Abstract
It has recently become clear that several pathogenic DNA viruses express virally encoded microRNAs in infected cells. In particular, numerous microRNAs have been identified in a range of human and animal herpesviruses, and individual microRNAs have also been identified in members of the polyoma- and adenovirus families. Although their functions remain largely unknown, it seems likely that these viral microRNAs play an important role in viral replication in vivo. Here we present an analysis of the microRNAs expressed in human cells during the latent and productive phases of the human papillomavirus genotype 31 (HPV31) replication cycle. Although over 500 cellular microRNAs were cloned and identified, not a single HPV31-specific microRNA was obtained. We therefore concluded that HPV31, and possibly human papillomaviruses in general, does not express viral microRNAs.
microRNAs (miRNAs) are a family of ∼22-nucleotide (nt) noncoding RNAs expressed by all animals and plants. The human genome encodes over 400 different miRNAs, and these may be expressed at several thousand copies per cell. miRNAs are thought to play key roles in differentiation and development by posttranscriptionally downregulating the function of cellular mRNA transcripts (1).
Cellular miRNA is transcribed by RNA polymerase II in the form of a long, capped, polyadenylated precursor RNA called a primary miRNA (pri-miRNA) (2, 16). The mature miRNA forms part of one arm of an ∼80-nt RNA hairpin within the pri-miRNA. The first step in miRNA processing is the nuclear cleavage of the pri-miRNA by the RNase III enzyme Drosha to give an ∼60-nt RNA hairpin bearing a 2-nt 3′ overhang, called a pre-miRNA (15). The pre-miRNA is transported to the cytoplasm by the karyopherin exportin 5, while the flanking RNA sequences are retained in the nucleus and eventually degraded (2, 22). In the cytoplasm, the pre-miRNA is recognized by a second RNase III enzyme, called Dicer, which removes the terminal loop, leaving a second 2-nt 3′ overhang (9, 12). One strand of the resultant miRNA duplex intermediate is then taken up by the RNA-induced silencing complex (11). The mature single-stranded miRNA functions as a guide RNA to target the RNA-induced silencing complex to complementary mRNAs, which are then degraded and/or translationally inhibited (6, 17, 21, 23).
Because pri-miRNAs can be quite short (≤200 nt) and because miRNAs are not antigenic, the ubiquitous miRNA regulatory pathway would seem to offer a perfect mechanism for viruses to downregulate the expression of cellular mRNAs encoding factors that could inhibit virus replication in vivo. However, the first step in cellular miRNA biogenesis occurs in the nucleus, and miRNA processing leads to the eventual degradation of the entire pri-miRNA, with the exception of the mature miRNA itself. As discussed in detail elsewhere (5), this suggests that viruses that replicate exclusively in the cytoplasm or that contain an RNA genome would be unlikely to express miRNAs. Moreover, the fact that miRNAs act at the mRNA rather than the protein level means that the phenotypic effect of miRNAs can be slow to reveal itself, especially if the relevant protein is quite stable. Therefore, miRNAs may not present an advantageous approach to the regulation of host cell gene expression in the case of viruses that undergo rapid, lytic replication cycles. On the other hand, the posttranscriptional downregulation of host cell mRNAs by virally encoded miRNAs would seem to be potentially very advantageous to nuclear DNA viruses that establish long-term latent or persistent infections. In fact, analyses of several members of the herpesvirus family, which invariably establish long-term latent infections in infected animals, have revealed virally encoded miRNAs in every herpesvirus species examined thus far (3, 4, 10, 18, 19). The largest number of miRNAs, a total so far of 23, has been reported for Epstein-Barr virus, but several other herpesviruses have also been shown to express multiple miRNAs in latently infected cells. Unfortunately, it has not yet proven possible to identify the mRNAs targeted by these miRNAs or to determine how they enhance the level of herpesvirus replication.
