Abstract
Replication of nepoviruses (family Comoviridae) occurs in association with endoplasmic reticulum (ER)-derived membranes. We have previously shown that the putative nucleoside triphosphate-binding protein (NTB) of Tomato ringspot nepovirus is an integral membrane protein with two ER-targeting sequences and have suggested that it anchors the viral replication complex (VRC) to the membranes. A second highly hydrophobic protein domain (X2) is located immediately upstream of the NTB domain in the RNA1-encoded polyprotein. X2 shares conserved sequence motifs with the comovirus 32-kDa protein, an ER-targeted protein implicated in VRC assembly. In this study, we examined the ability of X2 to associate with intracellular membranes. The X2 protein was fused to the green fluorescent protein and expressed in Nicotiana benthamiana by agroinfiltration. Confocal microscopy and membrane flotation experiments suggested that X2 is targeted to ER membranes. Mutagenesis studies revealed that X2 contains multiple ER-targeting domains, including two C-terminal transmembrane helices and a less-well-defined domain further upstream. To investigate the topology of the protein in the membrane, in vitro glycosylation assays were conducted using X2 derivatives that contained N-glycosylation sites introduced at the N or C termini of the protein. The results led us to propose a topological model for X2 in which the protein traverses the membrane three times, with the N terminus oriented in the lumen and the C terminus exposed to the cytoplasmic face. Taken together, our results indicate that X2 is an ER-targeted polytopic membrane protein and raises the possibility that it acts as a second membrane anchor for the VRC.
Many positive-strand RNA viruses replicate in association with membranes derived from the endoplasmic reticulum (ER) (37, 38). Various ER-derived structures, including spherules (Brome mosaic bromovirus) and membranous vesicles or rosettes (picornavirus superfamily), are found in infected cells (3, 4, 6, 32, 33, 41). Viral nonstructural proteins and RNA synthesis localize to these modified ER structures, suggesting that they are the sites of viral replication. Compartmentalization of viral RNA synthesis in the viral replication complexes (VRCs) provides an environment for increased local concentration of replication components and offers protection from RNA degradation by the host. The diverse nature of membranous replication complexes suggests highly specific interactions between viral proteins and intracellular membranes. Exogenous expression of viral nonstructural proteins individually or in combination has been used to investigate the role of these proteins in membrane association during VRC biogenesis. For example, the 1a protein of Brome mosaic bromovirus was shown to induce the formation of membranous spherules on the ER and to recruit the polymerase and viral RNA template to these structures, suggesting that it is a key organizer of the VRCs (8, 9). Similarly the 3AB, 2BC, and 2C proteins of poliovirus and other picornaviruses, the 6-kDa protein of potyviruses, and the 60-kDa and 32-kDa proteins of Cowpea mosaic comovirus (CPMV) target to the ER membranes in the absence of other viral proteins and induce modifications of intracellular membranes similar to these found in viral infection (7, 12, 13, 40). Membrane-binding domains have been identified in some viral membrane proteins, and these include transmembrane hydrophobic helices, amphipathic helices, and other less-well-defined sequences (16, 17, 46, 47, 52). However, the mechanisms of membrane binding and ER targeting are still poorly understood.
Tomato ringspot nepovirus (ToRSV) (a member of the family Comoviridae) has a bipartite genome (35, 36). Each RNA is first translated into a large polyprotein, which is subsequently cleaved into mature and intermediate proteins by a virus-encoded cysteine proteinase (Pro) (11, 20). RNA1 encodes proteins necessary for RNA replication, which include the RNA-dependent RNA polymerase, the proteinase, the genome-linked protein (VPg), and a putative nucleoside triphosphate-binding protein (NTB) (49). In vitro processing studies have also revealed the presence of two additional protein domains (X1 and X2) in the N-terminal region of the RNA1-encoded polyprotein (Fig. 1 A) (51). Similar to other plant picorna-like viruses, ToRSV infection induces severe morphological alterations of ER membranes, and ToRSV VRCs are associated with ER-derived membranes (19, 44). Several viral proteins containing the NTB domain have been detected in infected plants, including the mature NTB protein, the predominant NTB-VPg polyprotein, and a 90-kDa polyprotein which may correspond to the X2-NTB-VPg intermediate polyprotein (19). These proteins are tightly associated with ER membranes and cofractionate with ER-associated VRCs (19). When expressed independently of other viral proteins, the NTB-VPg protein localizes to ER membranes (52). ER binding of the protein is mediated by two regions present in the NTB domain, i.e., a C-terminal transmembrane helix and an N-terminal amphipathic helix (50, 52). These results led us to suggest that NTB and/or a polyprotein containing the NTB domain acts as a membrane anchor for the replication complexes. This suggestion is in agreement with the observation that the 60-kDa protein (equivalent to NTB-VPg) from CPMV, another member of the family Comoviridae, is an ER-targeted protein (7). The ToRSV X2 protein domain shares conserved amino acid motifs with the CPMV 32-kDa protein (35). Both proteins are highly hydrophobic and are situated immediately upstream of the NTB domain in the RNA1-encoded polyprotein. The CPMV 32-kDa protein is an ER-associated protein and has been suggested to play a key role in VRC assembly (7, 31).
FIG. 1.
Computer-assisted prediction of transmembrane helices (TM) in the ToRSV X2 protein. (A) Schematic representation of putative transmembrane helices within the X2 domain. The RNA1-encoded polyprotein is shown at the top of the panel with the indicated individual protein domains. Vertical lines represent the cleavage sites recognized by the ToRSV proteinase. The X2 protein domain is shown below the polyprotein diagram, with strongly and weakly predicted transmembrane helices represented by black and gray squares, respectively. The hydrophobicity plot of X2 is shown at the bottom of the panel. Hydrophobicity was calculated using the algorithm of Kyte and Doolittle with a window size of 17 amino acids (26). (B) Prediction of transmembrane helices within X2. The entire deduced amino acid sequence of X2 (GenBank accession number DQ469829) is shown at the top of the panel. Amino acids are numbered from the first amino acid of the X2 protein domain according to the previously proposed X1-X2 cleavage site (51). Predicted transmembrane domains are shown for each program (as indicated on the left of the panel). Uppercase letters indicate very high prediction scores, while lowercase letters indicate lower prediction scores. The amino acid sequences deleted in the TM1, TM2, TM3, and TM4 deletion mutants are underlined in the X2 sequence. A naturally occurring putative N-glycosylation site (NMS) is shown by the arrow.
In this study, we have investigated the membrane association of X2 in planta and in vitro. We show that X2 is targeted to the ER. ER binding is mediated by multiple domains, including two C-terminal transmembrane helices and a less-well-defined domain further upstream. In vitro glycosylation experiments confirm that X2 is a polytopic membrane protein that traverses the membrane at least three times.
MATERIALS AND METHODS
Plasmid construction.
