Abstract
West Nile virus, a member of the Flavivirus genus, causes fever that can progress to life-threatening encephalitis. The major envelope glycoprotein, E, of these viruses mediates viral attachment and entry by membrane fusion. We have determined the crystal structure of a soluble fragment of West Nile virus E. The structure adopts the same overall fold as that of the E proteins from dengue and tick-borne encephalitis viruses. The conformation of domain II is different from that in other prefusion E structures, however, and resembles the conformation of domain II in postfusion E structures. The epitopes of neutralizing West Nile virus-specific antibodies map to a region of domain III that is exposed on the viral surface and has been implicated in receptor binding. In contrast, we show that certain recombinant therapeutic antibodies, which cross-neutralize West Nile and dengue viruses, bind a peptide from domain I that is exposed only during the membrane fusion transition. By revealing the details of the molecular landscape of the West Nile virus surface, our structure will assist the design of antiviral vaccines and therapeutics.
West Nile virus (WNV), a member of the Flavivirus genus, causes a febrile illness that can lead to fatal meningitis or encephalitis in humans, horses, and birds (14). The ability of West Nile virus to infect birds, and to be carried between hosts in mosquitoes, has allowed it to spread at an alarming pace throughout the United States, where it was first detected in 1999, to Canada, the Caribbean, Central America, and Colombia (6, 16). West Nile virus is already indigenous to Africa, Europe, and Asia (6). There are no specific treatments or vaccines approved for clinical use against West Nile virus. Several other closely related flaviviruses are important human pathogens, including dengue, yellow fever, and Japanese encephalitis (JE) viruses.
Flaviviruses package their positive-strand RNA genome into particles consisting of a rigid outer protein shell and an underlying lipid membrane. The major envelope glycoprotein, E, and a small membrane protein, M, form the outer shell. C-terminal α-helical hairpins anchor E and M in the lipid membrane. As the principal envelope component, E is responsible for receptor binding. A candidate host cell receptor for West Nile virus is αVβ3 integrin (8, 19), but E may achieve initial attachment by binding glycosaminoglycans (18) or, like dengue virus E, by binding a carbohydrate recognition protein through a glycan on the viral surface (29, 37). Indeed, the C-type lectin DC-SIGNR has recently been reported to mediate cellular attachment of West Nile virus by specifically binding the glycan on West Nile virus E (9).
Receptor binding directs the virion to the endocytic pathway. Once flaviviruses reach an endosome, they must fuse their lipid membrane with the host cell membrane in order to deliver the viral genome into the cytoplasm for replication. The reduced pH of the endosome triggers a conformational rearrangement in E, which delivers the energy required for membrane fusion by bending the two apposed membranes towards each other, inducing them to fuse (24). Flavivirus E proteins belong to the structurally conserved “class II” fusion proteins, which are also found in alphaviruses. Crystal structures of three class II fusion proteins—dengue virus E (25-27), tick-borne encephalitis (TBE) virus E (4, 35), and Semliki Forest virus E1 (11, 20)—before and after their fusogenic conformational rearrangements provide us with a detailed molecular picture of the fusion mechanism of these viruses. First E (or E1) inserts a hydrophobic anchor, the so-called fusion loop, into the outer bilayer leaflet of the host cell membrane. Second, E folds back on itself, directing its C-terminal transmembrane anchor towards the fusion loop. This fold-back forces the host cell membrane (held by the fusion loop) and the viral membrane (held by the C-terminal transmembrane anchor) against each other, resulting in fusion of the two membranes. We now report the structure of a soluble fragment (residues 1 to 406) of the E protein (sE) from West Nile virus in its prefusion conformation. The sE fragment contains all but ∼50 residues of the E ectodomain.
MATERIALS AND METHODS
Expression and purification of WNV sE.
Nucleotides 925 to 2142 containing the E gene of the polyprotein of WNV strain 2741 (GenBank accession no. AF206518) (2) were inserted into the pPSC12 baculovirus cloning vector (Protein Sciences Corporation, Meriden, CT). ExpresSF+ cells from Spodoptera frugiperda (Protein Sciences Corporation) were infected with the recombinant baculovirus, causing them to secrete soluble E protein (sE) into the medium. The medium was concentrated on a Biomax 30 Pellicon XL concentrator (Millipore), adjusted to pH 7.1 with 1 N NaOH, and loaded on a preequilibrated immunoaffinity column. The immunoaffinity column was prepared by coupling the immunoglobulin G (IgG) fraction from West Nile virus sE-immunized horse serum to N-hydroxysuccinimide-activated HiTrap columns (Amersham Biosciences). The column was thoroughly washed with phosphate-buffered saline (PBS), and bound sE was eluted with 100 mM glycine-HCl, pH 2.7. Similar elution conditions were used to purify dengue virus sE in its prefusion conformation by immunoaffinity chromatography (25). Eluted fractions were neutralized with 100 mM Tris-HCl, pH 8.0, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Fractions containing sE were concentrated using an Ultra-4 ultrafiltration device (Millipore) and further purified by size-exclusion chromatography in 10 mM HEPES (pH 7.5)-0.1 M NaCl-17 mM n-octyl-β-d-glucoside. Purified sE was concentrated to 10 g liter−1 in Amicon Ultra centrifugal filters (Millipore).
Crystallization and data collection.
