). Likely candidates for such a role are endogenous retroviral long terminal repeats (LTRs) (12). Their structure harbors functional enhancers (40), promoters, and polyadenylation signals (43) normally used for retroviral gene expression. However, it was recently demonstrated that LTRs may drive the transcription of adjacent host genomic sequences (26, 45). In the human genome, LTRs may either flank endogenous retroviral “bodies” or exist in the form of solitary LTRs, arisen most probably due to homologous recombination between two identical retroviral LTRs (20, 34) (Fig. 1).
FIG. 1.
Schematic representation of solitary (left) and proviral (right) LTR expression. The transcription driven from 5′-proviral LTRs results in mRNAs of viral genes, whereas the expression of either solitary or 3′-proviral LTRs results in the transcription of host nonrepetitive genomic sequences flanking the 3′ ends of the retroelements.
In attempts to identify factors that might be involved in human-chimpanzee evolutionary divergence, we have focused our research on promoter activities of the HERV-K (HML-2) family of human endogenous retroviruses, the only retroviral group known to contain human-specific members (9, 35). Human-specific HERV-K (HML-2) LTRs share significant sequence identity and form a well-defined cluster (named the HS family) in a phylogenetic tree (9, 35). The members of this family, who have retained their transcriptional activity (4, 15, 16, 47), were found to be tissue-specifically methylated (23, 24, 28) and probably still keep some infectious potential (13, 17, 39, 44). The HS family is thought to be the most biologically active retroviral family in human cells. Several individual HS LTRs are polymorphic in human populations (3, 30, 33), which suggests their very recent integration. In the human genome, the HS family is represented by 156 mostly (∼86%) human-specific LTR sequences. The HS family members can be parts of full-sized HERV-K (HML-2) proviruses (12% of individual HS representatives), truncated proviruses (5%), or solitary LTRs (83%).
Recently, we developed a new technique, termed genomic repeat expression monitoring (GREM), for experimental genome-wide identification of promoter-active repetitive elements (7). The technique is based on hybridization of repeat 3′-flanking genomic DNA to pools of total cDNA 5′-terminal parts, followed by selective PCR amplification of the genomic DNA-cDNA heteroduplexes. The resulting library of cDNA/genomic DNA hybrids can be used as a source of tags for individual transcriptionally active repeats. GREM was shown to be adequate for tasks of both quantitative and qualitative analyses of promoter activity. In model experiments, we used GREM to create the first genome-wide map of HS elements that display promoter activity in the testis. Here we utilized GREM for the first comprehensive comparison of HS element promoter activities in healthy human tissue (testicular parenchyma) and in the corresponding cancer (seminoma) from the same patient. We found that at least 50% of HS LTRs were promoter active, and we mapped 20 new functional human-specific promoters. The transcription of many HS LTRs was up- or downregulated in the seminoma. The promoter strengths differed greatly among individual HS elements, and their transcript levels ranged from ∼3 to ∼0.001% of the marker beta-actin gene transcript level. We showed that the main factors affecting the LTR promoter activity were the LTR type (5′ proviral, 3′ proviral, or solitary) and location relative to genes.
MATERIALS AND METHODS
DNA sequence analysis.
The human-specific HERV-K LTR group (HS) consensus sequence was taken from our previous work (9). LTR flanking regions were investigated with the RepeatMasker program (http://www.repeatmasker.org; A. F. A. Smit and P. Green, unpublished data). Homology searches against GenBank were done using the BLAST web server of NCBI (http://www.ncbi.nlm.nih.gov/BLAST) (1). To determine genomic locations of LTR flanking regions, the UCSC genome browser and BLAT searches (http://genome.ucsc.edu/cgi-bin/hgGateway) were used.
Oligonucleotides.
Oligonucleotides were synthesized using an ASM-102U DNA synthesizer (Biosan, Novosibirsk, Russia). Their structures can be found in Table 1.
TABLE 1.
Genomic primer sets used for PCR amplification
| Primer name and type | Sequence (5′-3′) | Accession no.a |
|---|---|---|
| Oligonucleotides used for GREM procedure | ||
| LTR-specific primers | ||
| LTRfor1 | GTCTTGTGACCCTGACACATCC | |
| LTRfor2 | CCTCCATATGCTGAACGCTG | |
| LTRfor3 | GGGGCAACCCACCCCTAC | |
| Suppression adapter oligonucleotides | ||
| A1A2 | GTAATACGACTCACTATAGGGCAGTCGACGCGTGCCCGGTCCGAC | |
| A3 | GTCGGACCGGGC | |
| A1 | GTAATACGACTCACTATAGGGC | |
| A2 | AGTCGACGCGTGCCCGGTCCGAC | |
| A4 | TCGACGCGTGCCCGGTCCGACCT | |
| Oligonucleotides used for cap switch-based cDNA amplification | ||
| CDS | AAGCAGTGGTATCAACGCAGAGTAC(T)30 | |
| riboCS | TAACAACGCAGAGTACGCRGRGRG | |
| CS | TAACAACGCAGAGTACGCGG | |
| Primers used for RT-PCR experiments | ||
| LTR-specific primers | ||
| LTRfor1 | GTCTTGTGACCCTGACACATCC | |
| LTRfor2 | CCTCCATATGCTGAACGCTG | |
| LTRfor3 | GGGGCAACCCACCCCTAC | |
| Unique genomic primers specific to LTR 3′-flanking regions | ||
| gLTR62 | TAAGTGGATATAATTACTAAGTCCAGG | AC068381 |
| gLTR80 | CCAACATCTGTCTCTTCCCTG | AP002754 |
| gLTR81 | GACCATTTGCATGGACAAATC | AL451165 |
| gLTR99 | CCATCCCTTCCATGCCTTAG | AC069420 |
| gLTR100 | AGCTTTGTGGATTGTAATTTGG | AC072054 |
| gLTR108 | CTCAGTAAAGATGAAGGTATGACAAG | AL139421 |
| gLTR109 | GAGGCAGAGGTTGCAGTGAGCC | AC002400 |
| gLTR113 | ATAAAGGAGAAATCTTCCATGAAG | AC026424 |
| gLTR115 | TGTGACGGTATAATGGCCTCT | AC008648 |
| gLTR121 | TGCAATGTTCATGTTCGCTCC | AC012175 |
| gLTR129 | GGTTATGAATAAAGTTCCCTCGG | AC027750 |
| gLTR131 | AGAATAGAGCGAACAGACACAG | AL352982 |
| gLTR138 | AGGTTATTGATACATTGCATCGAC | AC023201 |
