Abstract
Picornaviridae are a large virus family causing widespread, often pathogenic infections in humans and other mammals. Picornaviruses are genetically and antigenically highly diverse, with evidence for complex evolutionary histories in which recombination plays a major part. To investigate the nature of recombination and selection processes underlying the evolution of serotypes within different picornavirus genera, large-scale analysis of recombination frequencies and sites, segregation by serotype within each genus, and sequence selection and composition was performed, and results were compared with those for other nonenveloped positive-stranded viruses (astroviruses and human noroviruses) and with flavivirus and alphavirus control groups. Enteroviruses, aphthoviruses, and teschoviruses showed phylogenetic segregation by serotype only in the structural region; lack of segregation elsewhere was attributable to extensive interserotype recombination. Nonsegregating viruses also showed several characteristic sequence divergence and composition differences between genome regions that were absent from segregating virus control groups, such as much greater amino acid sequence divergence in the structural region, markedly elevated ratios of nonsynonymous-to-synonymous substitutions, and differences in codon usage. These properties were shared with other picornavirus genera, such as the parechoviruses and erboviruses. The nonenveloped astroviruses and noroviruses similarly showed high frequencies of recombination, evidence for positive selection, and differential codon use in the capsid region, implying similar underlying evolutionary mechanisms and pressures driving serotype differentiation. This process was distinct from more-recent sequence evolution generating diversity within picornavirus serotypes, in which neutral or purifying selection was prominent. Overall, this study identifies common themes in the diversification process generating picornavirus serotypes that contribute to understanding of their evolution and pathogenicity.
The positive-stranded RNA virus family Picornaviridae comprises a diverse range of mammalian viruses classified into a total of nine genera that differ in host species, disease associations, and persistence (57). Members of Picornaviridae form small (30-nm) nonenveloped icosahedral virus particles comprising the structural proteins VP1 to VP4 assembled as pentameric subunits. Genome sizes range from approximately 7,000 to 8,800 bases, and genomes contain a single reading frame encoding a polyprotein that is posttranslationally cleaved into structural and nonstructural proteins (2A to 2C and 3A to 3D). Some picornaviruses have an additional leader (L) protein before the start of the structural gene region. Untranslated regions (UTRs) at the 5′ and 3′ ends of the genome have roles in virus RNA replication and translation.
Picornaviruses infecting humans comprise members of the Enterovirus, Rhinovirus, Parechovirus, Kobuvirus, and Hepatovirus genera. Other genera infecting domestic animals and rodents include foot-and-mouth disease virus (FMDV) and equine rhinitis A virus (ERAV) in the Aphthovirus genus and porcine enterovirus (Teschovirus genus), equine rhinitis B virus (Erbovirus genus), and Theiler's virus (Cardiovirus genus). Subgroupings within genera comprise species, such as species A to D among human enteroviruses (HEVs), and a further division of variants into serotypes. These possess highly specific susceptibilities to antibody-mediated neutralization and can be identified using panels of serotype-specific antisera (34). Serological identification is important because immunity from past infection or immunization is serotype specific and does not confer protection from infection with heterologous serotypes. Because neutralizing antibody is targeted to the virus capsid, serological classification is reflected by nucleotide (nt) and inferred amino acid sequence divergence in the VP1 region and other structural proteins (9, 39). The availability of nucleotide sequences from each of the classified human enterovirus serotypes now provides an alternative to cross-neutralization for virus identification, notwithstanding the fact that the classification remains serological in origin.
Nucleotide sequences of human enteroviruses are known to be predictive of serotype only in structural gene regions, and it is increasingly recognized that recombination has frequently occurred in their evolution (2, 11, 12, 14, 18, 31, 38, 41, 43, 43, 50, 51, 55). For example, comparison of complete genome sequences of echovirus 18 (EV18) provided evidence for a recombination event with EV9 with a breakpoint in the 2C protein (2). Recombination breakpoints been detected for coxsackievirus group B4 (CVB4), EV11, and EV19 sequences (30, 32) and in larger analyses at several positions in P2 and P3 among naturally occurring enteroviruses and poliovirus vaccine strains (14, 51). Uncoupling of sequence phylogenies between different parts of the genome has led to the concept of semi-independent evolution of structural and nonstructural regions (31, 40). In this model, enteroviruses circulating worldwide are conceptualized as a population of serologically distinct capsid gene sets recombining with a range of nonstructural genes with only transient epidemiological linkage between genome regions associated with episodic spread of individual recombinant forms. Through sampling enterovirus isolates collected over various time periods in the same geographical area, we demonstrated that HEV-A and HEV-B showed high time-correlated frequencies of recombination, where the circulation of individual recombinants within a human population was highly restricted, often to periods of 2 years or less in the case of HEV-B serotypes (55).