A second family of pathogenic DNA viruses that establish long-term latent or persistent infections in vivo is the human papillomaviruses (HPVs). HPVs infect cells in the basal epithelia and translocate to the nucleus, where the viral DNA genomes become established as episomes (7, 13). In infected basal epithelial cells, the viral genomes replicate coordinately with the cellular genome, early viral proteins are expressed at low levels, and the late, structural genes remain silent. After each cell division, one daughter cell moves towards the surface of the epithelial layer and begins to differentiate. Upon differentiation of the cells into suprabasal cells, viral DNA amplification occurs and late gene expression is induced, leading to virion production. At this time, the viral late promoter (P742 in HPV genotype 31 [HPV31]) is activated, leading to high-level expression of the E1^E4 and E5 open reading frames as well as L1 and L2, which encode the viral capsid proteins.
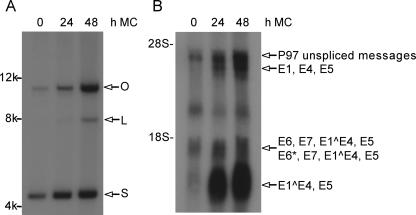
HPV31 is an oncogenic HPV that provides a well-established model for the analysis of HPV replication and gene expression. LKP1 cells stably maintain HPV31 episomes at about 50 copies per cell and mimic latently infected basal cells (8). These cells express viral early mRNAs, which primarily consist of transcripts encoding E6-E7-E1^E4-E5 and E6*-E7-E1^E4-E5, expressed from the early P97 promoter. HPV31-infected LKP1 cells can be induced, however, to undergo differentiation by suspension culture in 1.5% methylcellulose, as previously described (20). This induces amplification of the HPV31 viral genome (Fig. 1A) and concomitant expression of the viral late mRNAs (Fig. 1B).
FIG. 1.
Northern and Southern analyses of undifferentiated and methylcellulose-differentiated HPV31-positive LKP1 cells. LKP1 cells were harvested for analysis following growth in undifferentiated monolayer cultures (0 h) or following differentiation in 1.5% methylcellulose for 24 and 48 h. (A) Southern analysis of total LKP1 DNA showing HPV31 amplification following suspension in methylcellulose. Total DNA was sheared prior to being loaded into the gel and analyzed as previously described (8). Supercoiled (S), open circular DNA (O), and linear (L) forms of the HPV31 genome are indicated. (B) Northern analysis of total RNAs from undifferentiated and differentiated LKP1 cells showing induction of late gene expression following suspension in methylcellulose for 24 and 48 h. Early transcripts include E6-E7-E1^E4-E5 and E6*-E7-E1^E4-E5, and late transcripts include E1^E4-E5 and E1-E4-E5. Total RNA was isolated using a mirVANA isolation kit from Ambion Inc. (Austin, TX). The RNAs from the 24- and 48-h methylcellulose time points were combined for the miRNA cloning analysis.
Total RNA samples were obtained from undifferentiated and differentiated HPV31-infected LKP1 cells (Fig. 1B), using a mirVANA isolation kit from Ambion Inc. (Austin, TX). These samples were used to cDNA clone RNAs of ≥18 nt but ≤25 nt in length, as previously described (3, 4, 14). This approach is expected to clone all cellular miRNAs, any viral miRNAs that might be expressed during latent or productive HPV31 replication, and also any fragments of cellular mRNAs or noncoding RNAs that fall into this size range. RNAs isolated from LKP1 cells induced to differentiate for 24 and 48 h were combined for the miRNA analysis to increase the likelihood of detecting differentiation-induced virally encoded miRNAs. The cDNAs were concatenated, cloned, and subjected to DNA sequence analysis (3, 4, 14). The identities of the sequences obtained were then analyzed by comparison to the HPV31 genome and to databases covering cellular miRNAs, noncoding RNAs, and mRNAs.