| gLTR139 | CAATAACAGTCATTCTCACTGGAG | AC118278 |
| gLTR140 | GAGTTGGGATGTGGTCTTAGG | AL353588 |
| gLTR141 | CTCATGCTAAACTGTCTGATTATGC | AC105049 |
| gLTR145 | TTGTGCAAACTGTCTACAGCCA | BC001407 |
| gLTR149 | AACATACAGGTTGAGGCCAGG | AC016577 |
| gLTR152 | TTGTAGCTGACCAACAGCCTGC | AC068213 |
| gLTR155 | TTAGGCCAGGGTCTCACTGAG | U47924 |
| HERV-K (HML-2) proviral gag gene-specific primer | ||
| Gag rev | AATGGCCCAATCATTCCATA | |
| Unique primers specific to known human genes | ||
| DOCK2 for | CCCAGGCATGGTAGGACCTATATTAGAG | AC008648 |
| DOCK2 rev | CGGTAATCCAGCAGCTTCTCCAG | AC008648 |
| BICD1 for | GCAGTCCCGACGCAGCAAAG | AC021108 |
| BICD1 rev | CTGTAGGTAGGAAGCCCCTGAAGTC | AC021108 |
| SLC4A8 for | CTGTTCCTGTACTGTGCCTGCATGTC | AC027750 |
| SLC4A8 rev | TGTAGCAGACAAGGGAACTGGCATC | AC027750 |
| C9orf39 for | AGTCAGCAGACAGAGCTAAATCCGAG | AC015640 |
| C9orf39 rev | GATCACTTTCTTTCTGGTCTTTTCCATC | AC015640 |
| GPR74 for | GGGCTGCAGTTCCTCACACAGG | AC055844 |
| GPR74 rev | GCTGGAGCTGGAAAGCTTCTTGG | AC055844 |
| RPL8 for | CCCGTGTGAAGCTGCCCTCC | AC130332 |
| RPL8 rev | TTGTCAATTCGGCCACCTCCAG | AC130332 |
| KLRB1 for | ATTGTTCCACCAAAGAATCCAGCC | AC006432 |
| KLRB1 rev | CACAGGTGTTAGTTCTTTTTGGCAGATC | AC006432 |
| TA-PP2C for | TGGGGATGGAGGTGTTCAGTGC | AC002350 |
| TA-PP2C rev | AGACATAAAGCCACACCAATCATCATAAC | AC002350 |
| MMP24 for | ACCATCGGAGAGGAAACACGAGC | AL121753 |
| MMP24 rev | CACCGTCACCTCCTTAAACACCCA | AL121753 |
| SLC25A16 for | GCCTTATTAATGGAACAGTCCTC | AC037456 |
| SLC25A16rev | TGAGGATCAGACTGCTTCTTGC | AC037456 |
| C6orf211 for | AGGTTATTGATACATTGCATCGAC | AL590543 |
| C6orf211 rev | CACGGTGAGTAGAACCATCTTG | AL590543 |
| AND-1 for | CACCATCACTTCCACAAGTCAC | AL352982 |
| AND-1 rev | AGAATAGAGCGAACCAGACACAG | AL352982 |
| SLB for | CCAATCGAGCCAGCTCTA | AC074117 |
| SLB rev | TACCGTAAAGGCAACGCATT | AC074117 |
| ZNF432 for | CATTGATAGCAGGTTGCTGTAGA | AC011468 |
| ZNF432 rev | AGAGTATCCCAGGTGCTGGC | AC011468 |
| FLJ21816 for | AGTTCTGGCAGTTGTGTCATCC | AC002400 |
| FLJ21816 rev | GGAAGCCTGATGAACTCGTCT | AC002400 |
| CEBPZ for | CTTGCTTGGAACACGTCATCA | AC007390 |
| CEBPZ rev | GTGTCCAGGCTTTAATGTTGCT | AC007390 |
| LIPH1 for | ACCGTGGACTCAACTCTGCATTCTC | AC069420 |
| LIPH1 rev | TAGTCCCAGCAGCCGCCCTC | AC069420 |
| SENP2 for | TCTAGGGACAAACCTATCACATTTACTCG | AC069420 |
| SENP2 rev | TTGTGTGGGTTGAACAGTCCAAAGC | AC069420 |
GenBank accession numbers of corresponding LTRs from nonredundant and high-throughout genome sequence databases.
HS LTR insertion analysis.
Data on insertion polymorphisms of the HS family members in the human and chimpanzee genomes were partly previously reported by us and other authors (2, 6, 9, 11, 32, 35) and partly obtained using the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway, track “chimp”).
Tissue sampling.
A seminoma and normal testicular parenchyma were sampled from a surgical specimen containing a testicular germ cell tumor under non-neoplastic conditions. Representative samples were divided into two parts, with one being frozen immediately in liquid nitrogen and the other being formalin fixed and paraffin embedded for histological analysis.
RNA isolation and cDNA synthesis.
Total RNA was isolated from frozen tissues pulverized in liquid nitrogen using an RNeasy Mini RNA purification kit (QIAGEN). All RNA samples were further treated with DNase I to remove residual DNA. Full-length cDNA samples were obtained according to a cap switch effect-based SMART cDNA synthesis protocol (Clontech, BD Biosciences), using an oligo(dT)-containing primer (CDS), PowerScript reverse transcriptase (Clontech, BD Biosciences), and a riboCS oligonucleotide. When PowerScript reverse transcriptase reaches the 5′ end of an mRNA, the enzyme's terminal transferase activity adds a few additional deoxycytidine nucleotides to the 3′ end of the cDNA. The riboCS oligonucleotide, which contains three guanine ribonucleotide residues at its 3′ end, base pairs with the deoxycytidine stretch, creating an extended template. The reverse transcriptase then switches templates and continues replication to the end of the oligonucleotide. The resulting full-length single-stranded cDNA contains 5′-terminal sequences complementary to the riboCS oligonucleotide. An Advantage 2 polymerase mix (Clontech) and CS and CDS oligonucleotides were used to synthesize the second cDNA strands and to PCR amplify double-stranded cDNA. Prior to further hybridization in the GREM procedure, 1 μg of cDNA was digested with 10 units of AluI frequent-cutter restriction endonuclease (Fermentas) for 3 h at 37°C. This enzyme was used because the HS LTR consensus sequence lacks AluI recognition sites.
Selective amplification of genomic regions flanking HS LTRs.
Selective amplification of LTR 3′-flanking regions was based on the PCR suppression effect, described in detail elsewhere (6, 29, 41). Human genomic DNA (1 μg) was digested with 10 units of AluI (Fermentas) restriction endonuclease, ethanol precipitated, and dissolved in 20 μl of sterile water. One hundred picomoles of annealed suppression adapters (A1A2/A3) was ligated overnight to 300 ng of the digested DNA, using 3 units of T4 DNA ligase (Promega) at 16°C. The ligated DNA was purified using a Qiaquick purification column (QIAGEN) and eluted with 50 μl of water. One microliter of the eluted DNA was PCR amplified with the HS LTR-specific primer LTRfor1 and the adapter-specific primer A1, using the following cycling program: 72°C for 1 min, 95°C for 1 min, and 20 cycles of 95°C for 15 s, 65°C for 15 s, and 72°C for 1 min. PCR products were diluted 500-fold and used as templates for nested PCR with the downstream HS LTR-specific primer LTRfor2 and the adapter-specific primer A2 under the same cycling conditions, but for 22 cycles. The amplified LTR flanking sequences were treated with ExoIII exonuclease (Promega) to generate 5′-protruding termini exactly as described previously (6, 8).
GREM technique.