Recombination breakpoints between enterovirus serotypes have been found to be largely restricted to nonstructural regions of the genome, such as P2 and P3, and between the 5′ UTR and the capsid-encoding region (14, 42, 50, 51, 55). What limits the occurrence of recombination within the structural region is currently unexplained although much speculated on (31, 42, 55). While human enteroviruses remain the best-studied picornavirus group for recombination, it has become evident that changes in phylogeny relationship between serotypes in different regions of FMDV also occur, providing evidence for recombination in the evolution of serotypes within this virus group and potentially more widely in the picornavirus family (10).
The current study was designed to investigate the relationship between sequence phylogenies of other genera of picornaviruses as well those of as other positive-stranded mammalian RNA viruses in different genome regions with their serologically or genetically assigned classifications. Further analyses determined whether breakdown of segregation in nonstructural regions resulted from recombination and what other aspects of sequence diversity, selection pressures, and composition variables were associated with the phenomenon. This study identifies common trends and similarities in the underlying dynamics of, constraints on, and possible mechanisms for recombination in picornaviruses and other nonenveloped viruses that contrast with those for other mammalian positive-stranded RNA viruses.
MATERIALS AND METHODS
Nucleotide sequence alignments.
All sequence alignments were created between February and April 2006 using all available full-length genome sequences of different genera within the picornaviruses, as well as the majority of other available mammalian positive-stranded viruses as controls. Sequence annotations were screened to exclude patents, artificial mutants, and identical entries. Sequences were aligned using nucleotide- and codon-based methods implemented in ClustalW for noncoding and coding regions of the genome run within the Simmonic editor package (available from http://www.virus-evolution.org). Identical sequences or those showing less than 1% sequence divergence from each other were excluded. Virus groups selected for segregation and recombination analysis were those that could be unambiguously aligned, contained 10 or more different variants, two or more sero-/genogroups, and at least one group with more than one member (HEV-A, HEV-B, HEV-C, FMDV serotypes A to O and SAT1 to SAT3, and teschoviruses; Tables 1 and 2).
TABLE 1.
Virus sequence alignments
| Virusa | Subset | No. of sequencesb | Length (nt)c | No. of groupsd | Divergencee
|
|
|---|---|---|---|---|---|---|
| Intergroup | Intragroup | |||||
| Picornaviridae | ||||||
| FMDV | A-O | 103 | 8,386 | 4 (S) | 0.176 | 0.105 |
| SAT | 18 | 8,386 | 3 (S) | 0.220 | 0.139 | |
| Enterovirus | HEV-A | 28 | 7,656 | 3 (S) | 0.301 | 0.187 |
| HEV-C | 62 | 7,656 | 8 (S) | 0.277 | 0.210 | |
| HEV-C | 51 | 7,656 | 0.334 | 0.145 | ||
| Teschovirus | 31 | 7,127 | 11 (S) | 0.212 | 0.171 | |
| Flaviviridae | ||||||
| Flavivirus | JEV group | 30 | 11,117 | 3 (S) | 0.409 | 0.118 |
| Dengue virus | 83 | 10,787 | 4 (S) | 0.337 | 0.065 | |
| YFV | 7 | 10,862 | 4 (G) | 0.218 | 0.032 | |
| TBE group | 12 | 11,141 | 3 (S) | 0.365 | 0.138 | |
| HCV | 127 | 9,519 | 6 (G) | 0.437 | 0.189 | |
| HGV/GBV-C | 37 | 9,476 | 4 (G) | 0.144 | 0.113 | |
| Pestivirus | 44 | 12,596 | 4 (G/S) | 0.433 | 0.142 | |
| Alphaviridae | 47 | 10,707 | 4 (S) | 0.665 | 0.146 | |
| Caliciviridae | Noroviruses | 24 | 8,071 | 16 (S) | 0.618 | 0.039 |
| Astroviridae | Human | 10 | 6,882 | 6 (S) | 0.233 | 0.088 |
A full listing of sequences in each alignment is available from the author.