Plasmids psmRS-GFP (S-K), psmRS-GFP (B-K), and pER-dsRed2 have been described previously (52). Plasmids psmRS-GFP (S-K) and psmRS-GFP (B-K) have unique restriction sites for facilitating in-frame fusions to the C and N termini of green fluorescent protein (GFP), respectively. Plasmid pGFP-X2 was constructed by amplifying the entire coding region of X2 (GenBank accession number DQ469829) with Pfu polymerase (Stratagene) and primers 3 and 4, which include an SstI and a KpnI site, respectively (see Table S1 in the supplemental material for the sequences for all primers). The amplified cDNA fragment was digested with these enzymes and inserted into the corresponding sites of psmRS-GFP (S-K). Plasmid pX2-GFP was constructed in a similar manner by using primers 18 and 21, which include a BamHI and a KpnI site, respectively. The amplified fragments were digested with these enzymes and inserted into the corresponding sites of psmRS-GFP (B-K). Mutated derivatives of pX2-GFP were constructed as described above for pX2-GFP by using the following pairs of primers: 38/39 for TM1, 42/43 for mX2, 40/46 for TM2, 40/41 for TM2-3, 47/41 for TM3, and 44/21 for cX2. A PCR-based site-directed mutagenesis method (18) was used to generate deletion mutants of pX2-GFP by using the following pairs of primers: 36/37 for ΔTM1, 28/29 for ΔTM2, and 30/31 for ΔTM3. To construct the ΔTM1-2-3 mutant, two rounds of site-directed mutagenesis were conducted. In the first round, the TM2-3 region was deleted using primer pair 28/31, and in the second round, the TM1 region was deleted using primer pair 36/37. Agroinfiltration vectors pBIN-GFP-nN and pBIN-nN-GFP, which contain the SstI/KpnI or BamHI/KpnI sites, have been described previously (52). These restriction sites were used to insert cDNA fragments containing the GFP fusions mentioned above. Plasmid pBIN19-p19, containing the Tomato bushy stunt virus suppressor of gene silencing, has also been described previously (52). To construct pBIN-X2-HA, the entire coding region of X2 was amplified as described above using primers 13 (containing an NcoI site) and 79 (containing an Xbal site and the coding sequence for the hemagglutinin [HA] tag). The PCR fragment was digested with NcoI and XbaI and ligated into the corresponding sites of plasmid pBBI525. A KpnI-EcoRI fragment from the resulting plasmid was then transferred into the binary vector pBIN19.
To construct plasmid pT7-X2, fragments containing the X2 coding region were amplified as described above using primers 54 (containing an MscI site) and 53 (containing an XhoI site). The amplified fragments were digested with MscI and XhoI and introduced into the corresponding sites of plasmid pCITE-4a (+) (Novagen). Plasmid pT7-Gln-X2 was constructed in a similar manner by using primers 52 (containing an MscI site and the coding region for an introduced N-glycosylation site) and 53. To construct pT7-X2-Gln, the PCR-based site-directed mutagenesis method was used to mutate NGH to NGS in the N terminus of GFP in the pX2-GFP plasmid by using primers 56/57. This resulted in the introduction of an N-glycosylation site. The resulting plasmid was then used as a template to amplify a fragment with primer pair 13/55. The amplified fragment contained the entire X2 coding region and a small portion of the GFP-coding region, which includes the introduced glycosylation site. The fragments were digested with NcoI and inserted into the corresponding site of pCITE-4a (+). Other plasmids were produced by PCR-based mutagenesis. Plasmids pT7-X2ΔTM2-Gln, pT7-X2ΔTM3-Gln, pT7-X2ΔTM2-3-Gln, pT7-X2ΔTM1-Gln, and pT7-X2ΔN-Gln were obtained using pT7-X2-Gln as a template and primer pairs 28/29, 30/31, 28/31, 80/81, and 84/86, respectively. Similarly, pT7-X2ΔTM2-3-Gln was used as a template to produce pT7-X2ΔTM2-3-4 by using primers 82/83. Plasmids pT7-Gln-X2ΔTM1, pT7-Gln-X2ΔM, pT7-Gln-X2ΔN, pT7-Gln-X2ΔTM3, and pT7-Gln-X2ΔTM2-3 were constructed using template pT7-Gln-X2 and primer pairs 80/81, 85/86, 84/86, 30/31, and 28/31, respectively. Finally, plasmids pT7-X2ΔTM3, pT7-X2ΔTM2-3, and pT7-X2ΔTM1 were constructed using plasmid pT7-X2 as a template and primer pairs 30/31, 28/31 and 80/81, respectively.
Agroinfiltration of Nicotiana benthamiana plants and confocal microscopy.
Binary vectors containing the plant expression cassettes with the X2 fusion proteins were transformed into Agrobacterium tumefaciens LBA4044 (Invitrogen) by electroporation. The transformed bacteria were then used for agroinfiltration as previously described (52). Three days after agroinfiltration, GFP and dsRed2 fluorescence were analyzed with a confocal microscope (Leica) as described previously (52). The acquired images were processed with Leica confocal software and Photoshop 7.0 (Adobe).
Subcellular fractionation and membrane flotation assays.
Three to 4 days postagroinfiltration, plant tissues were extracted and fractionated into postnuclear (S3), soluble (S30), and membrane-enriched (P30) fractions as previously described (19, 40). The P30 fraction was resuspended in a volume of homogenization buffer equivalent to that used for the S30 fraction or treated with an equal volume of 1 M NaCl or 0.1 M Na2CO3 (pH 11). Membrane flotation assays were conducted essentially as described previously (52). Briefly, 800 μl of S3 or P30 fraction was adjusted to a final volume of 1.9 ml of 71.5% sucrose (wt/vol) in NTE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM EDTA) and overlaid with 7 ml of 65% sucrose in NTE and 3.1 ml of 10% sucrose in NTE. After centrifugation at 100,000 × g for 18 h, 12 1-ml fractions were collected from the bottom of the tube.
Separation of proteins by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunodetection were conducted as previously described (19) using a mouse monoclonal anti-GFP antibody (BD Biosciences), a rat anti-HA antibody (Roche), or a rabbit polyclonal anti-Bip antibody (donated by M. Chrispeels). The secondary antibodies were goat anti-mouse, goat anti-rat, or goat anti-rabbit immunoglobulin G conjugated with horseradish peroxidase (Bio/Can).
In vitro translation assays and deglycosylation assays.
Coupled in vitro transcription-translation reactions in the presence or absence of canine microsomal membranes and deglycosylation assays of translation products were conducted as previously described (50).
Computer-assisted multiple-sequence alignments and prediction of putative transmembrane helices and amphipathic helices.
Transmembrane helices in nepovirus and comovirus proteins were predicted using the following programs: PHDhtm (34), Sosui (22), Tmpred (23), TMAP (30), TMHMM (43), andHMMTOP (48). Prediction and projection of amphipathic helices were conducted using the Antheprot program (15). Multiple protein sequences were aligned using the ClustalW program (10).
RESULTS
Computer-assisted prediction of hydrophobic regions within X2.
