Abstract
F proteins from baculovirus nucleopolyhedrovirus (NPV) group II members are the major budded virus (BV) viral envelope fusion proteins. They undergo furin-like proteolysis processing in order to be functional. F proteins from different baculovirus species have a long cytoplasmic tail domain (CTD), ranging from 48 (Spodoptera litura multicapsid NPV [MNPV]) to 78 (Adoxophyes honmai NPV) amino acid (aa) residues, with a nonassigned function. This CTD is much longer than the CTD of GP64-like envelope fusion proteins (7 aa), which appear to be nonessential for BV infectivity. Here we have investigated the functional role of the CTD of Helicoverpa armigera single-capsid NPV (HearNPV), a group II NPV. We combined a newly constructed HearNPV f-null bacmid knockout-repair system and an Autographa californica MNPV (AcMNPV) gp64-null bacmid knockout-pseudotype system with mutation and rescue experiments to study the functional role of the baculovirus F protein CTD. We show that except for the 16 C-terminal aa, the HearNPV F CTD is essential for virus spread from cell to cell. In addition, the CTD of HearNPV F is involved in BV production in a length-dependent manner and is essential for BV infectivity. The tyrosine residue Y658, located 16 aa from the C terminus, seems to be critical. However, HearNPV F without a CTD still rescues the infectivity of gp64-null AcMNPV BV, indicating that the CTD is not involved in processing and fusogenicity. Altogether, our results indicate that the F protein is essential for baculovirus BV infectivity and that the CTD is important for F protein incorporation into BV.
The Baculoviridae family is a large group of enveloped, double-stranded DNA viruses which exclusively infect arthropods, predominantly insects of the order Lepidoptera (4). Baculoviruses are divided into two genera, Nucleopolyhedrovirus (NPV) and Granulovirus. The NPV genus can be subdivided phylogenetically into two subgroups, namely, group I and group II (15, 46). During an infection cycle, baculoviruses produce two virion phenotypes that play distinct roles. Virions of the occlusion-derived virus mediate the transmission of virus from insect to insect, whereas budded virus (BV) mediates the cell-to-cell spread of virus in infected insects and in cell culture.
BVs enter cells via the endocytic pathway, including two steps, namely, clathrin-mediated endocytosis and low-pH-dependent membrane fusion (21). GP64 is the major envelope fusion protein found in the BVs of Autographa californica multicapsid NPV (AcMNPV) and other members of the group I NPVs. Previous studies showed that GP64 is involved in BV binding to target cells, is necessary for low-pH-dependent membrane fusion to allow entry, and is important for efficient virion budding or egress from the cell surface (5, 25, 29). Recently, a new type of envelope fusion protein, named F, was identified from Lymantria dispar MNPV (32), Spodoptera exigua MNPV (16), and Helicoverpa amigera NPV (HearNPV) (20), and it is most likely present in all group II NPVs. The F protein is the major glycoprotein of group II BVs and is expressed first as a precursor (F0) and then cleaved into two disulfide bridge-linked subunits (F1 and F2) by a cellular furin-like convertase (16, 43). The cleavage is essential for activation of the F protein to a fusogenic form (43). In addition, group II NPV F proteins are able to rescue infectious BV production of a GP64 knockout mutant of AcMNPV (22). Therefore, these F proteins are functionally analogous to GP64. As such, baculovirus F proteins are similar in structure to envelope fusion proteins of vertebrate viruses (10).
The cytoplasmic tail domains (CTDs) of many viral envelope fusion proteins play an important role in virus assembly and budding through binding to the nucleocapsid protein (see references 37 and 40 for a review). The CTDs of virus glycoproteins are clearly involved in virus assembly, since they bind to the matrix protein of the nucleocapsid and collate the viral envelope and nucleocapsid (9, 12, 45). Some other commonly recognized functions of the CTDs of viral glycoproteins are roles in glycoprotein trafficking and subcellular localization (11, 14, 18, 31, 34, 36, 47) and in the assembly of mature virions (6, 7, 8, 27, 30). In addition, further roles of CTDs in virion morphology, in completion of membrane fusion, and in regulation have also been reported (1, 2, 13, 17, 24, 26, 33, 35, 42). Unlike GP64, the BV fusion protein of a group I baculovirus (AcMNPV) with a short CTD of 7 amino acids (aa), baculovirus group II NPV F proteins have comparatively long CTDs, ranging from 48 (Spodoptera litura MNPV) to 78 (Adoxophyes honmai NPV) aa residues. The CTD of AcMNPV GP64 is not essential for the production of infectious BV; removal of the CTD results in a reduction in budding efficiency (29). The role of the CTDs of baculovirus F proteins remains to be determined.
In this study, we investigated the functional role of the CTD (48 aa) of the HearNPV F protein as a representative of group II baculovirus F proteins. We determined the minimal length of the CTD that still renders infectious BV production by making C-terminal truncations of HearNPV F and found that the CTD can be truncated, removing up to 16 aa residues from the C terminus, without a loss of infectivity. However, further F protein CTD truncations are able to support infectious virus production of AcMNPV gp64-null mutants when pseudotyped. Our results indicate that the CTDs of F proteins of group II NPVs are essential for infectious BV production. Deletion of the F protein CTD does not affect F protein synthesis or fusogenicity.
MATERIALS AND METHODS
Cells and virus.
HzAM1 cells (23) and the Spodoptera frugiperda cell line IPLB-SF-21 (40) were cultured at 27°C in plastic tissue culture flasks (Nunc) in Grace's insect medium (pH 5.9 to 6.1; Invitrogen) supplemented with 10% fetal bovine serum. BVs of the HearNPV G4 isolate were used for infections and were propagated in HzAM1 cells (39).
Deletion of HearNPV f gene.
For the generation of an f gene deletion mutant by recombination mediated by the RecE and RecT proteins (ET recombination), the HearNPV bacmid HZ8 (41) was used as the parental bacmid (41). Long primers were designed with 50 nucleotides (nt) at the 5′ end corresponding to 50-nt flanking regions of the f gene in the HearNPV G4 genome. The forward primer was 5′-GGATTTGCCGTCGTAACAAATTAAACAAAAAAATTTTCATAAAAACAAATAAATTTAAGGGCACAATAACTG-3′ and contained the viral flanking sequences from nt 127755 to 127804 according to the HearNPV G4 genome sequence (GenBank accession no. AF271059). The reverse primer was 5′-CATACATTTATTAATCTAGTTATTTTTATTAAACCTTTCCGTAATAGTTATTCCTGTGCGCGACGGTTAC-3′, with viral flanking sequences from nt 129905 to 129954. The 3′ ends (underlined regions) of the primers anneal to the chloramphenicol resistance gene (cat) of pBeloBac11 (38).