Number of sequences in alignment.
Length of alignment (including alignment gaps).
Number of predefined groups. The classification method is indicated in parentheses. S, serologically defined; G, genotypic classification.
Mean pair-wise Jukes-Cantor-corrected distance between and within groups of complete genome sequences.
TABLE 2.
Other picornavirus genera
| Virusa | Subset | No. of sequencesb | Length (nt)c | Groupsd | Divergencee |
|---|---|---|---|---|---|
| HAV | 28 | 7,504 | 1 | 0.166 | |
| ERAV | 8 | 7,734 | NK | 0.109 | |
| Kobuvirus | 3 | 8,283 | NK | 0.090 | |
| HRV-A | 5 | 7,656 | 5 (S) | 0.332 | |
| Erbovirus | 2 (11) | 8,817 | 10 (G) | 0.311 | |
| Parechovirus | 5 | 7,329 | NK | 0.246 | |
| Enterovirus | BEV | 10 | 7,656 | 10 (G) | 0.240 |
| Cardiovirus | EMCV | 4 | 8,101 | NK | 0.197 |
| Theiler's virus | 4 | NK | 0.225 |
A full listing of sequences in each alignment is available from the author.
Number of sequences in alignment. Eleven sequences were available from the capsid gene of erboviruses and were included in the analysis.
Length of alignment (including alignment gaps).
Number of predefined groups. The classification method is indicated in parentheses. S, serologically defined; G, genotypic classification; NK, not known.
Mean pair-wise Jukes-Cantor-corrected distance between groups of complete genome sequences.
For recombination analysis, each data set contained an outgroup. Where possible, this was a more divergent sequence that could still be defensibly aligned with the remaining sequences (e.g., the poliovirus Leon sequence for alignments of HEV-A and HEV-B). Where such a sequence was not available (e.g., for hepatitis C virus [HCV]), a 50% consensus sequence was generated from a representative sequence from each assigned sero-/genogroup. Phylogenetic trees were constructed to ensure that the 50% consensus sequences occupied an ancestral group/outgroup position (data not shown).
Classification into serotypes and genotypes.
Classification of sequences within picornavirus genera followed published divisions of sequences into serotypes. For FMDV, published sequences were initially split into two groups containing serotypes A to O and SAT serotypes 1 to 3 and were analyzed separately. Enteroviruses were split into HEV-A, HEV-B, and HEV-C, and variants within each were assigned according to their serotypes. Teschovirus groups corresponded to the 11 serotypes described in the published classification (68).
Virus groups containing individual serotypes represented by only single examples or which lacked defined variants at the serotype level of divergence were excluded from recombination and segregation analyses (listed in Table 2). For these viruses, analysis was restricted to determination of sequence divergence and sequence composition. Virus groups analyzed in this way were HAV, ERAV, and kobuviruses, all of which show restricted diversity in the capsid-encoding region. The more divergent erboviruses, parechoviruses, and human rhinovirus A major group (HRV-A) were all virus groups that lacked multiple examples of individual serotypes or proposed serotypes. Bovine enterovirus (BEV) was originally classified as a separate species within the Enterovirus genus, with individual variants originally classified into two serotypes (8). However, these two groups are highly divergent, more typical of sequence divergence between species in other enterovirus groups (69). I therefore analyzed the two groups separately. Similarly, Theiler's virus and encephalomyocarditis virus (EMCV)/Mengo virus were split as species within the Cardiovirus genus, and variants within each were analyzed as separate groups.