The GREM technique includes hybridization of PCR-amplified genomic sequences flanking repetitive elements (HS LTRs in our case) with cDNA, followed by selective amplification and cloning of hybrid DNA duplexes. For each tissue (seminoma and testicular parenchyma), 100 ng of ExoIII-treated LTR flanking sequences, obtained as described above, was mixed with 300 ng of cDNA in 4 μl of hybridization buffer (0.5 M NaCl, 50 mM HEPES, pH 8.3, 0.2 mM EDTA), overlaid with mineral oil, denatured at 95°C for 5 min, and hybridized at 68°C for 14 h. The final mixture was diluted with 36 μl of dilution buffer (50 mM NaCl, 5 mM HEPES, pH 8.3, 0.2 mM EDTA), and 1 ng of the obtained DNA equivalent was PCR-amplified with 0.2 μM adapter-specific primer A2 and 0.2 μM cDNA 5′-end-specific primer CS under the following conditions: 72°C for 5 min to fill in the ends of DNA duplexes, followed by eight cycles of 95°C for 15 s, 65°C for 15 s, and 72°C for 1 min 30 s. The PCR products were diluted 500-fold and reamplified by nested PCR for 20 cycles (95°C for 15 s, 65°C for 15 s, and 72°C for 1 min 30 s) with 0.2 μM nested adapter-specific primer A4 and 0.2 μM HS LTR 3′-end-specific primer LTRfor3. The final PCR products were cloned into Escherichia coli by using a pGEM-T vector system (Promega) and sequenced by the dye termination method using an Applied Biosystems 373 automatic DNA sequencer.
RT-PCR.
All reverse transcription-PCR (RT-PCR) experiments were reproduced at least three times, using independent cDNA preparations. For RT-PCR control of the LTR transcriptional status, we used pairs of primers, one of which was specific for a 3′-terminal part of a particular HS LTR (see Table S4 in the supplemental material for sequences) and the other of which was specific for a unique sequence within the corresponding genomic LTR 3′-flanking region. Prior to RT-PCR analysis, the priming efficiencies of the primers were examined by genomic PCRs at various temperatures, depending on the primer combination used. These PCRs were done for 19, 22, 25, and 28 cycles, with 40 ng each of the human genomic DNA templates isolated from both tissues. RT-PCR was done with cDNA samples from a mature human seminoma and healthy testicular parenchyma, with an equivalent of 20 ng total RNA being used as a template in each PCR, performed in a final volume of 40 μl. Five-microliter aliquots of the reaction mixture after 21, 24, 27, 30, 33, 36, and 39 cycles of amplification were analyzed by electrophoresis on 1.5% agarose gels. To find out the transcriptional levels of selected known genes in both tissues under study, we performed another series of RT-PCR experiments with primers designed predominantly against neighboring constitutive exons in the middle part of the corresponding cDNA molecule. The cycling conditions of these reactions also varied depending on the particular primer combination used, and the PCRs were performed in a final volume of 40 μl as described above. In all cases, the transcriptional status was determined by the number of PCR cycles needed to detect a PCR product of the expected length, and the PCR product concentration was measured using a Photomat system and Gel Pro Analyzer software.
RESULTS AND DISCUSSION
GREM library construction and initial analysis.
We used GREM to compare the HS element promoter activities in normal testicular parenchyma and in a seminoma, a testicular germ cell tumor derived from surgical material taken from the same adult patient. A complete GREM experimental protocol was published and discussed in detail previously (7). Briefly, GREM is based on hybridization of a total pool of cDNA 5′-terminal parts with specifically PCR-amplified genomic DNA fragments flanking 3′ termini of repetitive elements (HS LTRs in our case), followed by selective amplification of the genome-cDNA heteroduplexes. The GREM outcome is a set of amplified cDNA/genomic DNA heteroduplexes, referred to below as expressed LTR tags (ELTs), which are further cloned and sequenced. Every particular ELT contains a 3′ HS LTR terminal portion and a fragment of the 3′-flanking genomic DNA. Importantly, the ELT content is determined by the transcripts appearing due to promoter activities of the corresponding LTRs but not to readthrough transcription. The ELT number is proportional to the concentration of the corresponding mRNA, thus making GREM a useful tool for quantitative analysis of transcripts directed by LTR promoters. Previously, we reported the construction of a GREM library for normal testicular parenchyma (7), and in this study we applied GREM to generate an ELT library for a seminoma (both tissue specimens were obtained from the same patient). The ELT libraries for testicular parenchyma (created previously) and the seminoma (obtained in this study) were analyzed further and compared in detail.
Five hundred ELT clones were sequenced for each tissue. After the removal of rearranged plasmid and low-quality sequences, 395 and 419 ELTs, for normal testicular parenchyma and the seminoma, respectively, were taken for analysis. An ELT analysis allowed us to unambiguously map the corresponding promoter-active solitary and 3′-proviral LTRs. However, such mapping was impossible in the case of 5′-proviral LTRs because the adjoining proviral regions are repetitive and identical in sequence (Fig. 1). The detailed results of ELT mapping are shown in Table S1 in the supplemental material. Apart from the data on HS LTR promoter activities, the table contains a description of every individual HS element's genomic neighborhood and the results of previously performed functional tests, such as RT-PCR and differential methylation analyses.
To test the applicability of GREM to the task of quantitative analysis of LTR promoter activity, we addressed the issue of whether there is a correlation between the LTR-directed transcript level, as measured by RT-PCR, and the frequency of the corresponding ELT occurrence in the GREM library. RT-PCR amplification was done with pairs of primers, one of which was specific for the 3′-terminal part of a particular LTR and directed outwards from the LTR and the other of which was directed towards the LTR, designed against a unique genomic locus located at a distance of 70 to 300 bp from the LTR 3′ end. Seminoma first-strand cDNAs were used as templates. Transcript levels were measured relative to the housekeeping beta-actin gene transcript level. For a sample of 20 HS LTRs, the frequencies of ELT occurrence in the seminoma correlated linearly with RT-PCR-measured transcript levels (Table 2), with a correlation coefficient of 0.92, as shown previously for a testicular parenchyma library (7). Such a correlation suggests that in this case, GREM was adequate for quantitative characterization of LTRs displaying promoter activity.
TABLE 2.
Relative LTR transcript levels and frequencies of occurrence of the corresponding ELTs in seminoma and normal testicular parenchyma libraries
| LTR no. | Parenchyma library
|
Seminoma library
|
||
|---|---|---|---|---|
| Transcript levela (% of beta-actin gene transcript level) | ELT frequency (%) | Transcript levela (% of beta-actin gene transcript level) | ELT frequency (%) | |
| 62 | 0.004 ± 0.001 | 0 | 0.014 ± 0.003 | 0.24 |
| 80 | 0.16 ± 0.04 | 0.25 | 0.17 ± 0.03 | 0.24 |
| 81 | 1.4 ± 0.4 | 15.19 | 0.15 ± 0.03 | 1.44 |
| 99 | 2.9 ± 0.2 | 32.66 | 0.45 ± 0.10 | 5.05 |
| 100 | 0 | 0 | 0.016 ± 0.007 | 0.24 |
| 108 | 0.26 ± 0.09 | 0.50 | 0.013 ± 0.005 | 0 |
| 109 | 0 | 0 | 0 | 0 |
| 113 | 0.17 ± 0.03 | 0.25 | 0.23 ± 0.05 | 0.24 |
| 115 | 0.02 ± 0.005 | 0.25 | 0.019 ± 0.006 | 0.24 |
| 129 | 0.24 ± 0.03 | 3.61 | 0.054 ± 0.007 | 0 |
| 131 | 0.013 ± 0.004 | 0 | 0.014 ± 0.002 | 0.24 |
| 138 | 0.032 ± 0.013 | 0.25 | 0 | 0 |
| 139 | 0.16 ± 0.05 | 1.77 | 0.12 ± 0.02 | 0.72 |
| 140 | 0.13 ± 0.04 | 0.25 | 0.085 ± 0.017 | 0 |
| 141 | 0.01 ± 0.003 | 0 | 0.014 ± 0.003 | 0.24 |
| 145 | 0.35 ± 0.06 | 2.79 | 0.23 ± 0.04 | 1.68 |
| 149 | 0.24 ± 0.03 | 1.52 | 0.054 ± 0.016 | 0.48 |
| 152 | 0.12 ± 0.02 | 0.76 | 0.92 ± 0.17 | 11.30 |
| 155 | 0.059 ± 0.014 | 0.76 | 0.44 ± 0.07 | 4.57 |
| 5′-proviral LTRs | 1.08 ± 0.09 | 14.94 | 5.58 ± 0.88 | 44.47 |
Relative transcript levels were measured by RT-PCR.