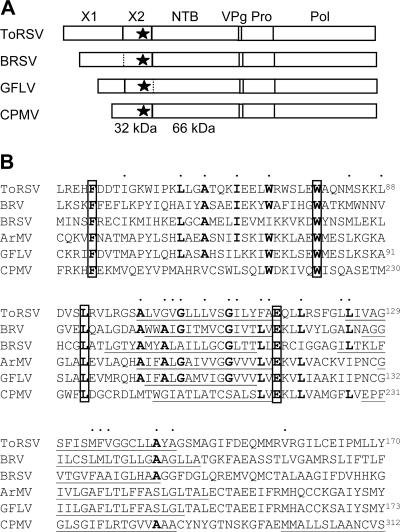
Computer analysis of the primary sequence of the ToRSV X2 protein revealed the presence of four hydrophobic regions (TM1, TM2, TM3, and TM4) as shown in the hydrophilicity profile (Fig. 1A). Two of these regions had very high hydrophobicity values (TM2 and TM3) and were predicted to be transmembrane helices by all programs, although the exact borders of the predicted transmembrane helices differed among programs (Fig. 1B). Two other possible transmembrane segments (TM1 and TM4) were less hydrophobic and were each predicted by only one of the six programs considered. The equivalent protein domains for four distinct nepoviruses and the CPMV 32-kDa protein were also examined for the presence of putative transmembrane helices (Fig. 2). A previously identified conserved sequence motif (F-X28-W-X11-L-X23-E) (35) was present in all sequences. The sequences aligned to the ToRSV TM2 and TM3 regions were also predicted to be transmembrane helices for all the proteins analyzed. In the case of the comovirus 32-kDa protein, a third possible transmembrane helix was also identified by the majority of the programs at a position corresponding to that of the weakly predicted ToRSV TM4 domain.
FIG. 2.
Multiple-protein-sequence comparison of the X2 protein domains of nepoviruses and the 32-kDa protein of comoviruses. (A) Schematic representation of the RNA1-encoded polyprotein of nepoviruses of subgroups C (ToRSV), B (Beet ringspot nepovirus [BRSV]), and A (Grapevine fanleaf nepovirus [GFLV]) and of a comovirus (CPMV). Vertical lines represent cleavage sites identified by in vitro processing experiments (21, 27). In the cases of BRSV and GFLV, two hypothetical cleavage sites are shown by the dashed lines. The star represents the highly conserved region shown in panel B. (B) Multiple-protein-sequence alignment of the conserved region present within the X2 protein domain of five nepoviruses belonging to subgroup C (ToRSV and Blackcurrant reversion nepovirus [BRV]), subgroup B (BRSV), and subgroup A (GFLV and Arabis mosaic nepovirus [ArMV]) and within the CPMV 32-kDa protein. The following GenBank accession numbers were used to retrieve the sequences from the database: NC003509 for BRV, NC003693 for BRSV, NC003615 for GFLV, NC006057 for ArMV, and P03600 for CPMV. The conserved amino acids present in the “protease cofactor conserved motif” (F-X28-W-X11-L-X23-E) are boxed (35). Dots above the sequence represent amino acids which are similar in all the sequences. Underlined sequences represent the core regions of transmembrane helices predicted as shown in Fig. 1B. Only transmembrane helices predicted by the majority of the programs are shown. Numbering of amino acids for ToRSV and GFLV is shown according to the proposed X1-X2 cleavage site (27, 51) and for CPMV according to the start codon. For other sequences, the X1-X2 cleavage site has not been identified and amino acids within the sequence were left unnumbered.
Subcellular localization of GFP-tagged X2 proteins.
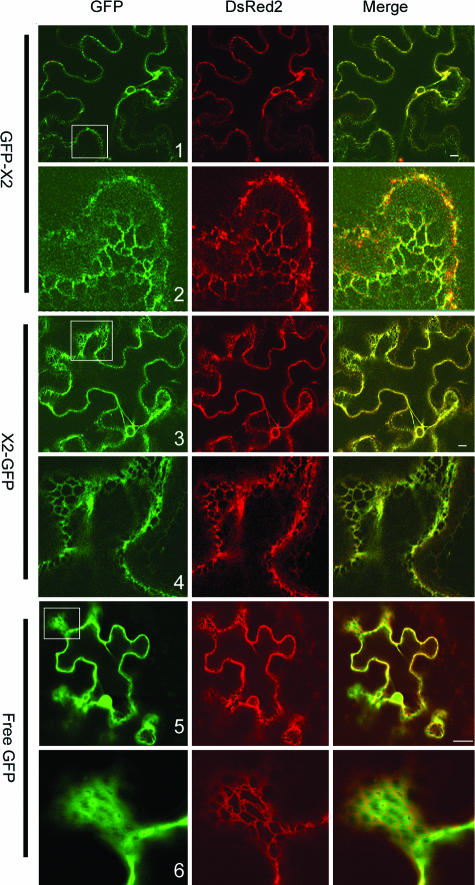
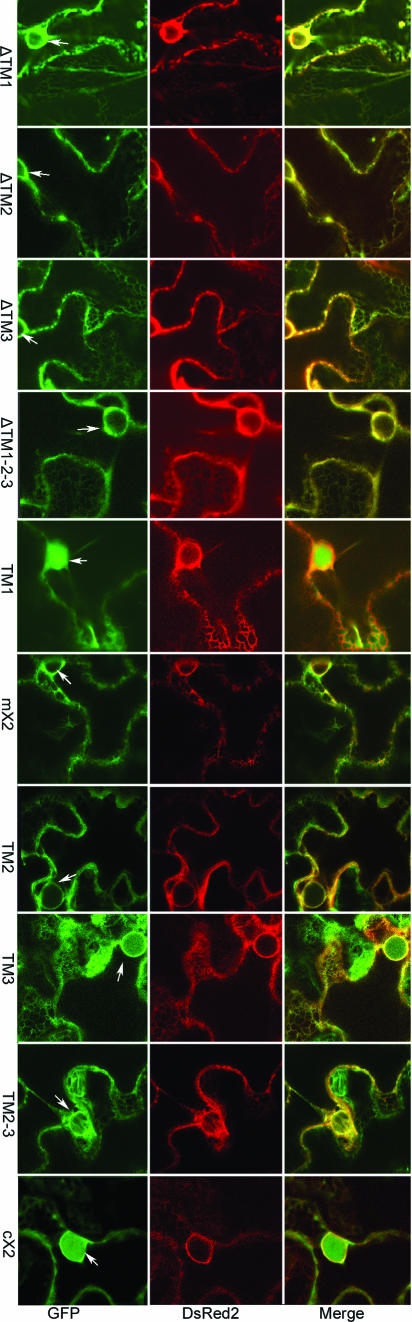
To determine whether X2 is able to associate with intracellular membranes in planta, GFP was fused in frame to either the N or the C terminus of X2 (Fig. 3A). The fusion proteins were expressed in N. benthamiana plants by using agroinfiltration. GFP fluorescence in transfected epidermal cells was analyzed using a confocal laser scanning microscope 3 days after agroinfiltration. A previously described ER marker (ER-dsRed2, in which the red fluorescent protein is targeted to the lumen of the ER) (52) was coexpressed with the GFP fusion proteins. Confocal images show that ER-dsRed2 labeled both the perinuclear area and the cortical ER network, a result consistent with our previous observations (Fig. 4, dsRed2). The fluorescence associated with free GFP was present in the cytosol (cytoplasm is usually pressed against the plasma membrane due to the presence of large vacuoles in mature epidermal cells which occupy most of the intracellular space) and inside the nucleus (Fig. 4, panel 5). The green fluorescence was sometimes found in proximity to the ER cortical network but did not coincide with the ER marker (Fig. 4, panel 6). In contrast, the fluorescence associated with GFP-X2 and X2-GFP overlapped with the ER marker in both the perinuclear area and the cortical ER network (Fig. 4, panels 1 to 4). Similar fluorescence patterns were observed when X2-GFP and GFP-X2 were transfected in cultured tobacco BY-2 cells by biolistic bombardment (data not shown). These results suggest that X2 is targeted to ER membranes, at least in the context of the GFP fusions.