For the flaviviruses, HCV was divided into genotypes 1 to 6, with a 50% consensus sequence as an outgroup. Pestiviruses were classified into four groups: bovine viral diarrhea virus types 1 and 2, border disease virus, and swine vesicular disease virus. Because nomenclature was inconsistent in published annotations, these assignments were verified by phylogenetic analysis. Hepatitis G virus (HGV)/GB virus C (GBV-C) variants were split into four groups, genotypes 1 to 4 (56), and the chimpanzee-derived sequence AF070476 was used as an outgroup. Compete genome sequences within the more divergent Flavivirus genus were divided into dengue viruses (four serotypes), the Japanese encephalitis virus (JEV) group (including West Nile virus [WNV]/Kunjin virus, and Murray Valley encephalitis virus; three serotypes), and the tick-borne encephalitis virus (TBE) group (including Powassan and Alkhurma serotypes). Finally, complete genome sequences of yellow fever virus (YFV) were classified into genotypes as previously described (35). Consensus sequences (50%) were used as outgroups in each.
Alphaviruses were divided into four groups (Semliki Forest virus, Sindbis virus, Venezuelan equine encephalitis virus, and Eastern equine encephalitis virus), and the more divergent Barmah Forest virus was used as an outgroup. Viruses in the Norovirus genus of Caliciviridae were classified into 14 serotypes falling into two genogroups (3), and results from groups I and II were combined. Finally, available complete genome sequences of human astroviruses were labeled according to their previously described serotype designations (37).
Recombination analysis.
The detection of recombination was achieved through generating phylogenetic trees from different region of viral genomes and examining them for incongruities in tree structure. This process was automated in the program TreeOrder Scan implemented in the Simmonic Sequence Editor version 1.5 (54, 55). Bootstrapped phylogenetic trees were generated by the programs SEQBOOT, DNADIST, NEIGHBOR, and CONSENSE in the PHYLIP v3.62 or v3.65 package and produced by successively generated sequence fragments from alignments of complete genome sequences of each species. Trees were compared after the sequences were reordered to match branching order as closely as possible; reordering of trees included branch rotation and movement of sequences or sequence groups, provided this occurred within groupings below the specified bootstrap threshold value (70% for all the analyses shown in this study). The compatibility of one tree with another was then computed by measurement of the number of times the phylogeny of one tree had to be violated (i.e., transfer of a sequence or group of sequences between different bootstrap-supported clades) to match tree orders. For the analysis described in this study, a bootstrap value of 70% has been used as the threshold for scoring phylogeny violations, as this is frequently used for assigning “robust” support for phylogenetic groups (24). Phylogenetic compatibility was computed separately for violations of tree order of sequences between predefined groups (group events) and for those occurring between sequences within groups (intragroup events).
Frequencies of phylogeny violations were analyzed by reference to the position in the genome where they occurred and also as summary values for a subgenomic region of a virus alignment. For the former, frequencies of phylogenetic violations were plotted for each pair-wise comparison of trees in the form of a half-diagonal matrix, where the x and y coordinates record the positions of the fragments from which the trees were obtained, and the “heat” scale shows violation frequencies. Because the latter quantity is influenced by the size and phylogenetic informativeness of each fragment, phylogeny violations were normalized by dividing by the minimum number of clades in the two trees being compared; regions without clades at the specified bootstrap level were excluded from analysis.
A further calculation was carried out to estimate the total number of phylogeny violations between or within preassigned groups with a particular genomic or subgenomic region (such as the structural gene region). This represents the sum of phylogeny violations upon comparison of each fragment with every other fragment divided by the expected representation of a phylogeny violation within the total set of pair-wise comparisons (1.5 times the number of comparisons/number of fragments). This figure was further normalized to enable comparison of data sets of different numbers and lengths of sequences and levels of phylogenetic informativeness by calculation of frequencies of phylogeny violations per thousand bases per bootstrap-supported clade.
Segregation analysis.
The segregation analysis method was used to investigate the extent to which a virus classification (such as into serotypes or genotypes) was congruent with the phylogeny of the nucleotide sequences within different regions of an alignment. Individual sequences were first labeled according to their designated serotypes/genotypes, and phylogenetic trees were constructed from sequentially generated fragments though the alignment (300 bases in length, incrementing by 50 bases). The order of sequences within a tree was permuted randomly to the extent permitted by the phylogeny of the sequences. Therefore, only sequences within clades with bootstrap support above the prespecified threshold(s) will remain grouped together in the output sequence order. This method is effective at identifying phylogenetically informative regions of a sequence alignment (where sequences of the same species or genotype group together); in regions without phylogenetic information or phylogenetic relationships that conflict with their assigned group, these groups will become randomly or differently dispersed in tree order. In the current study, the output from this analysis was plotted with the x axis representing the midpoint of each sequence fragment and the position in the tree represented on the y axis. Genotypes or species were color coded to allow direct visualization of the relationship between sequence group and tree position.