A total of 78 HS family members (50%) were found to be promoter active in at least one of the two tissues. For many individual LTRs, the ELT content differed significantly between the normal and cancerous tissues. It should be noted that many LTRs were poorly expressed and therefore represented by a small number of tags, thus making analysis of the tissue specificity of their expression problematic. In contrast, other HS elements were represented by larger ELT numbers sufficient for such an analysis (>10 ELTs). For instance, for seven individual HS LTRs and for a fraction of 5′-proviral LTRs, the ELT occurrence differed fivefold and greater between the testicular parenchyma and seminoma ELT libraries (Table 3, LTRs 81, 99, 116, 129, 147, 152, and 155 and all 5′-proviral LTRs). Among them, a fraction of 5′-proviral LTRs and four individual LTRs (116, 129, 152, and 155) were upregulated in the seminoma, whereas three other elements (LTRs 81, 99, and 147) were upregulated in testicular parenchyma. For LTRs 81, 99, 129, 152, and 155 and all 5′-proviral LTRs, we performed a series of RT-PCR experiments with cDNAs from the parenchyma and seminoma, using unique primers specific to the genomic flanks of the LTRs. For all LTRs, RT-PCR revealed a positive correlation of the transcript level with the data on up- or downregulation based on the relative ELT content. Two remaining LTRs (116 and 147) were flanked by low-level divergence genomic repeats that prevented the design of efficient genomic primers. Interestingly, 5′-proviral LTRs (those driving the transcription of viral genes) were strongly (approximately sixfold, according to RT-PCR data) upregulated in the seminoma, in good agreement with previous data suggesting preferential proviral gene expression in germ cell line tumors (18, 39, 46).
TABLE 3.
Relative ELT contents for promoter-active HS LTRs transcribed in normal testicular parenchyma and seminoma
| LTR no. | ELT content (%)a
|
LTR no. | ELT content (%)a
|
|||
|---|---|---|---|---|---|---|
| Testicular parenchyma | Seminoma | Testicular parenchyma | Seminoma | |||
| 3 | 0 | 0.24 | 100 | 0 | 0.24 | |
| 9 | 0 | 0.48 | 101 | 0.50 | 0.72 | |
| 10 | 1.01 | 1.20 | 105 | 0 | 0.24 | |
| 22 | 0 | 0.24 | 106 | 1.01 | 0 | |
| 24 | 0.25 | 0 | 107 | 0.25 | 0.48 | |
| 25 | 0.25 | 0.48 | 108 | 0.50 | 0 | |
| 27 | 0 | 0.24 | 110 | 0 | 0.48 | |
| 28 | 0.25 | 0 | 112 | 0.25 | 0.48 | |
| 30 | 0.50 | 0 | 113 | 0.25 | 0.24 | |
| 32 | 0.50 | 0 | 115 | 0.25 | 0.24 | |
| 33 | 0.25 | 0 | 116 | 0.50 | 3.61 | |
| 34 | 0.50 | 0 | 117 | 0.25 | 0 | |
| 35 | 0 | 0.48 | 118 | 0.25 | 0 | |
| 44 | 0 | 0.24 | 119 | 0 | 0.24 | |
| 45 | 0.50 | 0 | 120 | 1.52 | 0 | |
| 46 | 0.76 | 0.48 | 121 | 0.25 | 0 | |
| 51 | 0.25 | 0.24 | 122 | 0 | 0.24 | |
| 52 | 0.25 | 0 | 124 | 0.25 | 0.24 | |
| 53 | 0.50 | 0 | 129 | 0 | 3.61 | |
| 58 | 0.25 | 0 | 130 | 0.50 | 0 | |
| 61 | 1.27 | 1.44 | 131 | 0 | 0.24 | |
| 62 | 0 | 0.24 | 134 | 0 | 0.72 | |
| 65 | 0 | 0.48 | 138 | 0.25 | 0 | |
| 66 | 0.50 | 0.24 | 139 | 1.77 | 0.72 | |
| 76 | 0.50 | 0 | 140 | 0.25 | 0 | |
| 79 | 2.53 | 0.96 | 141 | 0 | 0.24 | |
| 80 | 0.25 | 0.24 | 142 | 1.27 | 0.48 | |
| 81 | 15.19 | 1.44 | 143 | 0.25 | 0 | |
| 82 | 0 | 0.24 | 144 | 0 | 0.48 | |
| 83 | 0.50 | 0 | 145 | 2.79 | 1.68 | |
| 84 | 0.50 | 0.96 | 147 | 5.57 | 1.44 | |
| 86 | 0 | 0.24 | 148 | 0 | 0.48 | |
| 89 | 0.25 | 0.48 | 149 | 1.52 | 0.48 | |
| 90 | 0 | 0.48 | 150 | 0.25 | 0 | |
| 91 | 0 | 0.24 | 152 | 0.76 | 11.30 | |
| 92 | 0.25 | 0 | 153 | 0.25 | 0.48 | |
| 93 | 0.50 | 0.72 | 154 | 0.50 | 0.48 | |
| 97 | 0 | 1.20 | 155 | 0.76 | 4.57 | |
| 98 | 1.01 | 1.20 | 5′-proviral LTRs | 14.94 | 44.47 | |
| 99 | 32.66 | 5.05 | ||||
The relative ELT content was calculated as the ratio of the number of tags for each individual HS LTR to the total number (395 and 419 for the testicular parenchyma and seminoma, respectively) of all ELTs in the sequenced library. For 5′ proviral LTRs, summarized values are given.
Qualitative analysis shows that the proportion of promoter-active HS LTRs is higher in gene-rich regions.
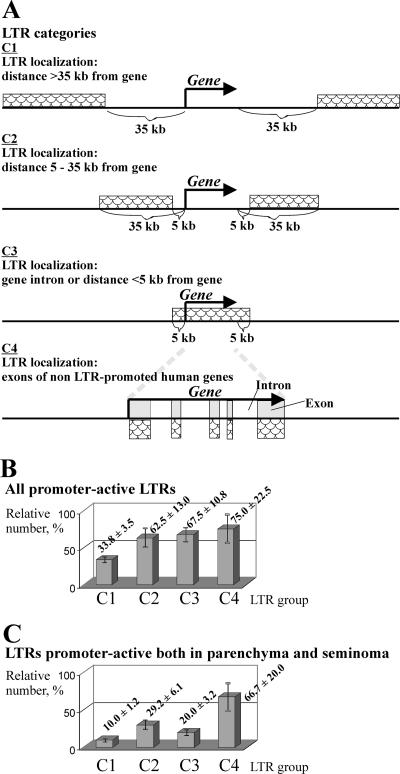
Fifty percent of all HS family members were found to be functional promoters in vivo in at least one tissue under study. Precise genomic positions of HS family members, found using the UCSC human genome web browser, allowed us to group them according to their distances (D) from known human genes or mapped cDNAs (shown schematically in Fig. 2A), as follows: group C1, D > 35 kb (80 HS elements); C2, 5 ≤ D ≤ 35 kb (24 elements); C3, HS elements located within gene introns or with D values of <5 kb (40 representatives); and C4, HS elements within exons of known non-LTR-promoted human cDNAs, and thus partly or wholly readthrough transcribed (12 representatives). For the last group, GREM makes it possible to detect promoter-active LTRs. Detailed information about LTR localization, neighboring genes, and mapped cDNAs is given in Table S1 in the supplemental material.