To induce homologous recombination, the helper plasmid pBAD-αβγ was electrotransferred into DH10B cells (Invitrogen) containing the HearNPV bacmid HZ8 (41). Electrocompetent cells were made according to the method of Muyrers et al. (28). PCR to generate linear fragments was performed by using the high-fidelity Expand long-template PCR system (Roche). The column-purified PCR product was digested with DpnI to eliminate template plasmid DNA. After digestion, the linear fragments were recovered from agarose gels and finally collected by ethanol precipitation. One microgram of linear fragments was used for electroporation into DH10B containing the bacmid HZ8, and the resulting f gene-deleted bacmid was designated HearNPV f-null. The deletion of the f gene from the f-null HearNPV bacmid was confirmed by EcoRI restriction profiling and PCR.
Donor plasmid and transposition.
The pFASTBAC Dual vector (Invitrogen) is the donor plasmid for the baculovirus insect cell expression system used for this study and carries the AcMNPV polyhedrin promoter and p10 promoter in opposite orientations. The enhanced green fluorescent protein (EGFP) gene was amplified by PCR, using the peGFP plasmid (Clontech) as template DNA. An upstream NheI restriction site and a downstream KpnI restriction site were introduced by subsequent primers for further cloning. The PCR products were first cloned into the pGEM-T Easy vector (Promega), giving pGFP. After a sequence check, the NheI-to-KpnI fragment of the GFP gene was cloned into the pFASTBAC Dual vector under the control of the p10 promoter, resulting in pFB-GFP.
The HearNPV f cassette containing the coding sequence of the HearNPV F protein plus the 300-nt promoter sequence (from nt 127811 to nt 130144) was amplified by PCR, using Pfu Ultra DNA polymerase (Stratagene). An upstream Bst1107I restriction site and a downstream HindIII restriction site (underlined) were introduced by the primers (forward primer, 5′-GGGGTATACCCRRRRAGAAAACCTTGGTACTGT-3′; and reverse primer, 5′-GCGAAGCTTACAATTTGTGTTCATCATATTGATC-3′) for further cloning. After cloning and sequencing verification, the Bst1107I-to-HindIII fragment of the HearNPV F cassette was cloned into pFB-GFP. The resulting vector, carrying the f gene cassette and the p10 promoter controlled by EGFP, was named pFB-F&GFP.
A unique SphI restriction site is located 425 nt upstream of the translational termination codon TAA (Fig. 1). To generate cytoplasmic tail truncation mutations, a forward primer (5′-ATCGCATGCAATTCGAGTTGGAAC-3′) covering the SphI restriction site (underlined) and reverse primers (Tr-6aa, 5′-GCGAAGCTTATTGATCGAAAGAATCTTTAG-3′; Tr-16aa, 5′-GCGAAGCTTAGGCTTTCAAATAGACGAGTTG-3′; Tr-19aa, 5′-GCGAAGCTTAATAGACGAGTTGACGATCGTTG-3′; Tr-29aa, 5′-GCGAAGCTTACGTTCGTGTGGGTCTTCGTCT-3′; Tr-40aa, 5′-GCGAAGCTTAGCACAAGTCCATACAACAAGATAC-3′; and Tr-49aa, 5′-GCGAAGCTTAACAAAAACATCGTAAAACTACAACAG-3′) located at different positions within the CTD sequence were designed. The translational termination codon TAA and a HindIII restriction site (underlined) were introduced by each truncation reverse primer to stop translation and to assist further cloning. PCR was performed by using the forward primer covering the SphI site and each reverse primer. All PCR products were cloned into the pGEM-T Easy vector. After sequence verification, the SphI-to-HindIII fragments were cloned into pFB-F&GFP, resulting in donor plasmids carrying f gene cassettes expressing F proteins with various truncations (6 to 49 aa from the C terminus) in the CTD. The donor plasmids were used both to construct the repair HearNPV bacmids and to pseudotype AcMNPV bacmids with truncated f.
FIG. 1.
Schematic diagram of the baculovirus group II f gene, showing the proteolytic cleavage site, fusion peptide (FP), heptad repeat (HR) regions, transmembrane region (TM), and cytoplasmic tail domain (CTD) of HearNPV F (A) and C-terminal truncations of F (B). Each donor plasmid carrying HearNPV f or a truncated f gene was used to construct a repair HearNPV bacmid and a pseudotyped AcMNPV bacmid.
Competent cells containing the f-null HearNPV bacmid and a gp64-null AcMNPV bacmid (provided by G. W. Blissard) were made according to the methods in the Bac-to-Bac manual (Invitrogen). Transpositions of inserts from donor plasmids to the f-null HearNPV and gp64-null AcMNPV bacmids were confirmed by diagnostic PCR using a gentamicin resistance gene-specific forward primer (5′-AGCCACCTACTCCCAACATC-3′) in combination with the M13 forward primer (5′-CCCAGTCACGACGTTGTAAAACG-3′) and by sizing of the products in 0.6% agarose gels.
Transfection of pseudotyped AcMNPV bacmids and HearNPV f repair bacmids into HZAM1 cells.
For the transfection of pseudotyped AcMNPV bacmids, Sf21 cells were transfected with approximately 1 μg bacmid DNA according to the methods in the Bac-to-Bac manual (Invitrogen). The supernatant was harvested at 5 days posttransfection (p.t.). For the transfection of repaired HearNPV bacmids, a modified transfection method was applied. Briefly, HzAm1 cells were seeded into six-well plates (Nunc) at 1 × 106 cells per well. After 24 h of incubation in Grace's insect medium supplemented with 10% fetal bovine serum, cell were washed twice with Grace's insect medium. Cells were then transfected with approximately 1 μg bacmid DNA, using 15 μl Lipofectin (Invitrogen). Supernatants containing BVs were harvested at 7 days p.t. from HaAM1 cells.
Western analysis.