The correspondence between the order of the sequences in the tree with their geno-/serogroup designations was formally quantified by counting the number of type transitions between sequences in the list order of the tree. Expected values for a perfectly segregated tree where sequences grouped by serotype would therefore correspond to one less than the number of serotypes in the data set, while the number in a tree where there was no relationship between serotype and phylogeny (i.e., randomly distributed in the tree) would lead to a much larger number of label transitions, depending on and computable from the number of assigned groups and numbers of members within each. Segregation values were expressed as values between 0% and 100% within the scale corresponding to these opposed outcomes.
Sequence diversity.
Sequence diversity was computed as mean pair-wise Jukes-Cantor (J-C)-corrected distances between nucleotide sequences or p distances for amino acid sequences between and within preassigned groups (intra- and intergroup divergence, respectively). For scanning variability across a genome, mean pair-wise J-C distances or p distances for amino acid sequences were calculated for successive 300-base fragments, incrementing by 50 (or 48 bases for amino acid distances) between fragments. Mean values for pair-wise comparison of sequences within subgenomic regions were also computed for comparison of sequence divergence in structural and nonstructural protein-encoding regions. For analysis of selection pressure on coding sequences, mean pair-wise J-C-corrected distances between preassigned groups were calculated separately at nonsynonymous and synonymous sites to calculate nonsynonymous (dN)/synonymous (dS) ratios. All computations were automated using scripts with the Simmonic2005 version 1.5 Sequence Editor.
Composition analysis.
Sequence compositions of structural and nonstructural coding regions of each virus were determined using the Simmonic2005 version 1.5 Sequence Editor. For each set of sequences, mean frequencies of mononucleotides (A, C, G, and U), of base combinations (i.e., S [G + C], R [A + G], and K [G + U]), and of dinucleotides were tabulated at each codon position separately and combined. Further columns recorded the ratios of observed dinucleotide frequencies to those expected from mononucleotide base frequencies (i.e., the frequency of AA [fAA] would be expected to correspond to fA · fA [61]). Dinucleotide frequencies were further normalized by comparison of observed values to those expected from the amino acid usage of the nucleotide sequence. For example, this procedure would be able to correct for overrepresentations of the frequencies of GG, GN (where N is any base), and NG dinucleotides at codon positions 1, 2, and 3 that resulted from an excess of glycine codons in the sequence.
For analysis of codon usage, sets of relative synonymous codon usage (RSCU) (53) values were calculated for each codon specifying a particular amino acid. RSCU values do not correct for base composition biases that would naturally skew codon usage. Therefore a new method was developed; by this new method, total (i.e., irrespective of direction) and net deviations of base frequencies away from expected ratios reflecting base composition at third or first codon positions were calculated individually for each amino acid to produce bias and variance values. Bias and variance values were also calculated for each base and base-pair combination (A, C, G, U, S, W, and K); these represent total and net deviations of frequencies of each base (pair) at synonymous positions from those expected from overall base composition of the sequence at the appropriate codon position. Finally, the value for the effective number of codons (ENc) was calculated for each coding sequence as previously described (66a). These values were compared to values expected from the sequences based on the G+C content of third codon positions.
RESULTS
Concordance between serotype classification and phylogeny in different genome regions.