FIG. 2.
Proportions of promoter-active LTRs in four groups differing by the distance of the LTRs from known human genes or mapped cDNAs. (A) LTRs were grouped according to their distances from known human genes into four categories (C1 to C4) (see the text for details). The relative content of promoter-active LTRs in a group was calculated as the ratio of the number of LTRs in the group, for which the corresponding ELTs were obtained, to the total number of all LTRs in the group. (B) LTRs which were promoter active in at least one of the two tissues studied. (C) LTRs which were promoter active both in testicular parenchyma and in the seminoma. Averages and standard errors of the means (error bars) are presented.
Since the numbers of representatives in these four groups varied substantially, we used relative rather than absolute values to characterize the promoter-active LTR distribution among them. Figure 2B shows the numerical ratios of the LTRs of a given category which were promoter active in at least one tissue under study to all LTRs in the category. It can be seen that the ratio for LTRs of group C1 was relatively low (34%), whereas that for LTRs located closer to known genes (group C2) was ∼1.8-fold greater (63%). For LTRs mapped within gene introns or in close proximity to genes (group C3), the ratio was slightly higher (68%). Finally, the largest proportion (75%) of promoter-active elements was observed for LTRs within exons (group C4).
Figure 2C shows the group distribution for promoter-active LTRs functioning both in the seminoma and in normal parenchyma. The proportions of such “ubiquitously” transcribed LTRs in groups C1, C2, C3, and C4 were different, i.e., 10, 29, 20, and 67%, respectively.
It can therefore be concluded that (i) the relative content of promoter-active LTRs in gene-rich regions is significantly higher than that in gene-poor genomic loci, (ii) this content is maximal for the HS elements from those regions where promoter-active LTRs “overlap” readthrough transcripts (group C4), and (iii) LTRs of group C4 most frequently serve as promoters in both tissues. At present, we cannot explain the clearly enhanced promoter activity of the group C4 representatives. This effect might suggest better accessibility of exon regions to transcription factors than to other genomic DNA.
Quantitative analysis shows that LTR promoter activities differ considerably, depending on the genomic neighborhood and the LTR status (solitary or proviral).
Counting of ELTs can be used to estimate the promoter activities of individual HS family members. By definition, promoter strength is the number of transcription initiation events per given time period. The GREM approach was used here to quantify the polyadenylated RNAs produced due to LTR promoter activity. Apart from promoter strength, the content of this RNA may also depend on other factors, such as RNA transfer from the nucleus, RNA stability, or polyadenylation. Since we are unable to estimate the contributions of these factors, the terms “promoter strength” and “promoter activity” should be understood as operational definitions throughout this report.
A counting of ELTs revealed quite different promoter activities (see below) for solitary and proviral LTRs, with the difference being dependent on the genomic neighborhood. The level of 5′-proviral LTR expression could not be measured properly because of the reasons mentioned above, so we focused on quantitative analysis of 3′-proviral and solitary LTR promoter activities in the four groups of HS elements (C1to C4). The relative promoter strength of a group of HS elements was calculated as the ratio of the relative content of the corresponding ELTs in the pool of all ELTs (except for those corresponding to 5′-proviral LTRs) to the relative content of the HS elements of this group (except for 5′-proviral LTRs) among HS elements of all groups (except for 5′-proviral LTRs).
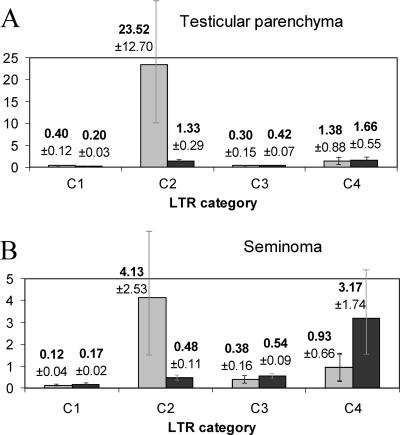
The diagrams in Fig. 3 show that 3′-proviral LTRs displayed similar transcriptional patterns in both tissues, with a low transcript level for group C1 members, a sharp ∼30- to 60-fold increase of this level for group C2, a relatively low level for group C3, and finally, an ∼2.5- to 5-fold increase for group C4 promoter-active HS elements located within exons. Solitary LTRs displayed different profiles: their average promoter activity was low for group C1 (LTRs located far from genes), moderate for groups C2 and C3 (closer to genes, or intronic locations), and finally, increased four- to sixfold for group C4. The maximal promoter activity of solitary LTRs is characteristic of the group C4 elements located within exons.
FIG. 3.
Relative promoter strengths of 3′-proviral (gray) and solitary (black) LTRs grouped according to their distances from genes (groups C1 to C4) (see the text for details). (A) Testicular parenchyma; (B) seminoma. Averages and standard errors of the means (error bars) are presented.
Also, these data suggest a selective suppression of proviral 3′-LTR transcription when the LTRs are in close proximity to genes. Such transcriptional suppression might be aimed at proviral gene silencing in gene-rich regions. Importantly, the relative promoter strengths of both solitary and 3′-proviral group C3 LTRs were significantly decreased in the normal tissue (testicular parenchyma). This may suggest a special cellular mechanism for selective suppression of “extra” (unwanted) promoters located within gene introns or very closely to genes. Such a mechanism might minimize possible destructive effects of background transcription, including the expression of antisense RNAs that could affect normal gene regulation mechanisms. It should be added that ∼90% of intronically located HS LTRs are inserted in the reverse orientation relative to the gene transcription direction and that their transcription could therefore create a pool of regulatory interfering RNAs (11). One more conclusion is that group C4 HS elements, whose transcripts “overlap” human readthrough RNAs, are enriched in promoter-active elements, thus again suggesting an interplay of readthrough and LTR-directed transcription.
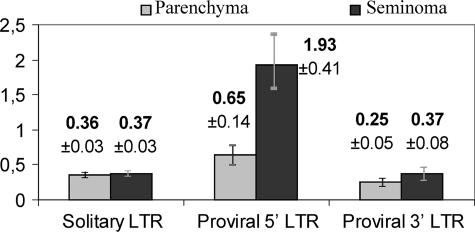
We further tried to compare average promoter activities for the 5′-proviral, 3′-proviral, and solitary LTR types (Fig. 4). The relative average promoter strength of a group of HS elements was calculated as the ratio of the relative content of the corresponding ELTs in the pool of all ELTs to the relative content of the HS elements of this group among HS elements of all groups. The results (Fig. 4) demonstrated that average promoter strengths of solitary and 3′-proviral LTRs were almost equal in both tissues under study. The promoter strength of 5′-proviral LTRs was approximately twofold higher in testicular parenchyma and approximately fivefold higher in the seminoma, in accord with extensive previous data in favor of an upregulation of HERV-K (HML-2) proviral gene expression in germ cell line tumors. It can be assumed that the proviral sequences contain some so far uncharacterized downstream regulatory elements that provide significantly more 5′-LTR expression, especially in the seminoma.