The expression and incorporation of the complete and truncated F proteins from the HearNPV f repair bacmids in BVs were examined by Western analysis, using polyclonal antibodies against F1 and F2 to probe sucrose-purified BVs (20). Briefly, sucrose-purified BVs were disrupted under reducing or nonreducing conditions and were then denatured for 10 min at 95°C. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to Western analysis as previously described (20). Antisera raised against the F protein in chickens were used at a 1:1,000 dilution, and the (truncated) F proteins were detected by treatment with horseradish peroxidase-conjugated rabbit anti-chicken immunoglobulin (Sigma) diluted 1:10,000 followed by ECL technology as described by the manufacturer (Amersham).
Budded virus production.
To investigate infectious BV production of bacmid-derived HearNPV repaired with full f and with CTD truncations, virus growth curves were generated and compared to those of wild-type HearNPV. HzAM1 cells were infected at a multiplicity of infection (MOI) of 5 50% tissue culture infective dose (TCID50) units/cell for 1 h. After infection, cells were washed with fresh medium once and then incubated in new medium. At 12, 24, 36, 48, 60, 72, 84, and 96 h postinfection (p.i.), the supernatants were collected. For each mutant virus and each time point, triplicate samples were collected for triplicate experiments. The quantity of infectious BVs from each sample was determined by an end-point dilution assay (EPDA) with HzAM1 cells. To analyze BV production of pseudotyped AcMNPVs compared to wild-type AcMNPV, Sf21 cells were infected at an MOI of 5. At 48 h p.i. for each treatment, samples were collected from triplicate supernatants, and the quantity of infectious BVs from each sample was determined by an EPDA with Sf21 cells. The results were exported to Microsoft Excel software, the averages of triple log-transformed TCID50 values were analyzed, and the standard deviations of the means were determined (see Fig. 3). For the infectivity of pseudotyped AcMNPVs (see Fig. 6), further one-way analysis of variance and post hoc tests were performed to analyze the significance of the differences in BV production among the pseudotyped viruses.
FIG. 3.
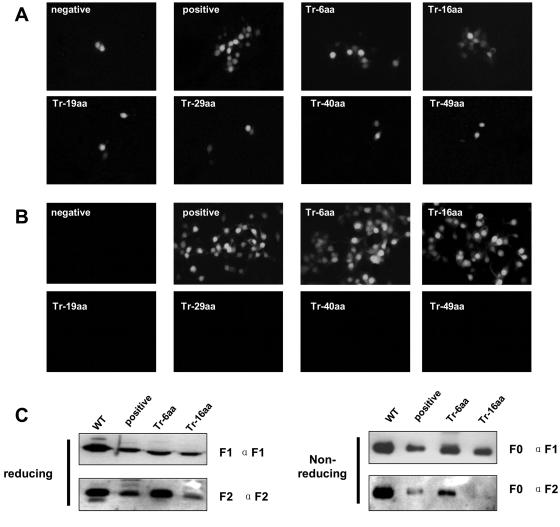
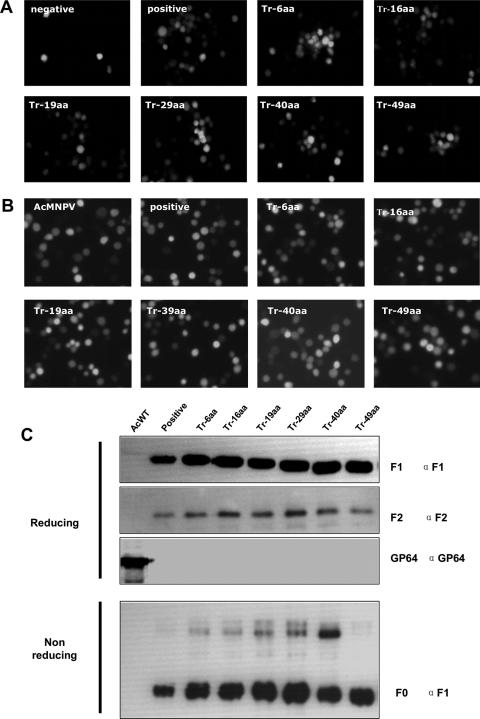
(A) Transfection of repair HearNPV bacmid. (B) Infection assay for viral propagation. (C) Western blot analysis of repair HearNPV BVs. HzAM1 cells were transfected with repaired HearNPV bacmids (positive control and Tr-6aa, Tr-16aa, Tr-19aa, Tr-29aa, Tr-40aa, and Tr-49aa mutants). GFP expression in each transfection was examined 5 days later (A). Supernatants were collected at 7 days p.t. After being clarified, the supernatants were used to infect new HzAM1 cells. GFP expression, indicating infectivity, was examined 3 days after infection (B). Western blot analysis was performed on purified wild-type HearNPV BVs (WT) and f-repaired HearNPV BVs (positive control and Tr-6aa and Tr-16aa mutants), using anti-F1 and anti-F2 under either reducing or nonreducing conditions.
FIG. 6.
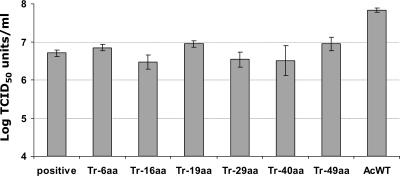
BV production of pseudotyped AcMNPVs. Sf21 cells were infected by F protein-pseudotyped AcMNPVs (Tr-6aa, Tr-16aa, Tr-19aa, Tr-29aa, Tr-40aa, and Tr-49aa) and wild-type AcMNPV. Each data point represents virion production at 48 h p.i. Data points represent means of triplicate infections and titrations, and error bars represent standard deviations from the means.
RNA analysis of HzAM1 cells infected with pseudotyped AcMNPV.
HzAM1 cells, which are susceptible but nonpermissive for AcMNPV, were challenged with pseudotyped AcMNPVs at an MOI of 5 TCID50 units per cell. Cells from each infection treatment were collected at 12 h p.i. Total RNAs were extracted from each cell sample by use of the Trizol reagent (Invitrogen). Early transcription of ie-1 was examined by 3′ rapid amplification of cDNA ends (RACE) analysis by using a RACE kit (Roche). In brief, first-strand cDNA synthesis was performed by using avian myeloblastosis virus reverse transcriptase and an oligo(dT) anchor primer according to the manufacturer's instructions. The cDNA was amplified by a PCR using the anchor primer and an ie-1-specific primer, ie-1-F (5′-GTACAAATACAGCAGCGTCGCTA-3). The PCR products were analyzed by agarose gel electrophoresis. Total RNA samples served as reverse transcription template controls.