Crystals were grown at 20°C by hanging drop vapor diffusion by mixing equal volumes of protein solution and the following reservoir solution: 15% (vol/vol) isopropanol, 0.1 M HEPES, pH 7.5, and 0.2 M sodium citrate. Crystals grew as rhomboids in space group P41212 with cell dimensions a = b = 93.26 Å and c = 159.32 Å. The asymmetric unit contains one molecule of sE. Crystals were transferred to a cryoprotective solution of 15% (vol/vol) isopropanol, 0.1 M HEPES, pH 7.5, 0.2 M sodium citrate, 17 mM n-octyl-β-d-glucoside, and 15% glycerol before being frozen in liquid nitrogen. Data were collected from a single crystal at 100 K on beamline X26C of the National Synchrotron Light Source, Brookhaven National Laboratory, Upton, NY. The data were processed with HKL2000 (33). Data collection statistics are presented in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parameter | WNV E native data set |
|---|---|
| Data collection | |
| Space group | P41212 |
| Cell dimensions | |
| a, b, c (Å) | 93.26, 93.26, 159.32 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å)a | 25.0-3.11 (3.23-3.11) |
| Rsym or Rmergea | 0.111 (0.721) |
| I/sIa | 14.5 (2.48) |
| Completeness (%)a | 94.8 (96.4) |
| Redundancya | 8.1 (8.1) |
| Refinement | |
| Resolution (Å) | 24.1-3.11 |
| No. of reflections | 11,858 |
| Rwork/Rfree | 0.206/0.268 |
| No. of atoms | |
| Protein | 3,043 |
| Ligand/ion | 38 |
| Water | 14 |
| B factors | |
| Protein (Å2) | 117 |
| Glycan (Å2) | 185 |
| Water (Å2) | 149 |
| RMSb deviations | |
| Bond length (Å) | 0.014 |
| Bond angle (°) | 1.676 |
Highest-resolution shell is shown in parentheses.
RMS, root mean square.
Structure determination.
The crystal structure of WNV E was determined by molecular replacement. Each domain was placed sequentially with AMoRe (30). Domain III from WNV (Protein Data Bank code 1ZTX) (31) was placed first, domain I from dengue virus type 3 (1UZG) (27) was placed second, and domain II from dengue virus type 2 (1OAN) (25) was placed last. The atomic coordinates of the three domains were then refined as rigid bodies with the Crystallography & NMR System (CNS) (5). Amino acids in the model were mutated to the West Nile virus sequence, and the model was rebuilt with O (17) based on 2Fo − Fc and Fo − Fc Fourier maps and density-modified maps. A sharpening B factor of −75 was applied to the structure factors to obtain the most informative maps. Coordinates were refined against data from 25- to 3.1-Å resolution by simulated annealing with torsion angle dynamics with CNS and then rebuilt with O in iterative cycles. Later cycles included restrained refinement of B factors for individual atoms and energy minimization against a maximum-likelihood target with CNS. In the final refinement cycles, carried out with REFMAC5 (39), the rigid-body motion of the protein molecules in the crystal was taken into account in terms of three tensors: one for translation (T), one for libration (L), and one for correlations of libration and translation (S) (39). This TLS refinement was alternated with cycles of restrained positional and B-factor refinement against a maximum likelihood target (see header of Protein Data Bank [PDB] entry 2I69 for refined TLS parameters). The final model contains residues 1 to 403, 14 water molecules (visible only with sharpening of the overall B factor), and one simple N-linked glycan on residue 154. The glycan contains three ordered sugar rings: two N-acetylglucosamines with a branched fucose. Based on the lack of side chain electron density and on the local backbone conformation, residue 28 was modeled as Gly instead of Asp. Refinement statistics are presented in Table 1. Ramachandran angles are good, with favored, additional, generous, and disallowed values of 81.2%, 17.7%, 1.2%, and 0%, respectively.
Measurement of scFv-Fc binding to sE peptides.
A set of overlapping 20-mer peptides spanning the entire sequence of West Nile virus sE was synthesized by Sigma-Genosys. The mutant, randomized-sequence, and dengue virus peptides (see Fig. 5) were synthesized at the Keck Biotechnology Resource Center at Yale University. Binding of scFv-Fcs 11, 71, 73, 79, and 95 (13) to West Nile and dengue virus E peptides was determined by enzyme-linked immunosorbent assay (ELISA). Ninety-six-well microtiter plates were coated with 1 μg peptide per well in 100 μl carbonate (pH 9.6) coating buffer, at 4°C overnight. Plates were blocked with either 1% (wt/vol) bovine serum albumin or 0.5% (wt/vol) nonfat dry milk in PBS containing 0.05% (vol/vol) Tween 20 (PBS-T) for 30 min at room temperature (r.t.) after they were washed three times with PBS-T. Antibodies were diluted in blocking solution and added to the antigen-coated plates previously blocked with the blocking solution and allowed to bind for 1 h at r.t. After being washed three times with PBS-T, the secondary antibody, goat anti-human IgG (1:500; Sigma), conjugated to alkaline phosphatase, was added to the plates and incubated for 45 min at r.t. Following washing and the addition of p-nitrophenylphosphate substrate (Sigma), color development was measured at 405 nm in a Labsystems Multiskan MS plate reader.
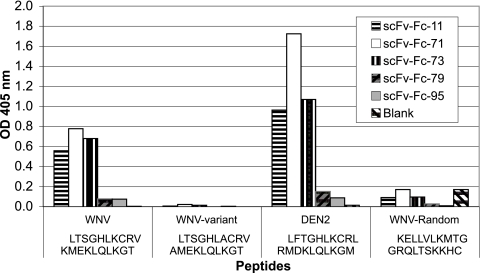
FIG. 5.
Partial epitope mapping of therapeutic recombinant antibodies previously selected by phage display (13). To map linear peptide sequences of scFv-Fcs 11, 71, 73, 79, and 95, we measured scFv-Fc binding to a set of overlapping 20-mer peptides spanning the entire sequence of West Nile virus sE by ELISA. scFv-Fcs 11, 71, and 73 bind a peptide consisting of West Nile virus residues 281 to 300. All three scFv-Fcs also recognize the homologous E peptide from dengue virus type 2 (DEN2) but not a peptide composed of the same amino acids in a randomized sequence. Furthermore, scFv-Fc binding was critically dependent on positively charged side chains (Lys or Arg) at positions 287 and 291 of West Nile virus E.