FIG. 4.
Comparison of relative promoter strengths of solitary, 5′-proviral, and 3′-proviral LTRs (per LTR). Gray and black bars represent the relative LTR promoter strengths in the testicular parenchyma and seminoma, respectively. For details of relative promoter strength calculation, see the text. Averages and standard errors of the means (error bars) are presented.
Regulation of HS element promoter activity.
According to microarray data obtained from the UCSC genome web browser, as many as 86 to 90% of genes located in close proximity to promoter-active LTRs in normal testicular parenchyma and seminomas are known to be transcribed in the testis. To investigate whether the promoter activities of the LTRs that mapped closely to genes correlated with the transcription of these genes, we used RT-PCR to check the transcriptional status of 16 genes located near randomly chosen HS family members. In both the parenchyma and seminoma, no clear-cut correlation was observed between transcriptional activities of genes and closely located LTRs (Table 4 [LTR locations relative to genes are given in Fig. S1 in the supplemental material]).
TABLE 4.
Relative transcript levels of HS LTRs and closely located human genes in testicular parenchyma and seminoma
| Gene no. | Gene name | Transcript level (% of beta-actin gene transcript level)a
|
LTR no. | Relative ELT content (%)
|
||
|---|---|---|---|---|---|---|
| Parenchyma | Seminoma | Parenchyma | Seminoma | |||
| 1 | DOCK2 | 0.06 ± 0.013 | 0.04 ± 0.013 | LTR 115 | 0.25 | 0.24 |
| 2 | BICD 1 | 0.04 ± 0.013 | 0.063 ± 0.017 | LTR 94 | 0 | 0 |
| 3 | SLC4A8 | 1.5 ± 0.2 | 1.37 ± 0.25 | LTR 129 | 0 | 3.61 |
| 4 | C9orf39 | 0.017 ± 0.004 | 0 | LTR 130 | 0.50 | 0 |
| 5 | GPR 74 | 0 | 0.013 ± 0.004 | LTR 136 | 0 | 0 |
| 6 | RPL 8 | 14.8 ± 3.1 | 11.2 ± 1.8 | LTR 154 | 0.50 | 0.48 |
| 7 | KLRB 1 | 0.013 ± 0.004 | 0.03 ± 0.01 | LTR 110 | 0 | 0.48 |
| 8 | TA-PP2C | 0.34 ± 0.06 | 0.34 ± 0.06 | LTR 112 | 0.25 | 0.48 |
| 9 | MMP 24 | 0.023 ± 0.004 | 0.025 ± 0.006 | LTR 146 | 0 | 0 |
| 10 | SLC25A16 | 0.025 ± 0.007 | 0.017 ± 0.004 | LTR 145 | 2.79 | 1.68 |
| 11 | C6orf211 | 0.093 ± 0.018 | 0.15 ± 0.04 | LTR 117 | 0.25 | 0 |
| 12 | AND-1 | 0.083 ± 0.020 | 0.64 ± 0.19 | LTR 131 | 0 | 0.24 |
| 13 | SLB | 0.027 ± 0.009 | 0.24 ± 0.06 | LTR 122 | 0 | 0.24 |
| 14 | ZNF 432 | 0.06 ± 0.02 | 0.016 ± 0.005 | LTR 119 | 0 | 0.24 |
| 15 | FLJ21816 | 0.12 ± 0.03 | 0.23 ± 0.04 | LTR 109 | 0 | 0 |
| 16 | CEBPZ | 0.008 ± 0.003 | 0.16 ± 0.05 | LTR 105 | 0 | 0.24 |
Relative transcript levels were measured by RT-PCR.
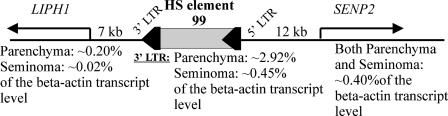
However, the transcript levels of eight tested genes did correlate with the promoter activities of the closely located LTRs in the seminoma and testicular parenchyma, including the following (LTR number/gene name): 115/DOCK2, involved in cytoskeletal rearrangements required for lymphocyte migration in the response to chemokines; 130/C9orf39, of unknown function; 154/RPL8, encoding the 60S ribosomal protein L8; 112/TA-PP2C, encoding a T-cell activation protein phosphatase; 145/SLC25A16, encoding a mitochondrial transfer protein; 131/AND-1, encoding an acidic nucleoplasmic DNA-binding protein of unknown function; 122/SLB, encoding a selective LIM binding factor homolog; and 105/CEBPZ, encoding CCAAT-enhancer binding protein zeta. On the other hand, LTR 129, which was strongly upregulated in the seminoma, is located within the fifth intron of the SLC4A8 gene, encoding a sodium bicarbonate solute carrier protein, which is expressed at essentially the same level in both tissues. An analysis of the genomic neighborhood of 3′-proviral LTR 99 (∼33% of all ELTs), which was greatly overexpressed in parenchyma and was transcribed at a sixfold lower (yet still rather high) level in the seminoma, revealed that provirus 99 was situated between two known human genes, i.e., 7 kb upstream of the LIPH1 gene, encoding a membrane-bound lipase precursor, and 12 kb upstream of the SENP2 gene, encoding a SUMO1-specific protease (Fig. 5). RT-PCR experiments demonstrated that SENP2 was transcribed in both tissues at a relatively high level of ∼0.4% of the beta-actin transcript level, whereas LIPH1 was upregulated in testicular parenchyma and significantly downregulated in the seminoma (0.2 and 0.02% of the beta-actin transcript level, respectively). Such a strong proviral 3′-LTR promoter activity might be due to the regulatory elements of both genes. SENP2 could provide a strong basal expression level, whereas LIPH1 could be responsible for the tissue specificity of the expression. Alternatively, HS element 99 and the LIPH1 gene could be colocalized within the same chromatin domain distinct from that containing SENP2. On the other hand, the observed SENP2 and LIPH1 transcription profiles could be significantly affected by numerous regulatory sequences of provirus 99 itself. We therefore concluded that multiple, sometimes contradictory, scenarios may take place in the transcriptional regulation of HS elements.
FIG. 5.
Schematic representation of HS element 99 localization relative to its LIPH1 and SENP2 gene neighbors and their transcript levels in testicular parenchyma and seminoma.
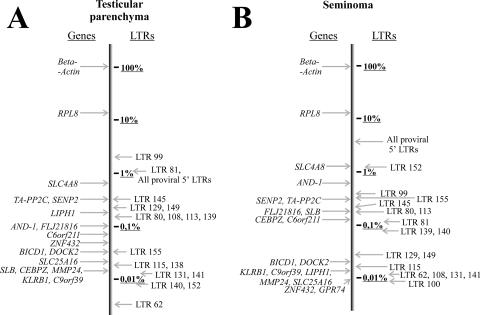
Finally, it should be mentioned that the abundances of transcripts varied ∼1,000-fold (Fig. 6) among expressed individual HS family members, from hardly detectable to levels comparable to those of housekeeping gene transcription. The high expression levels of certain LTRs capable of driving the transcription of host nonrepetitive genomic sequences in human tissues clearly suggest the possibility of their involvement in the formation of new functional genes and/or antisense regulation of preexisting genes.