RESULTS
Construction of f-null HearNPV bacmid.
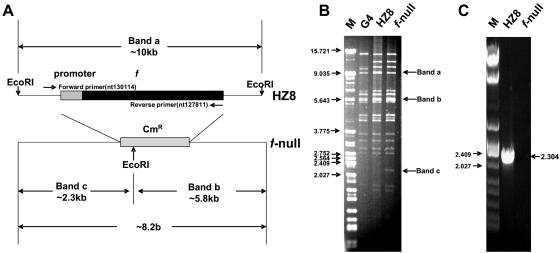
To study the functionality of the HearNPV f gene during virus infection, an f-null HearNPV bacmid was constructed by deleting the f gene from HearNPV bacmid HZ8 according to the method of Muyrers et al. (28). A 70-mer primer and a 72-mer oligonucleotide were used to PCR amplify a linear DNA fragment containing a cat gene flanked by 50-bp sequences homologous to the HearNPV genome. After DpnI treatment to eliminate the template plasmid, the PCR product was purified and transferred to recombinant competent cells. Bacterial colonies harboring the recombinant bacmid were selected on medium containing kanamycin and chloramphenicol. In the resulting f-null HearNPV bacmid, the f gene was correctly replaced by the cat gene (Fig. 2A). The deleted sequence was removed from 61 bp upstream of the translational start codon to 6 bp downstream of the stop codon (bp 127803 to 129904 according to the HearNPV G4 genome sequence). When the f gene was present, as in the parental bacmid HZ8 (41), a 10-kbp EcoRI band was present (band a), whereas two new EcoRI bands (bands b and c), of 5.8 and 2.3 kbp, respectively, were generated when the f gene was replaced by the cat gene (Fig. 2B). A PCR product the size of f was generated only in bacmid HZ8, not in the f-null HearNPV bacmid (Fig. 2C).
FIG. 2.
Strategy for generation of an f-null HearNPV bacmid by ET recombination in E. coli (A) and EcoRI restriction profiles (B) and PCR verification (C) of the f-null HearNPV bacmid. (A) At the HearNPV f gene (Ha133) locus in the HearNPV bacmid HZ8 (41), a PCR-amplified DNA fragment containing the chloramphenicol resistance gene (cat) flanked by 50-bp HearNPV sequences on both sides of the f gene was used to replace the f gene with a Cmr gene. (B) EcoRI restriction profiles of the wild-type HearNPV (G4 strain) genome, the parental HearNPV bacmid HZ8, and the f-null HearNPV bacmid. An ∼10-kb EcoRI restriction fragment containing the f gene from HZ8 was disrupted by ET recombination, resulting in two smaller fragments (with approximate lengths of 2.3 kb and 5.8 kb). (C) Diagnostic PCR of f-null HearNPV bacmid. Forward and reverse primers (panel A) were used for PCR. Bacmid HZ8 was used as a positive control for the presence of f. Sizes of markers (M) are indicated to the left of the gels (in kb).
Truncation in the HearNPV F CTD gene and construction of f repair HearNPV.
The egfp gene, controlled by the AcMNPV p10 promoter in association with the HearNPV f gene cassette, with and without truncation in the CTD, was transposed into the Tn7 transposition site of the f-null HearNPV bacmid according to a method in the Bac-to-Bac manual (Invitrogen). Bacmid DNA was then isolated from bacterial colonies after diagnostic PCR verification for correct insertion. Approximately 5 μg repair bacmid DNA from each transposed bacmid was transfected into 1 × 106 HzAM1 cells, with transfection of (f-null/egfp) as a negative control. At 7 days p.t., GFP expression in transfected cells was examined (Fig. 3A). The presence of EGFP in transfected cells signaled effective transfection. The production of infectious BVs from each transfectant was determined by infecting new HzAM1 cells with BVs in the transfection supernatant. At 5 days postinfection, EGFP expression in infected cells was examined (Fig. 3B). The repair bacmids with complete f (positive control) and the Tr-6aa and Tr-16aa recombinants were able to produce infectious BVs. The other recombinants, Tr-19aa, Tr-29aa, Tr-40aa, and Tr-49aa, showed only single infected cells (Fig. 3A) and were not able to produce infectious BVs (Fig. 3B). These results indicate that the CTD of F plays an important role in baculovirus BV propagation and that the CTD's length is critical for infectivity. The F protein CTD can be reduced to 32 aa residues, but further truncations of the CTD abort the generation of infectious HearNPV BVs.
To determine the expression of the f repair bacmid as a control and of the truncated recombinants Tr-6aa and Tr-16aa upon viral infection and incorporation into BVs, we performed Western analysis on purified BVs generated from the respective viruses. BVs from wild-type HearNPV were used as a positive control (Fig. 3C). Under reducing conditions, bands corresponding to a protein with the predicted molecular size of the F1 subunit (approximately 59 kDa) were detected for wild-type HearNPV and f repair BVs with an anti-F1 specific antibody. In the BVs from the Tr-6aa and Tr-16aa repair mutants, the F1 subunits were also detected by anti-F1 antibody, but they were smaller, being approximately 58 kDa and 57 kDa, respectively. Bands corresponding to F2 subunits with a molecular mass of 20 kDa were detected for wild-type HearNPV BVs and for the repair viruses (f repair, Tr-6aa, and Tr-16aa). These results show that F, Tr-6aa, and Tr-16aa protein precursors (F0) were expressed and cleaved by a furin-like convertase in infected HzAM1 cells and that these proteins were incorporated into BVs. To further investigate the processing of F, Tr-6aa, and Tr-16aa proteins, Western analysis was performed after SDS-PAGE under nonreducing conditions (Fig. 3C). Both anti-F1 and anti-F2 antibodies recognized the F0 bands. These results indicate that the F1 and F2 subunits in F, Tr-6aa, and Tr-16aa proteins in the repaired HearNPV were linked by disulfide bridges, like the case for wild-type HearNPV BVs (20).
BV production of f repair HearNPV.