Protein structure accession numbers.
The atomic coordinates of the West Nile virus sE crystal structure can be downloaded from the Protein Data Bank (rcsb.org) under accession code 2I69. Atomic coordinates for generating the entire outer glycoprotein layer of West Nile virus have been deposited under accession code 2I7J. Structure factors were deposited under accession code r2I69sf.
RESULTS
Molecular architecture of West Nile virus sE.
As expected from the 49% sequence identity shared with dengue virus type 3 sE (the most similar E protein whose structure has been determined), West Nile virus sE adopts the same three-domain fold as its homologs from dengue and TBE viruses (Fig. 1). Domain I organizes the structure, and its nine-stranded β-barrel core is superimposed well onto those of other flavivirus E structures (root mean square distance = 1.1 to 1.5 Å). Two long insertions in domain I form the elongated domain II, which is built mostly of β strands and bears the fusion loop at its tip. Domain III adopts a 10-stranded IgC-like fold (31) and is thought to contain the receptor binding site (19). While the overall flavivirus architecture is conserved in West Nile virus sE, there are significant differences between the West Nile virus sE structure and other flavivirus E structures. The most striking difference is that, unlike dengue and TBE virus sE, West Nile virus sE does not form dimers in the crystal. The elution volume of West Nile virus sE in size-exclusion chromatography suggests that it also does not form dimers in solution, at pH 5.0 to 8.5 (data not shown). Our structure provides a likely explanation for the failure of West Nile virus sE to dimerize. Domain II, which participates in all of the dimer contacts in the dimeric dengue and TBE virus sE structures, adopts a significantly different orientation relative to domains I and III in the monomeric West Nile virus sE structure. Surprisingly, the relative orientation of West Nile virus sE domains I and II is nearly identical to that found in the postfusion conformation of dengue virus sE (26). Indeed, when the postfusion dengue virus sE structure is superimposed onto the West Nile virus sE structure by using the core strands of domain I as the reference (root mean square distance = 1.9 Å), the orientations of domain II in each structure are then separated by a rotation of only 3°, which translates into a maximal difference in backbone atom positions of 5 Å at the fusion loop at the tip of domain II (Fig. 2A). For comparison, the mean backbone atom difference for domain III is 34.9 Å. In contrast, when the prefusion dengue virus sE and TBE virus sE structures are superimposed onto West Nile virus sE structure using domain I as the reference, the orientations of domain II in the sE structures are separated by a rotation of between 20° (to TBE virus sE, PDB entry 1SVB; Fig. 2B) and 13° (to dengue virus sE, PDB entry 1OKE; data not shown). The rotation is centered about a hinge point near Ile196 of West Nile virus E, at the interface between domains I and II (Fig. 2B). The altered domain orientation translates into a difference in backbone atom positions of 16 to 23 Å at the fusion loop. Domain III, however, adopts a nearly identical relative orientation relative to domain I in West Nile virus sE as in the prefusion virus TBE and dengue virus sE structures (Fig. 2B and data not shown). In summary, the rotation that relates domain II in West Nile virus sE to domain II in other prefusion flavivirus sE structures closely resembles the rotation undergone by domain II of dengue (26) and TBE (4) virus E, in what is believed to be one of the first steps of the fusogenic conformational rearrangement. Because the West Nile virus sE structure does not contain more hydrogen bonds or buried surface area at the domain I-domain II interface than the dengue or TBE virus prefusion sE structures, the rotation of domain II in West Nile virus sE from a prefusion-like to a postfusion-like conformation is not expected to contribute to the overall irreversibility of the fusion transition (which requires a lipid membrane and is driven by trimer formation [26]).
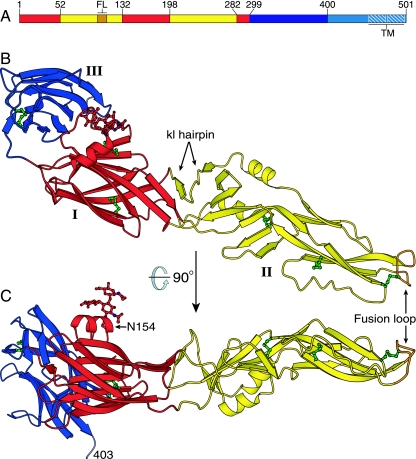
FIG. 1.
Structure of the West Nile virus sE monomer. (A) The three domains of West Nile virus E: domain I is red, domain II is yellow, and domain III is blue. The fusion loop (FL) is in orange. A 53-residue “stem” (cyan) links the ectodomain to a two-helix C-terminal transmembrane anchor (TM, white hatching). (B and C) The West Nile virus sE monomer, colored as in panel A, viewed in two perpendicular orientations. The last ordered residue in the sE structure (Ser403) and the kl hairpin, which forms the putative hydrophobic ligand-binding pocket, are labeled. The glycan at Asn154 and the six disulfide bonds are shown in ball-and-stick representation (in red and green, respectively).
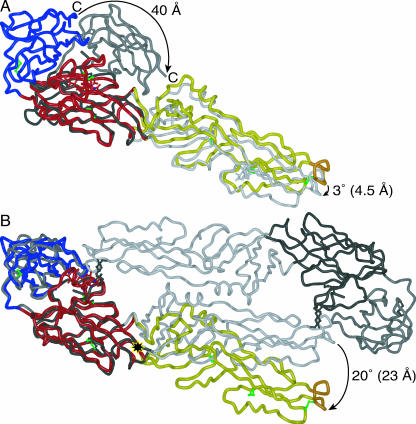
FIG. 2.