FIG. 6.
Relative transcript levels of some human genes and LTRs. Relative transcript levels of randomly chosen HS LTRs and those of known human genes were measured using RT-PCR. (A) Testicular parenchyma; (B) seminoma.
Concluding remarks.
We report here the first genome-wide comparison of in vivo promoter activities of a group of human-specific endogenous retroviruses in normal and cancerous germ line tissues. These were chosen because of the markedly high endogenous retroviral transcriptional activities in germ line cells, which are most probably needed to make de novo retroviral integrations inheritable (38). LTR promoter activity patterns in normal testicular parenchyma were compared with those in a seminoma (the corresponding tumor) sampled from the same patient. We found that at least 50% of HS elements possessed promoter activity. Individual LTRs were expressed at markedly different levels, ranging from ∼0.001 to ∼3% of the housekeeping beta-actin gene transcript level. Although HS elements formed several subclusters on a phylogenetic tree (5), no clear correlation between LTR primary structure and transcriptional activity was found in this study. In contrast, the LTR status (solitary or 5′ or 3′ proviral) was an important factor affecting LTR activity, as the promoter strengths of solitary and 3′-proviral LTRs were almost identical in both tissues, whereas 5′-proviral LTRs displayed higher promoter activities (approximately twofold and fivefold greater in the testicular parenchyma and seminoma, respectively). These data suggest that a proviral sequence harbors some as yet unknown downstream regulatory elements that provide significantly more 5′-LTR expression, especially in seminomas. Another important factor affecting promoter activity was the LTR distance from genes: the relative content of promoter-active LTRs in gene-rich regions was significantly higher than that in gene-poor genomic loci. Interestingly, in both tissues, this content was maximal for HS elements from those regions where promoter-active LTRs “overlapped” with readthrough transcripts; this effect might suggest better accessibility of exon regions to transcription factors than to other genomic loci. It should be mentioned that all HS elements overlapped with non-protein-coding regions of the corresponding transcripts. The observed preferable expression of “exonic” LTRs might be due to neighboring regulatory sequences, which are frequently present in untranslated exons. The detailed explanation of such a phenomenon is a matter of further studies.
Our data also suggest a selective suppression of transcription in both tissues for proviral 3′-LTRs located in gene introns. Such a transcriptional suppression might be aimed at silencing the proviral gene expression in gene-rich regions. In testicular parenchyma, the promoter strengths of intronically located solitary LTRs were also significantly decreased. This may suggest an as yet unknown mechanism(s) for selective suppression of “extra” promoters generated due to mutations or viral integrations and located within gene introns or very close to genes. Such a mechanism might minimize possible destructive effects of undesirable transcription. Many transcriptionally competent LTRs were mapped near known human genes, and as many as 86 to 90% of all genes located in close proximity to promoter-active LTRs are known to be transcribed in the testis. However, in general, no clear-cut correlation was observed between transcriptional activities of genes and closely located LTRs. The high expression levels of certain LTRs located in human gene introns might suggest the possibility of their involvement in antisense regulation of preexisting genes.
Finally, this is the first quantitative and qualitative comprehensive characterization of human promoters provided by a small particular group of endogenous retroviruses. An overwhelming majority of retroviral sequences, which occupy up to 8% of the human genome, still remain a subject of further investigations.
Supplementary Material
Acknowledgments
We thank Yuri Lebedev, Tatyana Vinogradova, and Lev Nikolayev (Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia) for fruitful discussions, Boris Glotov (Institute of Molecular Genetics, Moscow, Russia) for valuable comments on the manuscript, and Nadezhda Skaptsova (Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia) for the synthesis of oligonucleotides.
This work was supported by Russian Foundation for Basic Research grants 05-04-48682-a and 2006.20034, by grant MK-2833.2004.4 from the President of the Russian Federation, and by the Molecular and Cellular Biology Program of the Presidium of the Russian Academy of Sciences.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 3.Belshaw, R., A. L. Dawson, J. Woolven-Allen, J. Redding, A. Burt, and M. Tristem. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K (HML2): implications for present-day activity. J. Virol. 79:12507-12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buscher, K., U. Trefzer, M. Hofmann, W. Sterry, R. Kurth, and J. Denner. 2005. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 65:4172-4180. [DOI] [PubMed] [Google Scholar]
- 5.Buzdin, A., E. Gogvadze, E. Kovalskaya, P. Volchkov, S. Ustyugova, A. Illarionova, A. Fushan, T. Vinogradova, and E. Sverdlov. 2003. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 31:4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buzdin, A., K. Khodosevich, I. Mamedov, T. Vinogradova, Y. Lebedev, G. Hunsmann, and E. Sverdlov. 2002. A technique for genome-wide identification of differences in the interspersed repeats integrations between closely related genomes and its application to detection of human-specific integrations of HERV-K LTRs. Genomics 79:413-422. [DOI] [PubMed] [Google Scholar]
- 7.Buzdin, A., E. Kovalskaya-Alexandrova, E. Gogvadze, and E. Sverdlov. 2006. GREM, a technique for genome-wide isolation and quantitative analysis of promoter active repeats. Nucleic Acids Res. 34:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buzdin, A., S. Ustyugova, E. Gogvadze, Y. Lebedev, G. Hunsmann, and E. Sverdlov. 2003. Genome-wide targeted search for human specific and polymorphic L1 integrations. Hum. Genet. 112:527-533. [DOI] [PubMed] [Google Scholar]
- 9.Buzdin, A., S. Ustyugova, K. Khodosevich, I. Mamedov, Y. Lebedev, G. Hunsmann, and E. Sverdlov. 2003. Human-specific subfamilies of HERV-K (HML-2) long terminal repeats: three master genes were active simultaneously during branching of hominoid lineages. Genomics 81:149-156. [DOI] [PubMed] [Google Scholar]
- 10.Buzdin, A. A. 2004. Retroelements and formation of chimeric retrogenes. Cell. Mol. Life Sci. 61:2046-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buzdin, A. A., B. Lebedev Iu, and E. D. Sverdlov. 2003. Human genome-specific HERV-K intron LTR genes have a non-random orientation relative to the direction of transcription, and, possibly, participated in antisense gene expression regulation. Bioorg. Khim. 29:103-106. [DOI] [PubMed] [Google Scholar]