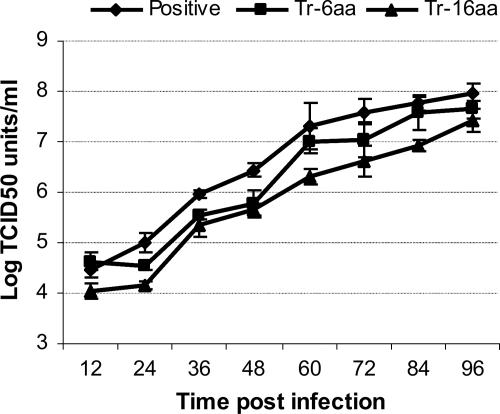
The F-, Tr-6aa-, and Tr-16aa-repaired f-null HearNPV bacmids produced infectious HearNPV BVs, and the further C-terminal truncation mutants did not. This result suggests that the CTD of baculovirus F protein is essential for the production of infectious BVs and that truncation of the CTD results in an inability to rescue the infectivity of an f-null HearNPV bacmid. To determine if and to what extent the 6- and 16-aa C-terminal truncations affect infectious BV production, we compared the f repair virus (control) and Tr-6aa and Tr-16aa HearNPV F mutants in one-step growth curve assays in HzAM1 cells (Fig. 4). Our results from one-step growth curve comparisons show that truncations of 6 aa and 16 aa from the C terminus of the F CTD reduce infectious BV production about threefold and eightfold, respectively.
FIG. 4.
Analysis of BV production by one-step growth curve analysis. The graph shows virus growth curves for the positive control (f repair) and Tr-6aa- and Tr-16aa-repaired HearNPVs. HzAM1 cells were infected in triplicate with HearNPVs at a multiplicity of 5.0 TCID50 units per cell. Each data point represents BV production at a different time point after infection. Data points represent means for triplicate infections and titrations, and error bars represent standard deviations from the means.
CTD truncations of HearNPV F and rescue of AcMNPV gp64-null bacmid infectivity.
Transfection and infection assays of HearNPV repaired bacmids containing complete f and CTD truncations indicated that the CTD of the HearNPV F protein plays an important role in infectious virion production. However, it was not clear how truncation of the CTD would influence infectious BV production. The possible consequences of the removal of the CTD from HearNPV F beyond the 16 C-terminal aa would be (i) on F protein processing prior to assembly into BVs, which would result in the impairment of subsequent viral endocytosis and low-pH-triggered envelope fusion in the next infection; and (ii) the absence of a linkage between HearNPV F and nucleocapsid proteins, leading to an inability to undergo proper BV formation. In the first case, noninfectious BVs would be formed, and in the latter case, no BVs would be formed.
The CTD of GP64 is not essential for infectious AcMNPV BV production (19), indicating that AcMNPV BV budding is not dependent on an intact CTD. This would suggest that CTD truncations of HearNPV F do not prevent infectious BV formation. Therefore, intact f, Tr-6aa, Tr-16aa, Tr-19aa, Tr-29aa, Tr-40aa, and Tr-49aa gene cassettes were inserted into the gp64-null AcMNPV bacmid system (20) to pseudotype gp64-null AcMNPV bacmids. The constructs were transposed into the bacmids with an AcMNPV p10 promoter-controlled egfp gene. Sf21 cells were transfected with pseudotyped gp64-null AcMNPV bacmids (Fig. 5A). A gp64-null AcMNPV bacmid carrying no HearNPV f gene served as a negative control. At 5 days p.i., the supernatant from each transfection treatment was used to infect new Sf21 cells (Fig. 5B). An AcMNPV virus carrying a p10 promoter-controlled egfp gene was used as a positive control for infection. Unlike the case for the HearNPV repair bacmid, all HearNPV f gene cassettes (f repair, Tr-6aa, Tr-16aa, Tr-19aa, Tr-29aa, Tr-40aa, and Tr-49aa) rescued the infectivity of the gp64-null AcMNPV bacmid mutants and produced infectious BVs. These results confirmed that the HearNPV f gene cassette was correctly expressed and processed in the AcMNPV-Sf21 system but show, in addition, that truncation of the CTD of the HearNPV F protein does not affect the formation of infectious AcMNPV BVs, indicating that the HearNPV F protein without a CTD is fusogenic and able to form infectious BVs.
FIG. 5.
(A) Transfection of gp64-null AcMNPV bacmid pseudotyped by the HearNPV f gene, with or without truncation in the CTD. (B) Infection assay for viral propagation. (C) Western blot analysis of pseudotyped AcMNPV BVs. Sf21 cells were transfected with HearNPV f gene-pseudotyped gp64-null AcMNPV bacmids (f repair, Tr-6aa, Tr-16aa, Tr-19aa, Tr-29aa, Tr-40aa, and Tr-49aa). GFP expression in each transfection was examined 5 days later. A gp64 gene-repaired gp64-null AcMNPV bacmid served as a positive control (A). Supernatants were collected at 7 days p.i. After clarification, the supernatants were used to infect new Sf21 cells. GFP expression, indicating infectivity, was examined at 3 days p.i. (B). Western blot analysis was performed on purified AcMNPV BVs and pseudotyped AcMNPV BVs (f repair, Tr-6aa, and Tr-16aa), using anti-F1, anti-F2, and anti-GP64 under either reducing or nonreducing conditions.
In addition to transfection and infection assays, we also examined the incorporation of the HearNPV F protein and F CTD truncation mutants into AcMNPV BVs (Fig. 5C). When separated by SDS-PAGE under reducing conditions and detected by F-specific antibodies, F1 and F2 were recognized separately in all pseudotyped AcMNPV (gp64-null/f repair, Tr-6aa, Tr-16aa, Tr-19aa, Tr-29aa, Tr-40aa, and Tr-49aa) mutant BVs. The molecular size of F1 in pseudotyped AcMNPV BVs gradually decreased from 60 kDa (for the full F protein) to 55 kDa (for a CTD truncation of 48 aa), whereas the molecular size of F2 stayed the same, at 20 kDa (Fig. 5C). The presence of GP64 in AcMNPV BVs and its absence in pseudotyped AcMNPV BVs confirmed the authenticity of rescue. Under nonreducing conditions, bands corresponding to F0 were detected in all pseudotyped AcMNPV BVs by anti-F1 antibody (Fig. 5C). Anti-F2 antibody also identified F0 in pseudotyped AcMNPV BVs (result not shown). These results indicate that HearNPV F proteins with truncations in the CTD are incorporated into AcMNPV BVs, like the intact HearNPV F protein. The incorporation of the HearNPV F protein and its truncated forms into AcMNPV suggests that the CTD is not important for proper F protein synthesis, folding, membrane trafficking, and fusogenicity.