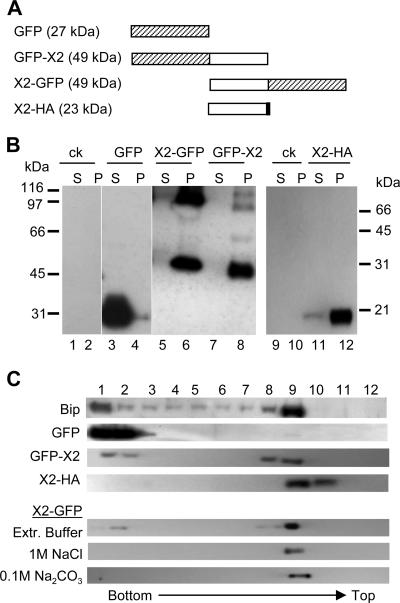
FIG. 3.
Subcellular fractionation of X2 fusion proteins. (A) Schematic representation of X2 fusion proteins. The white box represents the X2 domain, while the hatched and black boxes represent the GFP and HA domains, respectively. The predicted molecular mass of each fusion protein is indicated in parentheses. (B). Subcellular fractionation of X2 fusion proteins. Plant tissues expressing the various fusion proteins were fractionated into soluble (S) and membrane-enriched (P) fractions as described in Materials and Methods. Proteins were separated by SDS-PAGE (12% for lanes 1 to 8 and 15% for lanes 9 to 12) and detected by immunoblotting with anti-GFP (lanes 1 to 8) or anti-HA (lanes 9 to 12) monoclonal antibodies. Migration of molecular mass standards is indicated on the left (lanes 1 to 8) or right (lanes 9 to 12) side. ck, negative control transfected with pBin-p19 only. (C) Membrane flotation assays. Equal volumes of postnuclear (S3) fractions derived from plants expressing GFP-X2, X2-GFP, X2-HA, or unfused GFP were used for membrane flotation assays as described in Materials and Methods. Fractions were collected from the step sucrose gradient, and proteins present in each collected fraction (as indicated at the top of the panel) were separated by SDS-PAGE and immunodetected using anti-GFP, anti-HA, or anti-Bip antibody. Only the relevant parts of the gels are shown. In the case of X2-GFP, P30 fractions of X2-GFP were incubated for 30 min at 4°C in extraction buffer (Extr. Buffer) or in solutions of 1 M NaCl or 0.1 M Na2CO3 (pH 11) before the flotation assay.
FIG. 4.
Subcellular localization of GFP-X2 and X2-GFP. GFP fusions and ER-dsRed2 (an ER marker) were expressed in leaves of N. benthamiana by using agroinfiltration as described in Materials and Methods. Epidermal cells were examined 3 days after agroinfiltration by confocal microscopy. In the merge panel, the colocalization of the GFP fluorescence (green) and of the ER marker fluorescence (red) results in a yellow color. Panels 2, 4, and 6 are close-up views of regions included in the white squares in panels 1, 3, and 5. Bars on the merged images represent 10 μm.
Subcellular fractionations were performed to further study the membrane association of the fusion proteins. As described above, X2-GFP and GFP-X2 were expressed in N. benthamiana by using agroinfiltration. Three days after agroinfiltration, the leaves were extracted and soluble (S30) and membrane-enriched (P30) fractions were produced as described in Materials and Methods. Proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-GFP antibodies. In agreement with the confocal images, unfused GFP was detected mainly in the S30 fraction while the full-length GFP-X2 and X2-GFP (49 kDa) were detected only in the membrane-enriched P30 fractions (Fig. 3B, lanes 3 to 8). Larger forms of the proteins (about 100 kDa) were also detected in P30 fractions of X2-GFP and GFP-X2, which may correspond to dimers, as many membrane proteins can maintain their oligomeric forms in the presence of SDS (14). This possibility was not investigated further.
The presence of the GFP fusion proteins in P30 fractions may result from true membrane association or simply protein aggregation. To distinguish between these two possibilities, we used a membrane flotation assay. In this assay, total plant extracts (S3) are overlaid with a sucrose gradient and subjected to centrifugation. Low-density membranes and proteins associated with these membranes float to the upper part of the gradient while soluble proteins or aggregated proteins remain at the bottom. We used Bip (an endogenous ER luminal protein) (45) and unfused GFP as controls. As shown in Fig. 3C, Bip rose towards the top of the gradient (fractions 8 and 9) while GFP remained at the bottom of the gradient (fractions 1 and 2). GFP-X2 and X2-GFP were found in fractions 8 and 9, confirming that they are membrane associated.
To investigate the nature of the association of X2-GFP with membranes, we treated the P30 fractions containing X2-GFP with Na2CO3 (0.1 M, pH 11) and NaCl (1 M) and then conducted membrane flotation assays. These chemicals are known to release peripheral membrane proteins from the membranes but not integral membrane proteins (24, 39). X2-GFP was found in the membrane fraction (fraction 9) after both treatments, suggesting that it interacts directly with the lipid bilayer of the membrane (Fig. 3C).
To provide further evidence that X2 is a membrane-associated protein, we also fused the entire protein to a smaller epitope tag (X2-HA, in which the HA epitope tag is fused to the C terminus of X2) (Fig. 3A). X2-HA was mainly detected in the P30 fraction (Fig. 3B, lanes 11 and 12) and floated to the top of the gradient in a membrane flotation assay (Fig. 3C).
X2 contains multiple ER-targeting domains.