Comparison of West Nile virus sE to other flavivirus sE structures. (A) Structure of dengue virus type 2 sE in the postfusion conformation (PDB entry 1OK8, in shades of gray) superimposed on West Nile virus sE (colored as in Fig. 1), using domain I as the reference. The C termini (in domain III) of the structures are 40 Å apart, while the fusion loops (in domain II) are less than 5 Å apart. The view is rotated 25° relative to panel B and Fig. 1B to show domain III of dengue virus sE more clearly. (B) Structure of the TBE virus sE dimer in the prefusion conformation (PDB entry 1SVB, in shades of gray) superimposed on West Nile virus sE, using domain I as the reference. Domains III of the structures are superimposed well, but the orientations of domain II are separated by a 20° rotation about a point near residue 196 (marked with a black and yellow star). This rotation translates into a 23-Å displacement of the fusion loop.
The similarities between the structures of domain II in West Nile virus sE and in postfusion dengue virus sE extend beyond their relative orientations to domain I. Indeed, the last two strands of domain II, which form the so-called kl hairpin, are separated at the base of the hairpin in West Nile virus sE, just as they are in the postfusion sE structures. Also, the β-sheet hydrogen-bonding pattern formed by the l strand has the same register as in the postfusion dengue virus sE structure (26) but is shifted by two residues relative to the prefusion sE structures.
The postfusion-like conformation of domain II in West Nile virus sE results in a more elongated overall conformation of sE, which is incompatible with the mode of dimerization observed in dengue virus sE and TBE virus sE, as is evident in Fig. 2B. Thus, we propose that the preference for a postfusion-like orientation of domain II in West Nile virus sE is the principal reason why this sE protein does not form dimers in solution or in the crystal. Two additional factors could potentially prevent dimerization of West Nile virus sE: (i) the conformation of the glycan on Asn154 is incompatible with the formation of dimer contacts (unlike the homologous glycan in TBE virus, which forms dimer contacts), and (ii) one of the few residues that form a strong hydrogen bond at the dimer interface in the unliganded dengue virus type 2 sE structure (25), Gln316, is not conserved in West Nile virus, which instead has Gly319. The postfusion-like conformation of domain II in West Nile virus sE is also incompatible with structures of West Nile and dengue virus particles derived from electron cryomicroscopy (cryoEM) reconstructions (see Discussion).
The hydrophobic pocket.
Two structures of dengue virus type 2 sE revealed the presence of a pocket capable of binding hydrophobic ligands, such as the detergent n-octyl-β-d-glucoside (25). The pocket forms under a β-hairpin, the so-called kl hairpin, at the interface between domains I and II. Since significant rearrangements occur around the kl hairpin during the fusion transition, ligands that bind in the hydrophobic pocket could prove effective as inhibitors of membrane fusion and hence of viral entry (25, 26). To determine whether West Nile virus E would be capable of forming a similar hydrophobic pocket, we compared the 12 hydrophobic residues that line the pocket in dengue virus sE along with their West Nile virus homologs (Fig. 3). All of the hydrophobic residues either are identical in the two viruses or have hydrophobic side chains of similar lengths. We conclude that West Nile virus sE should be capable of forming a hydrophobic pocket similar to that of dengue virus sE. However, potential ligands can be expected to bind E only in mature West Nile virions. Indeed, once the initial step of the fusion transition has been initiated (and possibly also in immature virions), domain II will adopt its preferred postfusion-like conformation (Fig. 1 and 2), which is not capable of forming a hydrophobic pocket because the base of the kl hairpin is splayed open.
FIG. 3.
Structure-based alignment of the amino acid sequences of E proteins from West Nile virus (wn) strain 2741 (2), Japanese encephalitis virus (je) strain JaOArS982, dengue virus type 2 (d2) strain S1, and tick-borne encephalitis virus (tbe) strain Neudörfl. Dots indicate amino acid identities; dashes show gaps. The domains are indicated by a colored bar as in Fig. 1. The sequences are truncated at the last residue (406) of the soluble fragment (sE) of West Nile virus E, which we crystallized. The conserved glycosylation site in domain I is indicated by a red asterisk and red lettering. Residues lining the hydrophobic pocket in sE are shaded in gray. Residues that are exposed on the viral surface and are conserved in West Nile virus strains but not in other flaviviruses are shaded in magenta. Residues that are exposed on the viral surface and are conserved in wn, je, d2, and tbe viruses are shaded in orange.
Unique surface features of West Nile virus.
The individual domains of West Nile virus sE have a high degree of structural similarity to other flavivirus sE structures (25, 27, 31, 35, 42). Several loops in West Nile virus sE diverge in conformation. These include the loops in domain III that are exposed on the viral surface, in particular the DE (365 to 368) and FG (389 to 391) loops, and the glycosylated E0F0 loop (145 to 162) in domain I. Surface-exposed residues determine receptor specificity, vector preference, host range, and tropism of West Nile virus. A surprisingly small number of residues are responsible for all of these characteristics. Indeed, only 57 residues are conserved in West Nile virus strains but not conserved across flaviviruses (including JE virus). Of these residues, only 38 are exposed on the viral surface (Fig. 4).
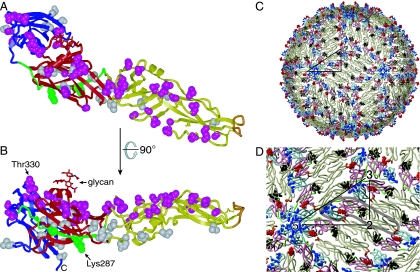
FIG. 4.