Infectivity of AcMNPV pseudotyped with HearNPV f.
To determine the effects of CTD truncations on the BV productivity of pseudotyped AcMNPV baculoviruses, BV production at 48 h p.i. was evaluated by EPDAs (Fig. 6). AcMNPV produced about 10 times more infectious BVs than the pseudotyped AcMNPVs (gp64-null/f repair, Tr-6aa, Tr-16aa, Tr-19aa, Tr-29aa, Tr-40aa, and Tr-49aa), and there was no significant difference observed among the pseudotyped AcMNPVs. This result demonstrates that the length of the CTD is not important for infectious BV production in AcMNPV.
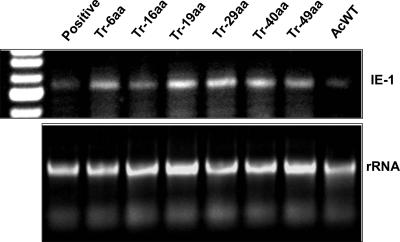
The previous results show that the CTD of F, except for the 16 C-terminal aa, is essential for HearNPV viral infectivity, whereas the CTD is not essential for fusogenicity. It is possible that (i) the HearNPV F protein without the 16 C-terminal aa of the CTD is incorporated into budded virions which are not infectious and, e.g., entry defective (defective in attachment to cell membranes and envelope fusion) in HzAM1 cells; or (ii) the F protein without CTD is not incorporated into virions. To investigate this, nonproductive infections were carried out in which HzAM1 cells were infected with pseudotyped AcMNPVs (gp64-null/f repair and the Tr-6aa, Tr-16aa, Tr-19aa, Tr-29aa, Tr-40aa, and Tr-49aa mutants). Successful viral entry occurred in all instances, as early transcripts of the AcMNPV ie-1 gene were detected at 12 h p.i. (Fig. 7). Thus, viral entry of pseudotyped AcMNPVs into HzAM1 cells was not affected by CTD truncations of the HearNPV F protein.
FIG. 7.
Detection of ie-1 gene early transcripts upon nonproductive infection of pseudotyped AcMNPVs in HzAM1 cells. HzAM1 cells were infected with gp64-null AcMNPVs pseudotyped with Tr-6aa, Tr-16aa, Tr-19aa, Tr-29aa, Tr-40aa, and Tr-49aa and with wild-type AcMNPV. At 12 h p.i., total RNA from infected cells of each infection was isolated. 3′ RACE analysis of the AcMNPV ie-1 gene was performed. PCR-amplified products were analyzed in a 1% agarose gel. Total RNA samples from each treatment were analyzed as template controls by reverse transcription.
DISCUSSION
A number of functional roles have been attributed to the CTDs of virus glycoproteins. These include regulation of the incorporation of viral glycoproteins into mature virions (8, 13, 35), determination of the site and efficiency of viral budding (21, 48), governance of the conformation of the glycoprotein's extracellular domain and processing, regulation of cell surface expression of glycoproteins (14, 34, 36), and control of virus envelope and cell membrane fusion (2, 6, 7, 26, 33). In the present study, we have investigated the role of the CTD of the major baculovirus group II envelope fusion protein F, more specifically the HearNPV F protein. Our results show that the CTD of the HearNPV F protein is critical for infectious BV production. In this respect, the role of the F CTD differs from that of GP64-like fusion proteins, the major envelope fusion proteins from NPV group I members. The minimal length of the CTD to support infectious HearNPV BV production was 32 aa; the C-terminal 16 aa were dispensable. For HearNPV F, however, the CTD is not essential for rescuing BV production and infectivity of a gp64-null AcMNPV mutant. This indicates that the CTD of HearNPV F is not involved in fusogenicity. Additionally, HearNPV F protein processing, including synthesis, folding, disulfide bridging, trafficking, and susceptibility to cellular convertase (furin-like) cleavage, is not affected by removing the CTD from the F protein.
The group II NPV F protein is a newly found baculovirus envelope fusion protein (16, 32). Common functional domains, such as a furin-like protease cleavage site (20, 43), a fusion peptide (44), a heptad repeat (G. Long, unpublished data), and a cytoplasmic tail, were identified. The arrangement of these domains is similar to the architecture of class I viral envelope fusion proteins from members of viral families infecting vertebrates. Unlike GP64, baculovirus F proteins have a long CTD, ranging from 52 aa to 78 aa. The CTD of GP64 (seven amino acids long) was reported to be dispensable for viral infectivity and not essential for efficient virion budding from the cell surface (29). Our experiments demonstrate that a CTD is not essential for infectious BV production by AcMNPV and possibly not for all group I baculoviruses.
However, our results demonstrate that in the case of HearNPV, only truncations of up to 16 aa from the C terminus of the CTD allow the repair of an f-null HearNPV bacmid, which underscores that the CTD is essential for viral infectivity. Other truncations (from Tr-19aa to Tr-49aa) were not able to rescue f-null HearNPV. The tyrosine residue (Y658, the last residue of Tr-19aa) is conserved in all baculovirus F proteins. This may suggest that this internal tyrosine-based signal is important for viral infectivity. There is evidence from other enveloped viruses that an internal tyrosine-based motif is crucial for the functional role of F-like proteins (3, 19, 48). If such a motif in the baculovirus F protein CTD has a similar function, it may explain why the Tr-16aa mutant can repair the f-null HearNPV bacmid, whereas the Tr-19aa mutant cannot (Fig. 3). Mutational analysis of the tyrosine residue in the HearNPV F CTD, which is also present in all other sequenced group II NPVs, should prove the importance of this aa in this process. Additionally, a slight decrease in infectious BV production was observed in association with the Tr-6aa and Tr-16aa truncations. This might suggest that these 16 C-terminal aa, representing the variable region in CTDs of baculovirus F proteins, stabilize the possible interaction between the F CTD and nucleocapsid in a length-dependent manner, thus guaranteeing the efficient incorporation of F into baculovirus group II BVs. A slight but significant decrease in infectious BV production was noted for AcMNPV when the seven C-terminal aa of the CTD were removed (29).