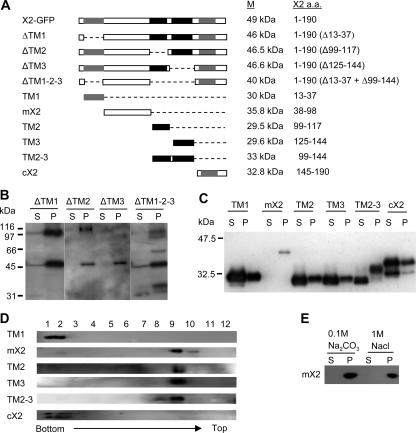
The tight association of X2-GFP fusion proteins with ER membranes prompted us to investigate sequence elements within X2 mediating the membrane association. We first generated several mutants of X2-GFP in which the hydrophobic segments TM1, TM2, and TM3 were deleted either individually or in combination (Fig. 5A, constructs ΔTM1, ΔTM2, ΔTM3, and ΔTM1-2-3). We found that all four X2 derivatives retained the ability to associate with the ER, i.e., they had patterns of fluorescence similar to those in the wild-type X2-GFP in confocal images (compare Fig. 4 and Fig. 6). They were also partitioned to membrane-enriched fractions in subcellular fractionation experiments (Fig. 5B). We then fused different portions of X2 (Fig. 5A, constructs TM1, TM2-3, TM2, TM3, cX2, and mX2) to the N terminus of GFP and tested whether any given fragment could target GFP to the ER. The fluorescence associated with the TM1 and cX2 fusion proteins did not overlap with that of ER-dsRed2 (Fig. 6). The proteins were detected in both the S30 and the P30 fractions (Fig. 5C). However, the presence of these proteins in the P30 fraction was probably due to protein aggregation rather than to membrane association, as the proteins remained at the bottom of the gradient in the membrane flotation assays (Fig. 5D). The highly hydrophobic TM2 and TM3 domains targeted the GFP to the ER membrane when fused to GFP individually (TM2 and TM3) or in combination (TM2-3) (Fig. 6). Targeting to the ER was partial when only one of the hydrophobic regions was included (as evidenced by the presence of some fluorescence within the nucleus with TM2 and TM3) (Fig. 6 and data not shown). The TM2 and TM3 proteins were partitioned in both the S30 and P30 fractions (Fig. 5C). The full-length, 33-kDa TM2-3 fusion protein was found predominantly in the P30 fraction. A 30-kDa truncated protein which may correspond to degradation products of the full-length protein was also detected in the S30 fraction. The TM2, TM3, and full-length TM2-3 fusion proteins present in the P30 fraction floated to the top of the gradient in membrane flotation assays, confirming that they are membrane associated (Fig. 5D). Surprisingly, although no hydrophobic sequence was predicted in this region, mX2 was found to associate with the ER in confocal pictures, fractionated with the membrane-enriched P30 fraction and partitioned with the membranes in the flotation assays (Fig. 6 and 5C and D). We treated the P30 fractions of mX2 with Na2CO3 (0.1 M, pH 11) and NaCl (1 M), which were separated subsequently into S30 and P30 fractions. We found that mX2 was present in the P30 fraction after the treatment, suggesting a direct interaction between mX2 and the lipid bilayer of the membranes (Fig. 5E). Taken together, these results suggested that X2 contains three ER-targeting domains, including two highly hydrophobic C-terminal regions and an additional domain further upstream.
FIG. 5.
Subcellular fractionation of X2-GFP mutant derivatives. (A) Schematic representation of X2-GFP derivatives. Only the X2 domains are shown in the panel. The GFP domain (not shown) is fused to the C terminus of each X2 derivative. The predicted transmembrane domains are shown with gray and black boxes as in Fig. 1. The predicted molecular mass (M) of each GFP fusion protein and the amino acids (a.a.) of X2 included in each fusion protein are shown on the right. (B and C) Subcellular fractionation of X2-GFP derivatives. Soluble (S) and membrane-enriched (P) fractions were prepared from plants expressing mutated X2-GFP proteins as described in Materials and Methods. Proteins were separated by SDS-PAGE (12%) and immunodetected with anti-GFP antibody. Migration of molecular mass standards is shown on the right of each gel. (D) Membrane flotation assays. For TM1, mX2, and cX2, postnuclear (S3) fractions were used for the flotation assays. In the cases of TM2, TM3, and TM2-3, P30 fractions were used. Fractions were collected from the step sucrose gradient, and proteins present in each collected fraction were separated by SDS-PAGE (12%) and immunodetected with the anti-GFP antibody. (E) Biochemical treatments of membrane-enriched fractions derived from plants expressing mX2. Membrane-enriched (P30) fractions from Fig. 4C were treated with 0.1 M Na2CO3 (pH 11) or 1 M NaCl for 30 min at 4°C. After separation of membrane-bound (P) and soluble (S) proteins, the presence of mX2 in these fractions was revealed by immunoblotting with the anti-GFP antibody.
FIG. 6.
Subcellular localization of X2-GFP derivatives in epidermal cells of N. benthamiana. Plants expressing X2-GFP derivatives and an ER marker (ER-dsRed2) were examined using confocal microscopy 3 days after agroinfiltration. Pictures represent portions of a single cell, including the nucleus (shown by the arrow) and the cortical ER network.
Topology of X2 in ER membranes inferred from the pattern of glycosylation in vitro.
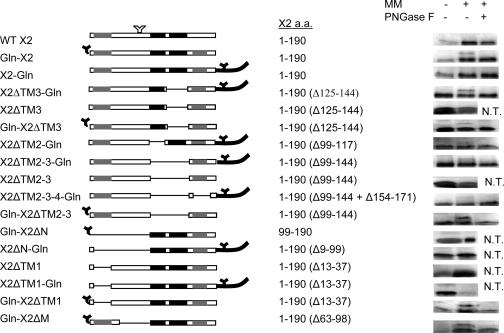
Our result indicating that X2 contains three ER-targeting domains suggests that it is a polytopic membrane protein. To investigate the topology of X2 in the ER membrane, we chose to examine the glycosylation patterns of X2 derivatives, into which N-glycosylation sites were introduced at different locations. Because the active site of the glycosyltransferase is situated on the luminal side of ER membranes, glycosylation of introduced N-glycosylation sites could occur only if they were translocated into the ER lumen. To test the glycosylation status of a protein, in vitro translations can be conducted in the presence or absence of microsomal membranes, which consist predominantly of ER membranes (Fig. 7). If glycosylation occurs, an additional slower-migrating band (about 3 kDa larger) should be detected with SDS-PAGE when translated in the presence of membranes. Glycosylation can be further confirmed by treatment of the reaction mixture with PNGase F, resulting in the disappearance of this additional protein. The wild-type X2 protein contains a possible N-glycosylation site (NMS) (Fig. 1). However, this sequence was not recognized in vitro (Fig. 7, construct X2). We then introduced an N-glycosylation site either at the N terminus (Gln-X2) or at the C terminus (X2-Gln) of the protein (Fig. 7). Gln-X2 was glycosylated, indicating that the N terminus of the protein is translocated into the ER lumen. In contrast, X2-Gln was not glycosylated. The observed translocation of the N terminus of the protein in the ER lumen confirms that X2 is a transmembrane protein.
FIG. 7.
In vitro glycosylation assays of wild-type or mutated X2. On the left is a schematic representation of the various X2 constructs. The predicted transmembrane domains are shown with gray and black boxes as in Fig. 1. Amino acids (a.a.) inserted at the N or C termini of the proteins are shown with dark lines. Introduced N-glycosylation signals are represented by black Y's. A naturally occurring putative N-glycosylation site is shown by the white Y, although this site was not recognized in any of the mutants tested. The dashed lines represent deleted regions within X2. The name of each construct is indicated on the left, and the amino acids of the X2 domain contained in each construct are indicated in the middle. In vitro glycosylation assays are shown on the right. Each protein was translated in the presence (+) or absence (−) of canine microsomal membranes (MM). The translation products were further treated with endoglycosydase F (PNGase F), separated by SDS-PAGE, and detected by autoradiography. Only the relevant portions of the gels are shown. N.T., not tested.