Distribution of West Nile virus-specific residues on sE. (A and B) Two perpendicular views of West Nile virus sE, with residues that are conserved in West Nile virus strains but not in other flaviviruses shown in space-filling representation. Residues that are exposed on the surface of the mature virus are in magenta; residues that are not exposed are in gray. The I0-strand peptide recognized by recombinant antibodies scFv-Fc 11, 71, and 73 (residues 281 to 300) is shown in green, with the two essential basic residues (Lys287 and Lys291) in space-filling representation. Most West Nile virus-specific neutralizing antibodies bind an epitope that includes Thr330 (21, 31, 36). The view in panel B is perpendicular to the viral surface, such that the outside of the virion is up. The views are the same as in Fig. 1B and C. (C) Atomic model of the West Nile virus outer protein shell based on the 9.5-Å-resolution electron cryomicroscopic reconstruction of dengue virus (41). E assembles into dimers in mature virions. The glycan of West Nile virus E is shown in red, residues lining the putative hydrophobic pocket in dark gray, residues 281 to 300 (a partial epitope of scFv-Fcs 11, 71, and 73) in green, and the epitope of therapeutic antibody E16 (32) in blue. The fusion loop is in orange. A black triangle connects the icosahedral symmetry axes. (D) Close-up of panel C, with the two-, three-, and fivefold icosahedral symmetry axes labeled. A single sE monomer is circled with a semitransparent gray line. The minimum separation between glycans (red) is ∼50 Å.
The loop bearing the only glycan of the outer protein shell of the mature West Nile virion is the most distinct from other sE structures. The N-linked glycosylation site is conserved in all flaviviruses except yellow fever virus and is located at Asn154, in a loop between the fifth and sixth strands of domain I, the so-called E0 and F0 strands. The E0F0 loop is several residues longer in West Nile virus E than in most other flaviviruses. Loss of the glycosylation site at position 154 in certain West Nile virus strains leads to strong attenuation, loss of neuroinvasiveness, and lower stability at mildly acidic pH (3). A similar phenotype is observed when the homologous glycan is lost in dengue virus (15). The glycosylated E0F0 loops in the dengue and TBE virus sE structures lack secondary structure and share no significant sequence identity with West Nile virus E. In contrast, in the West Nile virus sE structure, the E0F0 loop forms a compact fold around an α-helix (residues 154 to 162). The loop packs onto strand F0 and onto the N terminus, which adopts a slightly different conformation than in the dengue and TBE virus sE structures. The position of the glycan also shifts by 5 to 7 Å (radially outward relative to the dengue virus glycan and along the surface and away from the dimer interface relative to the TBE virus glycan). Despite this shift, the orientation of these glycans in dengue and West Nile virus E is such that they largely overlap and therefore localize to the same positions on the surface (Fig. 4C and D) (41). The slight differences between the dengue and West Nile virus cryoEM structures in the E0F0 loop region (28) are therefore most likely due to the additional bulk of the loop and its α-helix, rather than to the shift in the position of the glycan. We have modeled the glycan as a branched trisaccharide consisting of two N-acetylglucosamines with a fucose on the first N-acetylglucosamine [Asn→GlcNAc(Fucα16)β1-4GlcNAc]; however, the glycan contains additional, poorly ordered sugars. We note that the E0F0 loop of JE virus is likely to adopt a structure similar to that of West Nile virus E, since the E0F0 loop sequence is 71% identical in these two viruses. While the E0F0 loop may be largely protected from immune recognition by its glycan, its structure extends far enough from the glycan in West Nile virus that the E0F0 loop may constitute an attractive target for West Nile virus-specific neutralizing antibodies.
Recombinant antibody partial epitope mapping.
We recently used a phage display selection method to generate a panel of human single-chain variable region antibody fragments fused to an IgG1 Fc domain (scFv-Fcs). A similar approach has been used to generate cross-neutralizing recombinant antibodies against dengue virus (12, 22). Our recombinant antibodies neutralize both West Nile and dengue viruses by binding conserved epitopes in domains I and II (13). To map the epitopes of the scFv-Fcs more precisely, we measured the ability of five neutralizing scFv-Fcs to bind overlapping 20-mer peptides spanning the entire sequence of West Nile virus sE, by ELISA. Although this method does not allow conformational epitopes to be identified, we found that three of the scFv-Fcs with the best therapeutic properties (13), scFv-Fcs 11, 71, and 73, recognized a peptide spanning residues 281 to 300. The scFv-Fcs also recognize the homologous E peptide from dengue virus type 2 but not a peptide composed of the same amino acids in a randomized sequence (Fig. 5). Furthermore, scFv-Fc binding was critically dependent on positively charged side chains (Lys or Arg) at positions 287 and 291 of West Nile virus E.
DISCUSSION
Implications for viral assembly.
Structures of the mature West Nile (28) and dengue (41) virions have been determined by cryoEM image reconstruction, at 17-Å and 9.5-Å resolution, respectively. The two structures show that the structural assembly of E in the outer protein shell is very similar in the two viruses. The dengue virus cryoEM structure is of sufficient resolution to allow a highly accurate fitting of the atomic coordinates of the dengue virus sE crystal structure into the cryoEM structure (41). By superimposing our West Nile virus sE structure onto the cryoEM-fitted dengue virus sE atomic coordinates and applying icosahedral symmetry, we generated a pseudoatomic model of the entire outer protein shell of the West Nile virion (Fig. 4C and D). Interestingly, it is impossible to achieve a good fit for the 180 subunits of E in the cryoEM structures while avoiding steric clashes, with domain II in the same orientation relative to domain I as in our crystal structure. This suggests that the interface between domains I and II must be flexible in prefusion sE. This is consistent with observed variations of up to 10° in the relative orientations of domains I and II in dengue virus sE, despite the additional constraint of dimer formation in these dengue virus E proteins. However, in contrast to West Nile virus sE, these rotations in domain II of dengue virus sE are about an axis that is orthogonal to the axis of rotation of the fusion transition.