The GP64 protein of AcMNPV is necessary and sufficient for the low-pH-triggered membrane fusion which occurs during BV entry after endocytosis (5, 25). In this study, a gp64-null AcMNPV bacmid pseudotyping system was applied to exclude the possibility that the fusion ability of the HearNPV F protein is impaired by eliminating the CTD. HearNPV F proteins with sequential truncations in the cytoplasmic tail (from Tr-6aa to Tr-49aa) were all capable of pseudotyping a gp64-null AcMNPV bacmid (Fig. 5). This is also in agreement with the observation that the CTD of GP64 is not essential for viral infectivity (29).
Viruses use the host cellular machinery to translate viral proteins. Viral glycoproteins are synthesized in a similar fashion to that for cellular proteins directed to the secretory pathway. The CTDs of transmembrane proteins are exposed at the cytoplasmic side of the endoplasmic reticulum membrane and thus may interact with cellular proteins that regulate intracellular processing and trafficking. Deletion in the cytoplasmic portion of viral glycoproteins can have small, but also severe, effects on intracellular transport and surface expression (14, 18, 34, 36, 42). As we have shown here, truncation in the CTD of HearNPV F seems to have little or no negative effect on these processes. Truncation of the CTD in the HearNPV F protein has no influence on furin-like cellular convertase cleavage and subunit disulfide bridging. Successful incorporation into pseudotyped AcMNPV budded virions indicates proper folding of F without the CTD and trafficking to the viral budding destination. Moreover, the AcMNPV-pseudotyping competence of F without a CTD confirms our observation that truncations in the F CTD have no effect on the fusogenicity of the F protein.
Based on the evidence shown above, the CTD core up to the tyrosine residue (Y658) is proposed to be essential for HearNPV viral infectivity but not essential for the fusogenicity of the F protein. Three possible consequences of CTD deletion may lead to the failure to generate infectious HearNPV BVs, as follows: (i) HearNPV F without a CTD is incorporated into BVs, which are released from the infected cell but are not infectious because of some defect in viral entry (in cell attachment and envelope fusion) into HzAM1 cells; (ii) F without a CTD is unable to be incorporated into BVs as a consequence of impaired protein synthesis and transportation; and (iii) F without a CTD is properly synthesized and transported to budding sites in infected cells but is not able to be wrapped in mature and infectious BVs. The experiments described above showed that HearNPV F without a CTD is able to rescue the infectivity of a gp64-null AcMNPV bacmid. This indicates that F without a CTD is properly synthesized and fusion competent. Since AcMNPVs pseudotyped with F without a CTD were able to enter HzAM1 cells, as evidenced by the initiation of early gene transcription (Fig. 6), F without a CTD can properly fold and be transported. However, F without a CTD is apparently unable to generate infectious HearNPV BVs, underscoring the importance of the CTD in the incorporation of F into HearNPV BVs, and possibly into BVs of all group II baculoviruses. It is still possible that low levels of HearNPV BVs without F protein were produced, but they remained below the detection level.
In conclusion, we combined a newly constructed HearNPV f-null bacmid knockout-repair system (this paper) and an AcMNPV gp64-null bacmid knockout-pseudotype system to study the functional role of the CTD of baculovirus F proteins. We showed that the CTD of the HearNPV F protein determines infectious BV production in a length-dependent manner and that the CTD is essential for infectious BV production by HearNPV. The HearNPV F protein without a CTD still rescues the infectivity of a gp64-null AcMNPV, and thus the CTD is not involved in F protein processing and fusogenicity. It is possible that the CTD is important for proper incorporation of F into mature HearNPV BVs. Future experiments will determine which CTD domains or aa are involved in this interaction.
Acknowledgments
This work was supported financially by the Royal Academy of Sciences of The Netherlands (KNAW) (project 04-PSA-BD-02) and by a Ph.D. sandwich grant to G.L. from Wageningen University. The research was further supported by the 973 project (2003CB114202), PSA project 2004CB720404 from CAS, and an NSFC grant (30300012) to Gang Long.
We thank Hu Zhihong for fruitful discussions and continued interest in this research.
REFERENCES
- 1.Aguilar, J. C., W. F. Anderson, and P. M. Cannon. 2003. Cytoplasmic tail of Moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R peptide. J. Virol. 77:1281-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagai, S., and R. A. Lamb. 1996. Truncation of the COOH-terminal region of the paramyxovirus SV5 fusion protein leads to hemifusion but not complete fusion. J. Cell Biol. 135:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaufils, P., D. Choquet, R. Z. Mamoun, and B. Malissen. 1993. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 12:5105-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blissard, G., B. Black, N. Crook, B. A. Keddie, R. Possee, G. Rohrmann, D. Theilmann, and L. E. Volkman. 2002. Baculoviridae: taxonomic structure and properties of the family, p. 195-202. In M. H. V. Van Regenmortel et al. (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 5.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus GP64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 66:6829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brody, B. A., S. S. Rhee, and E. Hunter. 1994. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J. Virol. 68:4620-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cathoman, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christodoulopoulos, I., and P. M. Cannon. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J. Virol. 75:4129-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorfman, T., F. Mammano, W. A. Haseltine, and H. G. Gottlinger. 1994. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 68:1689-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hozie, R. W. Doms, and R. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation. J. Virol. 70:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gage, P. J., M. Levine, and J. C. Glorioso. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J. Virol. 67:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grange, M. P., V. Blot, L. Delamarre, I. Bouchaert, A. Rocca, A. Dautry-Varsat, and M. C. Dokhelar. 2000. Identification of two intracellular mechanisms leading to reduced expression of oncoretrovirus envelope glycoproteins at the cell surface. J. Virol. 74:11734-11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herniou, E. A., T. Luque, X. Chen, J. M. Vlak, D. Winstanley, J. S. Cory, and D. R. O'Reilly. 2001. Use of whole-genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.IJkel, W. F. J., M. Westenberg, R. W. Goldbach, G. W. Blissard, J. M. Vlak, and D. Zuidema. 2000. A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology 275:30-41. [DOI] [PubMed] [Google Scholar]