As mentioned above, computer predictions strongly suggest that TM2 and TM3 traverse the membrane and form a hairpin structure. Lack of glycosylation of X2-Gln suggests that if TM2 and TM3 form a hairpin structure in the membrane, the loop is in the ER lumen. To confirm this orientation, we deleted TM2 and TM3 individually or in combination. We hypothesized that deleting the second transmembrane domain of the hairpin (X2ΔTM3-Gln mutant) would result in the translocation of the C terminus of the protein in the lumen of the membrane. As expected, glycosylation of this mutant readily occurred in the presence of the membrane. This glycosylation was not due to the recognition of the internal NMS sequence, as the control X2ΔTM3 mutant remained unglycosylated. Introduction of this mutation in the Gln-X2 protein did not alter its state of glycosylation, suggesting that deletion of TM3 did not affect the orientation of the N-terminal region of the protein (compare mutant Gln-X2ΔTM3 to Gln-X2). Similarly, deletion of TM2 from the X2-Gln protein resulted in the reorientation of the C terminus of the protein in the lumen (X2ΔTM2-Gln mutant). These results provide support for the suggestion that TM2 and TM3 form a hairpin in the membrane. To confirm this, we deleted both domains from the X2-Gln protein (X2ΔTM2-3-Gln). Unexpectedly, glycosylation was still observed, although it was much reduced compared to that of the X2ΔTM3-Gln mutant. As described above, this glycosylation was not due to the recognition of the internal NMS sequence, as the X2ΔTM2-3 mutant was not glycosylated. To determine whether the putative TM4 domain played a role in the translocation of the C terminus of the protein in the membrane lumen, we constructed a triple mutant in which TM2, TM3, and TM4 were deleted. Low levels of glycosylation were still observed in this new mutant, suggesting that TM4 was not a primary determinant of the membrane topology. We conclude that an additional domain upstream of TM2 is likely responsible for the low level of glycosylation observed in the X2ΔTM2-3-Gln and X2ΔTM2-3-4-Gln proteins.
To investigate which region of X2 is responsible for the translocation of the N terminus of the protein in the lumen, we introduced a series of mutations in the Gln-X2 protein. First, we deleted both TM2 and TM3 (Gln-X2ΔTM2-3). Glycosylation of this mutant was still observed, suggesting that a region of X2 present between the N terminus of the protein and the TM2 domain acts as a transmembrane domain (Fig. 7). We then deleted the entire N-terminal region of X2 (Gln-X2ΔN). Translocation of the N terminus of the protein was eliminated, confirming the presence of a transmembrane segment in this region. This result is consistent with the in planta observation that an ER-targeting domain is present in the mX2-GFP fusion protein. TM1 is the only hydrophobic region predicted by computer. However, deletion of TM1 in the context of Gln-X2 (Gln-X2ΔTM1) did not prevent the glycosylation. The observed glycosylation was due to the recognition of the introduced N-terminal glycosylation site, as the control X2ΔTM1 mutant remained unglycosylated. A stretch of 35 amino acids located immediately upstream of the TM2 domain was also deleted (mutant Gln-X2ΔM). Glycosylation of this mutant was still observed. Based on these results, we tentatively suggest that a region confined within amino acids 38 to 62 may be involved in the membrane association and in the translocation of the N terminus of X2 in the membrane lumen. Finally, the ΔN and ΔTM1 deletions were also introduced in the X2-Gln protein. The X2ΔN-Gln and X2ΔTM1-Gln mutants remained unglycosylated, suggesting that deletion of the N-terminal region of the protein did not affect the orientation of its C terminus.
DISCUSSION
In this study, we used GFP fusion proteins to show that the ToRSV X2 protein contains several ER-targeting sequences. We acknowledge that our experimental system differs from a natural viral infection in several important aspects. First, the protein was translated from an mRNA rather than produced through polyprotein processing. As a result, a methionine was inserted at the N terminus of the protein. Second, fusion of X2 to GFP may affect the biological function of X2 and/or its intracellular localization in planta. However, it should be noted that both N- and C-terminal fusions of the protein to GFP resulted in similar fluorescence patterns. Also, several independent ER-targeting elements were identified within X2 by using in vivo GFP fusion assays and the presence of these membrane association domains was confirmed by in vitro glycosylation assays. Finally, the membrane association of X2 in vivo was confirmed using a smaller epitope tag (HA). We have previously shown that ER-derived membranes play a key role in ToRSV replication (19). X2 shares many sequence similarities with the C-terminal region of the 32-kDa protein of CPMV, which is also targeted to the ER when fused to GFP (7). In infected plants, the CPMV 32-kDa protein is found at or near ER-derived membrane vesicles which contain VRCs (31). The CPMV 32-kDa protein has been suggested to act as a second membrane anchor for the replication complex in addition to the 60-kDa protein (7). By analogy, it is tempting to suggest that the ToRSV X2 protein is also associated with ER-bound VRCs in infected plants, although we are unable to confirm this suggestion at this time, due to difficulties encountered in producing antibodies against this very hydrophobic protein.
Although the ToRSV X2 protein and the CPMV 32-kDa protein both target GFP to ER membranes, the fluorescence patterns of these proteins are somewhat different. Fluorescence associated with the ToRSV X2-GFP fusion protein is evenly distributed in the cortical ER network and in the perinuclear ER. No obvious membrane proliferation or alteration of membrane morphology was observed. In contrast, the CPMV 32-kDa protein-GFP fusion is specifically targeted to the cortical ER (7). It also induces aggregation of cortical ER and formation of small bodies near the nucleus. One possibility is that the different behaviors of the two fusion proteins are due to intrinsic properties of the two proteins, possibly modulated by divergent sequences outside the conserved motif. Alternatively, the differences observed could be due to the experimental system used. In this study, the fusion proteins were expressed by agroinfiltration, while in the CPMV study, the fusion proteins were expressed from a viral vector.
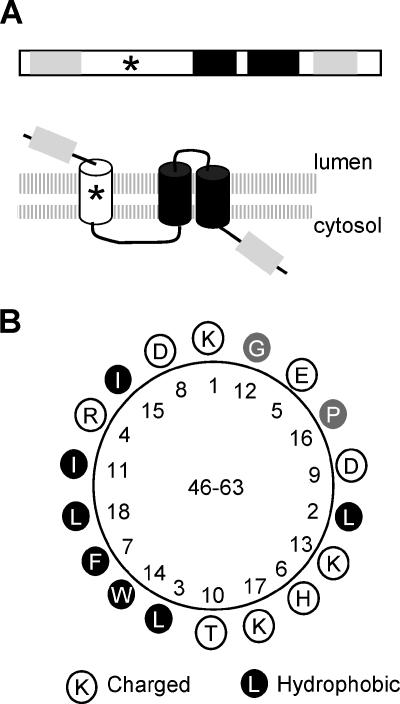
In this study, we have identified three distinct membrane-association domains within X2, i.e., two C-terminal transmembrane helices (TM2 and TM3) and a third, less-well-defined domain within the mX2 region. Each of the three elements could direct GFP to the ER membranes independently (Fig. 6). This observation is further supported by our in vitro glycosylation study, which suggests that all three domains have the ability to traverse the membranes (Fig. 7). Based on these results, we propose a model for the topology of X2 in the membrane (Fig. 8A). In this model, the N terminus of X2 is oriented in the lumen while the C terminus is cytosolic. The protein traverses the membrane three times.