Implications for membrane fusion.
A possible explanation for the inability of West Nile virus sE to form dimers in solution is that domain II prefers the postfusion-like orientation observed in the crystal structure over the prefusion-like orientation observed in the mature virion. The physical strain imposed on sE by the icosahedral assembly might serve to “spring-load” sE, allowing some of the energy required for membrane fusion to be stored in the metastable mature virus particle. However, we cannot rule out that the acidic conditions used to elute sE from our immunoaffinity column (see Materials and Methods) have irreversibly altered the native prefusion structure of sE, causing it to adopt a more postfusion-like monomeric structure.
Since the pH threshold of membrane fusion is approximately 6.5 in all flaviviruses including West Nile virus (1, 3, 26), we conclude that spring-loading of sE, if it exists, is not coupled to the low-pH trigger that initiates the fusion transition. It is not known precisely how E senses low pH. Two conserved histidines at the interface between domains I and III have been proposed to form part of the pH sensor of sE in all flaviviruses (4). Protonation of these histidines at pH < 6.5 is likely to be an important factor in the destabilization of the domain I-domain III interface. This could be the initial low-pH trigger that allows domains I and II to rotate out of the viral surface and expose the fusion loop. In West Nile virus sE, the two histidines (His144 and His320) form similar interactions with domain I as in TBE virus (35), supporting the notion that the low-pH trigger mechanism for the fusion transition is conserved across flaviviruses. However, domain II, which packs on top of the single nontransmembrane α-helix of the minor envelope protein, M (41), may also contain a pH-sensing element.
Implications for immune recognition.
Our atomic model of the outer glycoprotein layer of the West Nile virus particle reveals the location on the viral surface of potential receptor binding sites and of the epitopes of several previously described neutralizing antibodies. As expected, the epitope of therapeutic antibody E16 maps to a patch on domain III that is fully exposed on the viral surface (and also on the postfusion form of E) (31). Remarkably, the epitopes of all other strongly neutralizing West Nile virus-specific antibodies localize to the same patch on domain III (32, 36, 38). Indeed, most neutralizing antibodies against dengue and JE viruses also map to this region and are also serotype or strain specific. Within this epitope, the N-terminal loop of domain III (residues 302 to 309) and the BC loop (330 to 333) have a dominant role in flavivirus neutralization (21, 31, 32, 36, 38, 40). From the data presented above, we conclude that binding of West Nile virus-specific antibodies is likely to involve a subset of the 38 West Nile virus-specific residues that are exposed on the viral surface (Fig. 4). Of these residues, eight are in domain III (all in the dominant epitope described above), and five have in fact already been shown to bind neutralizing antibodies directly (residues 306, 308, 330, 366, and 391) (31, 36). The remaining 27 West Nile virus-specific residues are distributed fairly evenly throughout domains I and II, although there are none in the area around the fusion loop, which is highly conserved across flaviviruses (Fig. 3).
While neutralizing antibodies raised against West Nile virus particles invariably bind variable epitopes in domain III, our recombinant scFv-Fc neutralizing antibodies bind conserved epitopes in domains I and II (13). Thus, unlike conventional antibodies, some of our scFv-Fcs cross-neutralize dengue virus type 2 (36). Since the scFv-Fcs were selected for their ability to bind monomeric sE, their epitopes are not necessarily exposed on the viral surface. Three of the scFv-Fcs with the best therapeutic properties, scFv-Fcs 11, 71, and 73 (13), recognize a peptide spanning residues 281 to 300 and the homologous peptide from dengue virus type 2 (Fig. 5). This peptide sequence localizes mostly to a β-strand in domain I, strand I0. While the full epitope is likely to extend into neighboring strands, the entire β-sheet containing strand I0 is buried in the mature virion (Fig. 4). Most of the β-sheet, including strand I0, is also buried in the trimeric postfusion conformation of sE (26), so the strand will be exposed only briefly, during the fusogenic conformational rearrangement. Based on the location and conserved structure of this partial epitope, we conclude that a likely mechanism of neutralization for the cross-neutralizing scFv-Fcs is inhibition of the membrane fusion transition.
Implications for receptor binding.
There is preliminary evidence that αVβ3 integrin may serve as the host cell receptor for West Nile virus, by binding a region in domain III (8, 19). αVβ3 integrin binds ligands with RGD/E sequences. West Nile virus sE contains an RGE motif in domain III (residues 388 to 390), which is exposed on the viral surface and also forms part of the dominant neutralizing antibody epitope. The RGE motif does not, however, appear to be essential for αVβ3 integrin binding (8). Given the limited area of domain III that is exposed on the viral surface, we expect that high-affinity αVβ3 integrin binding would preclude binding by all or most neutralizing West Nile virus-specific antibodies, and vice versa. Thus, inhibition of receptor binding is the most likely mechanism of neutralization for these antibodies. Since antibody E16 is protective even when administered after cellular attachment has occurred (31), the virus may achieve initial attachment by binding glycosaminoglycans (18) or, like dengue virus E, by binding a carbohydrate recognition protein through a glycan on the viral surface (37). The latter forms of initial cellular attachment may not be sufficient for infection, and they would probably not interfere with the binding neutralizing antibodies such as E16, because both the glycan and a conserved residue cluster proposed to bind glycosaminoglycans in dengue virus are located in domain I (7).