- 17.Januszeski, M. M., P. M. Cannon, D. Chen, Y. Rozenberg, and W. F. Anderson. 1997. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J. Virol. 71:3613-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lodge, R., L. Delamarre, J.-P. Lalonde, J. Alvarado, D. A. Sanders, M. C. Dokhelar, E. A. Cohen, and G. Lemay. 1997. Two distinct oncornaviruses harbor an intracytoplasmic tyrosine-based basolateral targeting signal in their viral envelope glycoprotein. J. Virol. 71:5696-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lodge, R., J.-P. Lalonde, G. Lemay, and E. A. Cohen. 1997. The membrane proximal intracytoplasmic tyrosine residue of HIV-1 envelope glycoprotein is critical for basolateral targeting of viral budding in MDCK cells. EMBO J. 16:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long, G., H. Wang, M. Westenberg, J. M. Vlak, and Z. Hu. 2006. Function, oligomerization and N-linked glycosylation of the Helicoverpa armigera single nucleopolyhedrovirus envelope fusion protein. J. Gen. Virol. 87:839-846. [DOI] [PubMed] [Google Scholar]
- 21.Long, G., X. Pan, R. Kormelink, and J. M. Vlak. 2006. Functional entry of baculovirus into insect and mammalian cells is dependent on clathrin-mediated endocytosis. J. Virol. 80:8830-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lung, O., M. Westenberg, J. M. Vlak, D. Zuidema, and G. W. Blissard. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 76:5729-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McIntosh, A. H., and C. M. Ignoffo. 1983. Characterization of five cell lines established from species of Heliothis. Appl. Entomol. Zool. 18:262-269. [Google Scholar]
- 24.Melikyan, G. B., R. M. Markosyan, S. A. Brener, Y. Rozenberg, and F. S. Cohen. 2000. Role of the cytoplasmic tail of ecotropic Moloney murine leukemia virus Env protein in fusion pore formation. J. Virol. 74:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monsma, S. A., and G. W. Blissard. 1995. Identification of a membrane fusion domain and an oligomerization domain in the baculovirus GP64 envelope fusion protein. J. Virol. 69:2583-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulligan, M. J., G. V. Yamshchikov, G. D. Ritter, F. Gao, M. J. Jin, C. D. Nail, C. P. Spies, B. H. Hahn, and R. W. Compans. 1992. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J. Virol. 66:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murakami, T., and E. O. Freed. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. USA 97:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muyrers, J. P., U. Zhang, G. Testa, and A. F. Stewart. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27:1555-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oomens, A. G. P., and G. W. Blissard. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254:297-314. [DOI] [PubMed] [Google Scholar]
- 30.Owens, R. J., and J. K. Rose. 1993. Cytoplasmic domain requirement for incorporation of a foreign envelope protein into vesicular stomatitis virus. J. Virol. 67:360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmarini, M., N. Maeda, C. Murgia, C. De-Fraja, A. Hofacre, and H. Fan. 2001. A phosphatidylinositol 3-kinase docking site in the cytoplasmic tail of the Jaagsiekte sheep retrovirus transmembrane protein is essential for envelope-induced transformation of NIH 3T3 cells. J. Virol. 75:11002-11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson, M. N., C. Groten, and G. F. Rohrmann. 2000. Identification of the Lymantria dispar nucleopolyhedrovirus envelope fusion protein provides evidence for a phylogenetic division of the Baculoviridae. J. Virol. 74:6126-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragheb, J. A., and W. F. Anderson. 1994. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J. Virol. 68:3220-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raviprakash, K., L. Rasile, K. Ghosh, and H. P. Ghosh. 1990. Shortened cytoplasmic domain affects intracellular transport but not nuclear localization of a viral glycoprotein. J. Biol. Chem. 265:1777-1782. [PubMed] [Google Scholar]
- 35.Rein, A., J. Mirro, J. G. Haynes, S. M. Ernst, and K. Nagashima. 1994. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J. Virol. 68:1773-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauter, M. M., A. Pelchen-Mattchews, R. Bron, M. Marsh, C. C. LaBranche, P. J. Vance, J. Romano, B. S. Haggarty, T. K. Hart, W. M. Lee, and J. A. Hoxie. 1996. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoprotein on the cell surface. J. Cell Biol. 132:795-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt, A. P., and R. A. Lamb. 2004. Escaping from the cell: assembly and budding of negative-strand RNA virus. Curr. Top. Microbiol. Immunol. 283:145-196. [DOI] [PubMed] [Google Scholar]
- 38.Shizuya, H., B. Birren, U. J. Kim, V. Mancino, T. Slepak, Y. Tachiiri, and M. Simon. 1992. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl. Acad. Sci. USA 89:8794-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun, X., G. Zhang, Z. Zhang, Z. Hu, J. M. Vlak, and B. M. Arif. 1998. In vivo cloning of Helicoverpa armigera single nucleocapsid nuclear polyhedrosis virus genotypes. Virol. Sin. 13:83-88. [Google Scholar]
- 40.Vaughn, J. L., R. H. Goodwin, G. J. Tompkins, and P. McCawley. 1977. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro 13:213-217. [DOI] [PubMed] [Google Scholar]
- 41.Wang, H., F. Deng, G. P. Pijlman, X. Chen, X. Sun, J. M. Vlak, and Z. H. Hu. 2003. Cloning of biologically active genomes from a Helicoverpa armigera single-nucleocapsid nucleopolyhedrovirus isolate by using a bacterial artificial chromosome. Virus Res. 97:57-63. [DOI] [PubMed] [Google Scholar]
- 42.Waning, D. L., C. J. Russell, T. S. Jardesky, and R. A. Lamb. 2004. Activation of a paramyxovirus fusion protein is modulated by inside-out signaling from the cytoplasmic tail. Proc. Natl. Acad. Sci. USA 101:9217-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westenberg, M., H. Wang, W. F. IJkel, R. W. Goldbach, J. M. Vlak, and D. Zuidema. 2002. Furin is involved in baculovirus envelope fusion protein activation. J. Virol. 76:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westenberg, M., F. Veenman, E. C. Roode, R. W. Goldbach, J. M. Vlak, and D. Zuidema. 2004. Functional analysis of the putative fusion domain of the baculovirus envelope fusion protein F. J. Virol. 78:6946-6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyma, D. J., A. Kotov, and C. Aiken. 2000. Evidence for a stable interaction of GP41 with Pr55 (Gag) in immature human immunodeficiency virus type 1 particles. J. Virol. 74:9381-9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zanotto, P. M., B. D. Kessing, and J. E. Maruniak. 1993. Phylogenetic interrelationships among baculoviruses: evolutionary rates and host associations. J. Invertebr. Pathol. 62:147-164. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao, H., B. Lindqvist, H. Garoff, C. H. von Bonsdorff, and P. Liljestrom. 1994. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 13:4204-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]