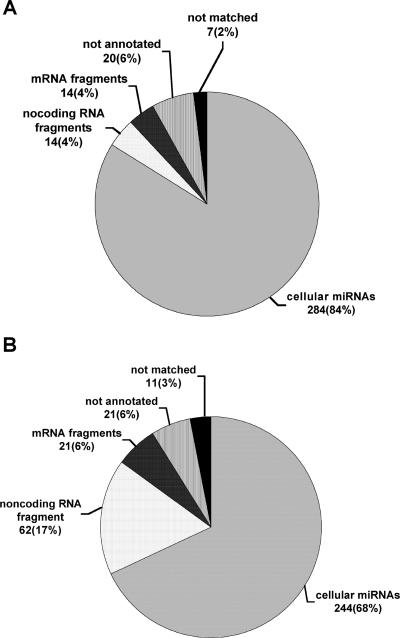
In Fig. 2, we provide a breakdown of the origins of the 339 18- to 25-nt RNAs that were cDNA cloned from undifferentiated LKP1 cells (panel A) and the 359 18- to 25-nt RNAs cloned from differentiated LKP1 cells (panel B). Of the 339 sequences obtained from undifferentiated cells, 284 (84%) represented miRNAs, while of the 359 sequences obtained from differentiated LKP1 cells, 244 (68%) represented miRNAs. Remarkably, however, all 528 miRNAs cloned in this analysis were of cellular origin. No cDNA sequences of HPV31 origin, either miRNA or otherwise, were identified, even though viral mRNAs were readily detected in the source RNA preparations derived from undifferentiated and differentiated LKP1 cells (Fig. 1B). The other 170 18- to 25-nt sequences that were obtained were all of cellular origin, mostly representing fragments of known mRNAs or noncoding RNAs. In Table 1, we present a compendium of the 56 distinct cellular miRNAs that were cloned from either differentiated or undifferentiated LKP1 cells. The patterns of miRNA expression in these cells were quite similar, regardless of differentiation state, with let-7b, miR-16, miR-17-5p, miR-21-5p, miR-27a, miR-31-5p, miR-93, miR-99b, miR-125b, and miR-221 being particularly well represented. We do not currently know if the small differences in miRNA expression pattern observed between differentiated and undifferentiated LKP1 cells are real or represent sampling variations (Table 1). It is also possible that some of the small number of “not annotated” or “not matched” sequences recorded in this analysis (Fig. 2) represent additional, currently uncharacterized miRNAs of cellular origin.
FIG. 2.
Origins of 18- to 25-nt sequences cloned from undifferentiated (A) and differentiated (B) LKP1 cells. RNAs of between 18 and 25 nt were cDNA cloned from total RNA preparations derived from undifferentiated and differentiated LKP1 cells, as shown in Fig. 1B. The resultant short cDNAs were concatenated, cloned, and sequenced. The origin of each of the 339 distinct sequences obtained from undifferentiated LKP1 cells (A) and the 359 sequences obtained from differentiated cells (B) was deduced by computer analysis and is shown. No HPV31-derived sequences of any kind were obtained.
TABLE 1.
Identities of cellular miRNAs cloned from HPV31-infected undifferentiated and differentiated LKP1 cells
| miRNA | No. of clones derived from LKP1 cells
|
|
|---|---|---|
| Undifferentiated | Differentiated | |
| let-7a | 5 | 1 |
| let-7b | 10 | 8 |
| let-7c | 1 | |
| let-7f | 1 | 3 |
| let-7i | 2 | 5 |
| miR-15b | 6 | 5 |
| miR-16 | 13 | 22 |
| miR-17-5p | 14 | 11 |
| miR-17-3p | 1 | |
| miR-19b | 1 | 3 |
| miR-20a | 1 | |
| miR-21-5p | 66 | 60 |
| miR-21-3p | 3 | 2 |
| miR-23 | 3 | 2 |
| miR-24-3p | 3 | 3 |
| miR-25 | 3 | 2 |
| miR-27a | 20 | 8 |
| miR-29a | 3 | 1 |
| miR-29b | 1 | 2 |
| miR-30a-5p | 1 | |
| miR-30c | 1 | |
| miR-31-5p | 15 | 14 |
| miR-31-3p | 8 | 2 |
| miR-34a-5p | 7 | 9 |
| miR-34a-3p | 1 | |
| miR-92a | 2 | 3 |
| miR-93 | 13 | 9 |
| miR-99a | 1 | 2 |
| miR-99b | 30 | 19 |
| miR-100 | 2 | |
| miR-103-3p | 1 | |
| miR-125a | 1 | |
| miR-125b | 17 | 8 |
| miR-130a | 1 | 1 |
| miR-138 | 5 | 2 |
| miR-140-3p | 1 | 2 |
| miR-141-3p | 1 | |
| miR-149 | 1 | |
| miR-152 | 1 | |
| miR-181b | 1 | |
| miR-188 | 1 | 1 |
| miR-191-5p | 2 | 2 |
| miR-192 | 1 | |
| miR-200a-3p | 1 | |
| miR-203 | 1 | |
| miR-205 | 1 | |
| miR-210 | 3 | 2 |
| miR-214 | 2 | |
| miR-218 | 1 | |
| miR-221 | 9 | 11 |
| miR-222 | 3 | 4 |
| miR-324-3p | 2 | |
| miR-338 | 1 | |
| miR-342 | 1 | |
| miR-424 | 1 | |
| miR-503 | 1 | |
| Total | 284 | 244 |