FIG. 8.
Topological model of X2 in ER membranes. (A) Proposed topological model of X2. On the top of the panel is the linear representation of membrane association domains of X2. The light gray regions represent hydrophobic domains (TM1 and TM4, as in Fig. 1) that do not traverse the membranes. Transmembrane α-helices TM2 and TM3 are shown by the black boxes as in Fig. 1. The star represents a putative amphipathic helix. Below the domain diagram is the topological model of X2 in ER membranes. The double-lipid layer of the membranes is represented by the two shaded horizontal lines. The predicted orientations of the various transmembrane domains within the membrane are shown. (B) Helical wheel projection of a putative amphipathic helix located between amino acids 46 and 63.
The highly hydrophobic TM2 and TM3 regions were strongly predicted to form a hairpin in the membrane not only in the ToRSV X2 protein domain but also in the equivalent protein domains of other nepoviruses and in the CPMV 32-kDa protein (Fig. 1 and 2). Our in vitro results suggest that the hairpin loop resides in the lumen of the membrane. This proposed topology is supported by the following evidence. First, the C terminus of the wild-type protein is exposed to the cytosolic face of the membrane. Second, deletion of TM3 or TM2 reversed the orientation of the C terminus of X2 from cytosol to lumen. Third, deletion of TM2 and TM3 resulted in a reduction of the translocation of the C terminus of the protein in the lumen, although it did not completely eliminate it (see below for a possible interpretation of this result). Finally, the N terminus of the Gln-X2ΔN protein, which includes the TM2 and TM3 domains but not the upstream membrane association domain, is oriented towards the cytosolic face of the membrane.
The observation that the central region of X2 (mX2) contains an ER-targeting sequence and probably traverses the membrane at least in vitro is surprising, as this region is largely hydrophilic and is not predicted to contain transmembrane domains (Fig. 1). A putative amphipathic helix is present between amino acids 46 and 63 and may be responsible for the translocation of the N terminus of the protein in the lumen (Fig. 8B). Similar putative amphipathic helices were also found at equivalent positions in the X2 protein domains of other nepoviruses and in the CPMV 32-kDa protein (data not shown). Amphipathic helices are initially oriented parallel to the membrane, with their hydrophobic faces towards the membrane and their hydrophilic faces towards the cytosol. Translocation of amphipathic helices across membranes usually involves the oligomerization of the amphipathic helix at the membrane surface followed by insertion of the oligomers into the membrane in a posttranslational manner through the barrel stave mechanism (2, 42). The X2 putative amphipathic helix may be inserted in the membrane in either orientation, providing a possible explanation for our observation that both Gln-X2ΔTM2-3 and X2ΔTM2-3-Gln are glycosylated. In fact, dual orientation of transmembrane segments has been documented (29). In the context of the wild-type X2 protein, the presence of the TM2 and TM3 domains may force the putative amphipathic helix to adopt a type I topology (in-out) (Fig. 8A). A similar situation was reported for the human band 3 protein, in which a downstream transmembrane domain dictated the orientation of upstream transmembrane segments (28). Further experimentation will be required to confirm the role of the proposed amphipathic helix in membrane association.
In this study, the topology of the mature X2 protein was analyzed. However, the protein is initially produced as a polyprotein in which X2 is located immediately upstream of the NTB domain. Also, intermediate polyproteins containing both the X2 and the NTB domains are likely to be present in infected cells. In fact, in addition to the NTB and NTB-VPg proteins, a 90-kDa membrane-associated protein containing the NTB domain, which may correspond to the X2-NTB-VPg polyprotein, was previously detected in infected plants (19). Previous analysis of the topology of NTB-VPg in ER membranes by in vitro glycosylation assays revealed that the N terminus of NTB is translocated into the ER lumen (52). This would be in apparent contradiction with the results presented here indicating that the C terminus of the mature X2 protein is oriented towards the cytosolic face of the membrane. One possible explanation is that in the context of the polyproteins, the C terminus of X2 or the N terminus of NTB adopts an orientation different from that observed with the mature proteins. Dual topology has been observed for the p7 protein of hepatitis C virus, in which case the protein adopts a different orientation when it is present within a larger polyprotein that also includes the E2 domain (E2-p7) (25). We have previously shown that the translocation of the N terminus of NTB-VPg in the lumen is directed by a putative amphipathic helix, which probably requires oligomerization to traverse the membrane (52). Although it is tempting to suggest that this process is inhibited in the context of larger polyproteins that contain the X2 domain, further experimentation will be required to resolve this issue.
The polytopic nature of X2 is reminiscent of that of the 2B protein of poliovirus. Both proteins are located immediately upstream of the NTB domain (2C in the case of poliovirus). The 2B protein of poliovirus has been shown to increase membrane permeability by forming a pore in the membrane (1). Recent evidence suggests that pore formation regulates the calcium concentrations of endoplasmic reticulum membranes and may play a role in preventing defensive apoptotic host cell response (5). It will be interesting to investigate whether X2 has the ability to modify membrane permeability or not.
Supplementary Material
Acknowledgments
We thank M. Chrispeels (University of California, San Diego) for anti-Bip antibodies and M. Weiss (PARC-Summerland) for help with the confocal microscope. S. C. Zhang and J. Chisholm are gratefully acknowledged for critical reading of the manuscript.
This work was supported in part by an NSERC discovery grant awarded to H.S.