The tetrameric C-type lectin DC-SIGNR has recently been reported to mediate cellular attachment of West Nile virus by specifically binding the high-mannose N-linked glycan on West Nile virus E (9). A related lectin, DC-SIGN, performs a similar function for dengue virus (29, 37). Although DC-SIGN is generally able to bind a broader range of glycans than DC-SIGNR, the former does not facilitate attachment of West Nile virus (9). This may be because DC-SIGN appears to require two adjacent glycans approximately 18 Å apart for optimal binding, as illustrated by the binding pattern of DC-SIGN on dengue virus particles (34). West Nile virus lacks the second glycosylation site that mediates DC-SIGN binding in dengue virus. Based on our structure, the spacing between glycans on the surface of West Nile virus is 50 Å around the fivefold icosahedral symmetry axis and 49 Å around the threefold axis (Fig. 4C and D). This is close to the 54-Å separation between carbohydrate recognition domains in the crystal structure of the DC-SIGNR tetramer (10), suggesting that the lectin could bind multiple (two to four) viral glycans simultaneously. While the reason for the apparent preference of West Nile virus for DC-SIGNR is still unclear, the spacing between glycans, as well as their precise composition, could favor binding to DC-SIGNR over DC-SIGN. Indeed, the specificity of carbohydrate recognition of the two lectins depends largely on whether glycans can bind all four carbohydrate recognition domains simultaneously (10, 23).
Conclusions.
We find, unexpectedly, that the West Nile virus sE structure shares similarities in the relative orientation of its three domains with both pre- and postfusion structures of dengue and TBE virus sE. It is still unclear whether the West Nile virus sE structure represents a true mechanistic intermediate in the membrane fusion transition, but the incompatibility of the structure with image reconstructions of intact viruses suggests that E may not be in its preferred conformation in the environment of a mature virion. Does the resulting mechanical energy stored in the outer protein shell, as we propose, serve to drive early steps of membrane fusion in the endosome? The answer to this question will require careful measurements of kinetic and energetic parameters of flavivirus membrane fusion and assembly. As anticipated, the known epitopes of West Nile virus-specific neutralizing antibodies map to an area of domain III that is exposed on the viral surface. West Nile virus may achieve initial cellular attachment through its evenly spaced domain I glycans (9); however, domain III is believed to mediate binding to a “true receptor” (possibly αVβ3 integrin [8, 18]), which determines cell tropism and targets the virus to the endocytic pathway. Antibodies against domain III would prevent binding of this type of receptor. In contrast, we show here that our recombinant antibodies recognize an epitope in domain I, which is only briefly exposed during the fusion transition and is at least partly conserved in dengue virus. We therefore expect that our recombinant antibodies act by inhibiting the fusion transition. Our analysis of the domain organization of West Nile virus sE and of the molecular landscape of the viral surface offers new insight into the membrane fusion mechanism, into likely modes of receptor binding, and into mechanisms of antibody neutralization. The detailed understanding of specific mechanisms of the viral life cycle gained from our structure provides a framework for the rational design of antiviral vaccines and therapeutics.
Acknowledgments
We thank staff at the X26C and X25 beamlines of the National Synchrotron Light Source at the Brookhaven National Laboratory.
This work was supported by NIH grants U01-AI061361 and R41-AI060217 to R.A.K. and grant R41-AI068154-01 to Y.M. and by Yale University.
Footnotes
Published ahead of print on 30 August 2006.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. F., T. G. Andreadis, C. R. Vossbrinck, S. Tirrell, E. M. Wakem, R. A. French, A. E. Garmendia, and H. J. Van Kruiningen. 1999. Isolation of West Nile virus from mosquitoes, crows, and a Cooper's hawk in Connecticut. Science 286:2331-2333. [DOI] [PubMed] [Google Scholar]
- 3.Beasley, D. W., M. C. Whiteman, S. Zhang, C. Y. Huang, B. S. Schneider, D. R. Smith, G. D. Gromowski, S. Higgs, R. M. Kinney, and A. D. Barrett. 2005. Envelope protein glycosylation status influences mouse neuroinvasion phenotype of genetic lineage 1 West Nile virus strains. J. Virol. 79:8339-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bressanelli, S., K. Stiasny, S. L. Allison, E. A. Stura, S. Duquerroy, J. Lescar, F. X. Heinz, and F. A. Rey. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, G. L., A. A. Marfin, R. S. Lanciotti, and D. J. Gubler. 2002. West Nile virus. Lancet Infect. Dis. 2:519-529. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y., T. Maguire, R. E. Hileman, J. R. Fromm, J. D. Esko, R. J. Linhardt, and R. M. Marks. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 3:866-871. [DOI] [PubMed] [Google Scholar]
- 8.Chu, J. J., and M. L. Ng. 2004. Interaction of West Nile virus with alpha v beta 3 integrin mediates virus entry into cells. J. Biol. Chem. 279:54533-54541. [DOI] [PubMed] [Google Scholar]
- 9.Davis, C. W., H. Y. Nguyen, S. L. Hanna, M. D. Sanchez, R. W. Doms, and T. C. Pierson. 2006. West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J. Virol. 80:1290-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg, H., Y. Guo, D. A. Mitchell, K. Drickamer, and W. I. Weis. 2005. Extended neck regions stabilize tetramers of the receptors DC-SIGN and DC-SIGNR. J. Biol. Chem. 280:1327-1335. [DOI] [PubMed] [Google Scholar]
- 11.Gibbons, D. L., M. C. Vaney, A. Roussel, A. Vigouroux, B. Reilly, J. Lepault, M. Kielian, and F. A. Rey. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320-325. [DOI] [PubMed] [Google Scholar]