It has been suggested that nuclear DNA viruses that establish long-term latent or persistent infections would be particularly likely to express miRNAs, as these could downregulate host cell factors that increase the likelihood that latently infected cells would eventually be eliminated by host innate or adaptive immune responses (5). Papillomaviruses establish long-term latent infections in humans and animals, and we therefore examined whether the oncogenic HPV31 isolate would express viral miRNAs in latently or lytically infected human cells. In fact, we failed to detect any virally encoded miRNAs in latently or lytically HPV31-infected LKP1 cells, although 528 cellular miRNAs were successfully cloned. While these data indicate that HPV31 is unlikely to express any viral miRNAs, it remains formally possible that HPV31 could express a viral miRNA during the very early phase of the viral replication cycle, immediately after infection, that is not present once latency is established. Note that Pfeffer et al. (18) previously reported a computer analysis of the genome of HPV18 that predicted that HPV18 also does not express any viral miRNAs. While it remains possible that other human or animal papillomaviruses have indeed evolved to express one or more miRNAs in infected cells, it seems highly unlikely that viral miRNAs are involved in the transformation of cells by the common human oncogenic viral strains HPV31 and HPV18.
Acknowledgments
This research was sponsored by grants from the National Institutes of Health to B.R.C. (GM071408) and L.A.L. (CA59655). G.L. was supported by a fellowship from the Illinois Department of Public Health.
REFERENCES
- 1.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 2.Cai, X., C. H. Hagedorn, and B. R. Cullen. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10:1957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, X., A. Schäfer, S. Lu, J. P. Bilello, R. C. Desrosiers, R. Edwards, N. Raab-Traub, and B. R. Cullen. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen, B. R. 2006. Viruses and microRNAs. Nat. Genet. 38:S25-S30. [DOI] [PubMed] [Google Scholar]
- 6.Doench, J. G., C. P. Petersen, and P. A. Sharp. 2003. siRNAs can function as miRNAs. Genes Dev. 17:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores, E. R., and P. F. Lambert. 1997. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 71:7167-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frattini, M., B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic papillomaviruses requires viral episomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. USA 93:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 10.Grundhoff, A., and D. Ganem. 2001. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:1857-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 12.Hutvágner, G., J. McLachlan, A. E. Pasquinelli, É. Bálint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 13.Laimins, L. A. 1996. Human papillomaviruses target differentiating epithelia for virion production and malignant conversion. Semin. Virol. 7:305-313. [Google Scholar]
- 14.Lau, N. C., L. P. Lim, E. G. Weinstein, and D. P. Bartel. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858-862. [DOI] [PubMed] [Google Scholar]
- 15.Lee, Y., A. Chiyoung, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Rådmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 16.Lee, Y., M. Kim, J. Han, K.-H. Yeom, S. Lee, S. H. Baek, and V. N. Kim. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23:4051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez, J., A. Patkaniowska, H. Urlaub, R. Lührmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563-574. [DOI] [PubMed] [Google Scholar]
- 18.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grässer, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 19.Pfeffer, S., M. Zavolan, F. A. Grässer, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 20.Ruesch, M., and L. A. Laimins. 1998. Human papillomavirus oncoproteins alter differentiation-dependent cell cycle exit upon suspension in semi-solid media. Virology 250:19-29. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz, D. S., G. Hutvágner, B. Haley, and P. D. Zamore. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10:537-548. [DOI] [PubMed] [Google Scholar]
- 22.Yi, R., Y. Qin, I. G. Macara, and B. R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng, Y., R. Yi, and B. R. Cullen. 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. USA 100:9779-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]