- 12.Coffin, J. M. 1996. Retroviridae: the viruses and their replication, p. 1767-1847. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 13.Dewannieux, M., S. Blaise, and T. Heidmann. 2005. Identification of a functional envelope protein from the HERV-K family of human endogenous retroviruses. J. Virol. 79:15573-15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewannieux, M., C. Esnault, and T. Heidmann. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35:41-48. [DOI] [PubMed] [Google Scholar]
- 15.Ehlhardt, S., M. Seifert, J. Schneider, A. Ojak, K. D. Zang, and Y. Mehraein. 2006. Human endogenous retrovirus HERV-K (HML-2) Rec expression and transcriptional activities in normal and rheumatoid arthritis synovia. J. Rheumatol. 33:16-23. [PubMed] [Google Scholar]
- 16.Frank, O., M. Giehl, C. Zheng, R. Hehlmann, C. Leib-Mosch, and W. Seifarth. 2005. Human endogenous retrovirus expression profiles in samples from brains of patients with schizophrenia and bipolar disorders. J. Virol. 79:10890-10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galli, U. M., M. Sauter, B. Lecher, S. Maurer, H. Herbst, K. Roemer, and N. Mueller-Lantzsch. 2005. Human endogenous retrovirus rec interferes with germ cell development in mice and may cause carcinoma in situ, the predecessor lesion of germ cell tumors. Oncogene 24:3223-3228. [DOI] [PubMed] [Google Scholar]
- 18.Herbst, H., M. Sauter, C. Kuhler-Obbarius, T. Loning, and N. Mueller-Lantzsch. 1998. Human endogenous retrovirus (HERV)-K transcripts in germ cell and trophoblastic tumours. APMIS 106:216-220. [DOI] [PubMed] [Google Scholar]
- 19.Hughes, J. F., and J. M. Coffin. 2001. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat. Genet. 29:487-489. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, J. F., and J. M. Coffin. 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl. Acad. Sci. USA 101:1668-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 22.Kazazian, H. H., Jr., and J. L. Goodier. 2002. LINE drive. Retrotransposition and genome instability. Cell 110:277-280. [DOI] [PubMed] [Google Scholar]
- 23.Khodosevich, K., Y. Lebedev, and E. Sverdlov. 2004. The tissue-specific methylation of human-specific endogenous retroviral LTRs. Bioorg. Khim. 30:493-498. [DOI] [PubMed] [Google Scholar]
- 24.Khodosevich, K., Y. Lebedev, and E. D. Sverdlov. 2004. Large-scale determination of the methylation status of retrotransposons in different tissues using a methylation tags approach. Nucleic Acids Res. 32:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kidwell, M. G., and D. Lisch. 1997. Transposable elements as sources of variation in animals and plants. Proc. Natl. Acad. Sci. USA 94:7704-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovalskaya, E., A. Buzdin, E. Gogvadze, T. Vinogradova, and E. Sverdlov. 2006. Functional human endogenous retroviral LTR transcription start sites are located between the R and U5 regions. Virology 346:373-378. [DOI] [PubMed] [Google Scholar]
- 27.Landry, J. R., and D. L. Mager. 2003. Functional analysis of the endogenous retroviral promoter of the human endothelin B receptor gene. J. Virol. 77:7459-7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavie, L., M. Kitova, E. Maldener, E. Meese, and J. Mayer. 2005. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K (HML-2). J. Virol. 79:876-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavrentieva, I., N. E. Broude, Y. Lebedev, I. I. Gottesman, S. A. Lukyanov, C. L. Smith, and E. D. Sverdlov. 1999. High polymorphism level of genomic sequences flanking insertion sites of human endogenous retroviral long terminal repeats. FEBS Lett. 443:341-347. [DOI] [PubMed] [Google Scholar]
- 30.Macfarlane, C., and P. Simmonds. 2004. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J. Mol. Evol. 59:642-656. [DOI] [PubMed] [Google Scholar]
- 31.Mager, D. L., D. G. Hunter, M. Schertzer, and J. D. Freeman. 1999. Endogenous retroviruses provide the primary polyadenylation signal for two new human genes. Genomics 59:255-263. [DOI] [PubMed] [Google Scholar]
- 32.Mamedov, I., A. Batrak, A. Buzdin, E. Arzumanyan, Y. Lebedev, and E. D. Sverdlov. 2002. Genome-wide comparison of differences in the integration sites of interspersed repeats between closely related genomes. Nucleic Acids Res. 30:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mamedov, I., Y. Lebedev, G. Hunsmann, E. Khusnutdinova, and E. Sverdlov. 2004. A rare event of insertion polymorphism of a HERV-K LTR in the human genome. Genomics 84:596-599. [DOI] [PubMed] [Google Scholar]
- 34.Mayer, J., T. Stuhr, K. Reus, E. Maldener, M. Kitova, F. Asmus, and E. Meese. 2005. Haplotype analysis of the human endogenous retrovirus locus HERV-K(HML-2.HOM) and its evolutionary implications. J. Mol. Evol. 61:706-715. [DOI] [PubMed] [Google Scholar]
- 35.Medstrand, P., and D. L. Mager. 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 72:9782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 37.Pickeral, O. K., W. Makalowski, M. S. Boguski, and J. D. Boeke. 2000. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prudhomme, S., B. Bonnaud, and F. Mallet. 2005. Endogenous retroviruses and animal reproduction. Cytogenet. Genome Res. 110:353-364. [DOI] [PubMed] [Google Scholar]
- 39.Rakoff-Nahoum, S., J. P. Kuebler, J. J. Heymann, E. M. Sheehy, M. G. Ortiz, S. G. Ogg, J. D. Barbour, J. Lenz, A. D. Steinfeld, and D. F. Nixon. 2006. Detection of T lymphocytes specific for human endogenous retrovirus K (HERV-K) in patients with seminoma. AIDS Res. Hum. Retrovir. 22:52-56. [DOI] [PubMed] [Google Scholar]
- 40.Ruda, V. M., S. B. Akopov, D. O. Trubetskoy, N. L. Manuylov, A. S. Vetchinova, L. L. Zavalova, L. G. Nikolaev, and E. D. Sverdlov. 2004. Tissue specificity of enhancer and promoter activities of a HERV-K(HML-2) LTR. Virus Res. 104:11-16. [DOI] [PubMed] [Google Scholar]
- 41.Siebert, P. D., A. Chenchik, D. E. Kellogg, K. A. Lukyanov, and S. A. Lukyanov. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23:1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sverdlov, E. D. 1999. Retroviral regulators of gene expression in the human genome as possible factors for its evolution. Bioorg. Khim. 25:821-827. [PubMed] [Google Scholar]
- 43.Sverdlov, E. D. 2000. Retroviruses and primate evolution. Bioessays 22:161-171. [DOI] [PubMed] [Google Scholar]
- 44.Turner, G., M. Barbulescu, M. Su, M. I. Jensen-Seaman, K. K. Kidd, and J. Lenz. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531-1535. [DOI] [PubMed] [Google Scholar]
- 45.van de Lagemaat, L. N., J. R. Landry, D. L. Mager, and P. Medstrand. 2003. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 19:530-536. [DOI] [PubMed] [Google Scholar]
- 46.Vinogradova, T., L. Leppik, E. Kalinina, P. Zhulidov, K. H. Grzeschik, and E. Sverdlov. 2002. Selective differential display of RNAs containing interspersed repeats: analysis of changes in the transcription of HERV-K LTRs in germ cell tumors. Mol. Genet. Genomics 266:796-805. [DOI] [PubMed] [Google Scholar]
- 47.Vinogradova, T. V., L. P. Leppik, L. G. Nikolaev, S. B. Akopov, A. M. Kleiman, N. B. Senyuta, and E. D. Sverdlov. 2001. Solitary human endogenous retroviruses-K LTRs retain transcriptional activity in vivo, the mode of which is different in different cell types. Virology 290:83-90. [DOI] [PubMed] [Google Scholar]
- 48.Weiner, A. M., P. L. Deininger, and A. Efstratiadis. 1986. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu. Rev. Biochem. 55:631-661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.