Footnotes
Published ahead of print on 23 August 2006.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Agirre, A., A. Barco, L. Carrasco, and J. L. Nieva. 2002. Viroporin-mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem. 277:40434-40441. [DOI] [PubMed] [Google Scholar]
- 2.Bechinger, B. 1999. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta 1462:157-183. [DOI] [PubMed] [Google Scholar]
- 3.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 4.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campanella, M., A. S. de Jong, K. W. Lanke, W. J. Melchers, P. H. Willems, P. Pinton, R. Rizzuto, and F. J. van Kuppeveld. 2004. The coxsackievirus 2B protein suppresses apoptotic host cell responses by manipulating intracellular Ca2+ homeostasis. J. Biol. Chem. 279:18440-18450. [DOI] [PubMed] [Google Scholar]
- 6.Carette, J. E., K. Guhl, J. Wellink, and A. Van Kammen. 2002. Coalescence of the sites of cowpea mosaic virus RNA replication into a cytopathic structure. J. Virol. 76:6235-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carette, J. E., J. van Lent, S. A. MacFarlane, J. Wellink, and A. van Kammen. 2002. Cowpea mosaic virus 32- and 60-kilodalton replication proteins target and change the morphology of endoplasmic reticulum membranes. J. Virol. 76:6293-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chisholm, J., A. Wieczorek, and H. Sanfacon. 2001. Expression and partial purification of recombinant tomato ringspot nepovirus 3C-like proteinase: comparison of the activity of the mature proteinase and the VPg-proteinase precursor. Virus Res. 79:153-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 13.Datta, U., and A. Dasgupta. 1994. Expression and subcellular localization of poliovirus VPg-precursor protein 3AB in eukaryotic cells: evidence for glycosylation in vitro. J. Virol. 68:4468-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeGrado, W. F., H. Gratkowski, and J. D. Lear. 2003. How do helix-helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. Protein Sci. 12:647-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deleage, G., C. Combet, C. Blanchet, and C. Geourjon. 2001. ANTHEPROT: an integrated protein sequence analysis software with client/server capabilities. Comput. Biol. Med. 31:259-267. [DOI] [PubMed] [Google Scholar]
- 16.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Echeverri, A. C., and A. Dasgupta. 1995. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208:540-553. [DOI] [PubMed] [Google Scholar]
- 18.Fisher, C. L., and G. K. Pei. 1997. Modification of a PCR-based site-directed mutagenesis method. BioTechniques 23:570-571, 574. [DOI] [PubMed] [Google Scholar]
- 19.Han, S., and H. Sanfacon. 2003. Tomato ringspot virus proteins containing the nucleoside triphosphate binding domain are transmembrane proteins that associate with the endoplasmic reticulum and cofractionate with replication complexes. J. Virol. 77:523-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hans, F., and H. Sanfacon. 1995. Tomato ringspot nepovirus protease: characterization and cleavage site specificity. J. Gen. Virol. 76:917-927. [DOI] [PubMed] [Google Scholar]
- 21.Hemmer, O., C. Greif, P. Dufourcq, J. Reinbolt, and C. Fritsch. 1995. Functional characterization of the proteolytic activity of the tomato black ring nepovirus RNA-1-encoded polyprotein. Virology 206:362-371. [DOI] [PubMed] [Google Scholar]
- 22.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 347:166. [Google Scholar]
- 24.Howell, K. E., and G. E. Palade. 1982. Hepatic Golgi fractions resolved into membrane and content subfractions. J. Cell Biol. 92:822-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isherwood, B. J., and A. H. Patel. 2005. Analysis of the processing and transmembrane topology of the E2p7 protein of hepatitis C virus. J. Gen. Virol. 86:667-676. [DOI] [PubMed] [Google Scholar]
- 26.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 27.Margis, R., M. Viry, M. Pinck, N. Bardonnet, and L. Pinck. 1994. Differential proteolytic activities of precursor and mature forms of the 24K proteinase of grapevine fanleaf nepovirus. Virology 200:79-86. [DOI] [PubMed] [Google Scholar]
- 28.Ota, K., M. Sakaguchi, G. von Heijne, N. Hamasaki, and K. Mihara. 1998. Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol. Cell 2:495-503. [DOI] [PubMed] [Google Scholar]
- 29.Ott, C. M., and V. R. Lingappa. 2002. Integral membrane protein biosynthesis: why topology is hard to predict. J. Cell Sci. 115:2003-2009. [DOI] [PubMed] [Google Scholar]
- 30.Persson, B., and P. Argos. 1994. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J. Mol. Biol. 237:182-192. [DOI] [PubMed] [Google Scholar]
- 31.Pouwels, J., J. E. Carette, J. Van Lent, and J. Wellink. 2002. Cowpea mosaic virus: effects on host cell processes. Mol. Plant Pathol. 3:411-418. [DOI] [PubMed] [Google Scholar]
- 32.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritzenthaler, C., C. Laporte, F. Gaire, P. Dunoyer, C. Schmitt, S. Duval, A. Piequet, A. M. Loudes, O. Rohfritsch, C. Stussi-Garaud, and P. Pfeiffer. 2002. Grapevine fanleaf virus replication occurs on endoplasmic reticulum-derived membranes. J. Virol. 76:8808-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rost, B., R. Casadio, and P. Fariselli. 1996. Refining neural network predictions for helical transmembrane proteins by dynamic programming. Proc. Int. Conf. Intell. Syst. Mol. Biol. 4:192-200. [PubMed] [Google Scholar]
- 35.Rott, M. E., A. Gilchrist, L. Lee, and D. Rochon. 1995. Nucleotide sequence of tomato ringspot virus RNA1. J. Gen. Virol. 76:465-473. [DOI] [PubMed] [Google Scholar]
- 36.Rott, M. E., J. H. Tremaine, and D. M. Rochon. 1991. Nucleotide sequence of tomato ringspot virus RNA-2. J. Gen. Virol. 72:1505-1514. [DOI] [PubMed] [Google Scholar]
- 37.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285:139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanfacon, H. 2005. Replication of positive-strand RNA viruses in plants: contact points between plant and virus components. Can. J. Bot. 83:1529-1549. [Google Scholar]
- 39.Sankaram, M. B., and D. Marsh. 1993. Protein-lipid interactions with peripheral membrane proteins, p. 127-162. In A. Watts (ed.), Protein-lipid interactions. Elsevier Science Publishers B. V., Amsterdam, The Netherlands.
- 40.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 42.Shai, Y. 1999. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1462:55-70. [DOI] [PubMed] [Google Scholar]
- 43.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 44.Stace-Smith, R. 1984. Tomato ringspot virus. CMI/AAB descriptions of plant viruses no. 290, Association of Applied Biologists, Wellesbourne, United Kingdom.
- 45.Staehelin, L. 1997. The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 11:1151-1165. [DOI] [PubMed] [Google Scholar]
- 46.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 48.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: applications to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 49.Wang, A., K. Carrier, J. Chisholm, A. Wieczorek, C. Huguenot, and H. Sanfacon. 1999. Proteolytic processing of tomato ringspot nepovirus 3C-like protease precursors: definition of the domains for the VPg, protease and putative RNA-dependent RNA polymerase. J. Gen. Virol. 80:799-809. [DOI] [PubMed] [Google Scholar]
- 50.Wang, A., S. Han, and H. Sanfacon. 2004. Topogenesis in membranes of the NTB-VPg protein of Tomato ringspot nepovirus: definition of the C-terminal transmembrane domain. J. Gen. Virol. 85:535-545. [DOI] [PubMed] [Google Scholar]
- 51.Wang, A., and H. Sanfacon. 2000. Proteolytic processing at a novel cleavage site in the N-terminal region of the tomato ringspot nepovirus RNA-1-encoded polyprotein in vitro. J. Gen. Virol. 81:2771-2781. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, S. C., G. Zhang, L. Yang, J. Chisholm, and H. Sanfacon. 2005. Evidence that insertion of Tomato ringspot nepovirus NTB-VPg protein in endoplasmic reticulum membranes is directed by two domains: a C-terminal transmembrane helix and an N-terminal amphipathic helix. J. Virol. 79:11752-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.