- 12.Goncalvez, A. P., R. H. Purcell, and C. J. Lai. 2004. Epitope determinants of a chimpanzee Fab antibody that efficiently cross-neutralizes dengue type 1 and type 2 viruses map to inside and in close proximity to fusion loop of the dengue type 2 virus envelope glycoprotein. J. Virol. 78:12919-12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gould, L. H., J. Sui, H. Foellmer, T. Oliphant, T. Wang, M. Ledizet, A. Murakami, K. Noonan, C. Lambeth, K. Kar, J. F. Anderson, A. M. de Silva, M. S. Diamond, R. A. Koski, W. A. Marasco, and E. Fikrig. 2005. Protective and therapeutic capacity of human single-chain Fv-Fc fusion proteins against West Nile virus. J. Virol. 79:14606-14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granwehr, B. P., K. M. Lillibridge, S. Higgs, P. W. Mason, J. F. Aronson, G. A. Campbell, and A. D. Barrett. 2004. West Nile virus: where are we now? Lancet Infect. Dis. 4:547-556. [DOI] [PubMed] [Google Scholar]
- 15.Guirakhoo, F., A. R. Hunt, J. G. Lewis, and J. T. Roehrig. 1993. Selection and partial characterization of dengue 2 virus mutants that induce fusion at elevated pH. Virology 194:219-223. [DOI] [PubMed] [Google Scholar]
- 16.Hayes, E. B., and D. J. Gubler. 2006. West Nile virus: epidemiology and clinical features of an emerging epidemic in the United States. Annu. Rev. Med. 57:181-194. [DOI] [PubMed] [Google Scholar]
- 17.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 18.Lee, E., R. A. Hall, and M. Lobigs. 2004. Common E protein determinants for attenuation of glycosaminoglycan-binding variants of Japanese encephalitis and West Nile viruses. J. Virol. 78:8271-8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J. W., J. J. Chu, and M. L. Ng. 2006. Quantifying the specific binding between West Nile virus envelope domain III protein and the cellular receptor alphaVbeta3 integrin. J. Biol. Chem. 281:1352-1360. [DOI] [PubMed] [Google Scholar]
- 20.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 21.Li, L., A. D. Barrett, and D. W. Beasley. 2005. Differential expression of domain III neutralizing epitopes on the envelope proteins of West Nile virus strains. Virology 335:99-105. [DOI] [PubMed] [Google Scholar]
- 22.Men, R., T. Yamashiro, A. P. Goncalvez, C. Wernly, D. J. Schofield, S. U. Emerson, R. H. Purcell, and C. J. Lai. 2004. Identification of chimpanzee Fab fragments by repertoire cloning and production of a full-length humanized immunoglobulin G1 antibody that is highly efficient for neutralization of dengue type 4 virus. J. Virol. 78:4665-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell, D. A., A. J. Fadden, and K. Drickamer. 2001. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 276:28939-28945. [DOI] [PubMed] [Google Scholar]
- 24.Modis, Y. 2006. Class II fusion proteins. In S. Pöhlmann and G. Simmons (ed.), Virus entry into cells. Landes Bioscience, Georgetown, Tex.
- 25.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA. 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 27.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79:1223-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 29.Navarro-Sanchez, E., R. Altmeyer, A. Amara, O. Schwartz, F. Fieschi, J. L. Virelizier, F. Arenzana-Seisdedos, and P. Despres. 2003. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 4:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navaza, J. 2001. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D 57:1367-1372. [DOI] [PubMed] [Google Scholar]
- 31.Nybakken, G. E., T. Oliphant, S. Johnson, S. Burke, M. S. Diamond, and D. H. Fremont. 2005. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437:764-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliphant, T., M. Engle, G. E. Nybakken, C. Doane, S. Johnson, L. Huang, S. Gorlatov, E. Mehlhop, A. Marri, K. M. Chung, G. D. Ebel, L. D. Kramer, D. H. Fremont, and M. S. Diamond. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 34.Pokidysheva, E., Y. Zhang, A. J. Battisti, C. M. Bator-Kelly, P. R. Chipman, C. Xiao, G. G. Gregorio, W. A. Hendrickson, R. J. Kuhn, and M. G. Rossmann. 2006. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell 124:485-493. [DOI] [PubMed] [Google Scholar]
- 35.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez, M. D., T. C. Pierson, D. McAllister, S. L. Hanna, B. A. Puffer, L. E. Valentine, M. M. Murtadha, J. A. Hoxie, and R. W. Doms. 2005. Characterization of neutralizing antibodies to West Nile virus. Virology 336:70-82. [DOI] [PubMed] [Google Scholar]
- 37.Tassaneetrithep, B., T. H. Burgess, A. Granelli-Piperno, C. Trumpfheller, J. Finke, W. Sun, M. A. Eller, K. Pattanapanyasat, S. Sarasombath, D. L. Birx, R. M. Steinman, S. Schlesinger, and M. A. Marovich. 2003. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 197:823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volk, D. E., D. W. Beasley, D. A. Kallick, M. R. Holbrook, A. D. Barrett, and D. G. Gorenstein. 2004. Solution structure and antibody binding studies of the envelope protein domain III from the New York strain of West Nile virus. J. Biol. Chem. 279:38755-38761. [DOI] [PubMed] [Google Scholar]
- 39.Winn, M. D., G. N. Murshudov, and M. Z. Papiz. 2003. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374:300-321. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, S., L. Li, S. E. Woodson, C. Y. Huang, R. M. Kinney, A. D. Barrett, and D. W. Beasley. 2006. A mutation in the envelope protein fusion loop attenuates mouse neuroinvasiveness of the NY99 strain of West Nile virus. Virology 353:35-40. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Y., W. Zhang, S. Ogata, D. Clements, J. H. Strauss, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2004. Conformational changes of the flavivirus E glycoprotein. Structure (Cambridge) 12:1607-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]