The initial step of the investigation was to determine how well phylogenetic relationships between nucleotide sequences from different parts of the genomes of picornaviruses and other positive-stranded viruses correlated with their classification into different sero-/genogroups. For picornaviruses, initial analysis concentrated on human enterovirus, FMDV, and teschovirus genera, because each contained groups of sequences containing multiple serotype groups and multiple sequences within each to enable segregation analysis (Table 1). Group assignment for the segregation analysis comprised the division of HEV-A, HEV-B, and HEV-C into serotypes. Teschoviruses were split into 11 serotypes as previously described (68), while complete FMDV genome sequences were first divided into separate A-to-O and SAT sequence data sets and then labeled into serotypes A to O and SAT1 to SAT3. For comparison, members of other positive-stranded RNA virus families, both enveloped (HCV, HGV/GBV-C, dengue virus, and JEV/WNV groups of flaviviruses and alphaviruses) and nonenveloped (noroviruses in the family Caliciviridae and human astroviruses), were similarly divided into standard serologically or genetically defined groupings (see Materials and Methods) (Table 1). The final two columns of Table 1 record the mean pair-wise distances between and within each of the previously defined sero/genogroups.
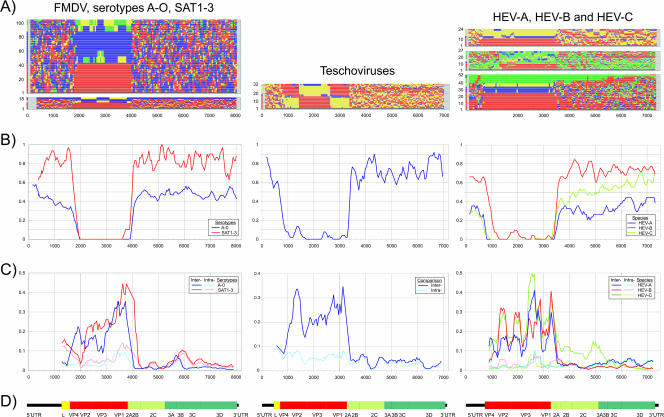
The extent to which segregation into these defined groups corresponded with nucleotide sequence relationships in different regions of the genome was determined using the program TreeOrder Scan (Fig. 1) (see Materials and Methods). The representation can be thought of as a stack of trees generated from sequential sequence fragments viewed “end on,” where color coding records the positions of sequences within the tree of each of the predefined groups. Viruses within all three picornavirus genera showed marked differences in phylogeny in different regions of the genome. Both groups of FMDV viruses, human enteroviruses, and teschoviruses all showed phylogenies congruent with their serological classification only in the capsid-encoding region of the genome, where phylogenetic trees recreated the order of sequences by their serological classification. Elsewhere, this congruence broke down, often almost entirely, as indicated by the scatter of serotype labels within phylogenetic trees. For each of the virus groups examined, the structural gene region delineated a sharp transition zone between congruent and noncongruent phylogenies. The leader proteins of FMDV and teschoviruses were as noncongruent as downstream nonstructural regions of the genome.
FIG. 1.
(A) Ordering (y axis) of variants assigned to different serotypes in phylogenetic trees generated from consecutive 300-base fragments across complete genome sequence alignments of aphthoviruses (left), teschovirus (middle), and human enteroviruses (right). Serotypes are color labeled as follows. For serotypes A to O of FMDV, serotype A is red, B is yellow, C is green, and O is blue; for SAT serotypes, SAT-1 is red, SAT-2 is yellow, and SAT-3 is blue; and for teschoviruses, serogroup 1 is red (serotypes 1, 3, 10, and 11), serogroup 2 is yellow (serotypes 9, 7, and 5), and serogroup 3 is blue (serotypes 6, 2, 4, and 8). For human enteroviruses, the following serotypes were labeled. For HEV-A, enterovirus 71 is red, CAV-16 is blue, and others are yellow; for HEV-B, echovirus 11 is red, echovirus 9 is blue, CBV-5 is yellow, and others are green; for HEV-C, PV1 is red, PV2 is blue, PV3 is green, and others are blue. (B) Segregation scores for consecutive fragments across genomes, where 0 (y axis) represents perfect phylogenetic segregation by assigned group (serotypes) and 1 represents the absence of association between phylogeny and group assignment. Similar results for slightly different sequence subsets of enteroviruses HEV-A to HEV-C have been presented previously (55) and are included for comparison. (C) Mean pair-wise amino acid sequence distances between sequences in consecutive 300-base fragments across the genome. Values are averaged over a window size of 3. Separate mean values were calculated for sequence comparisons within and between groups (pale and dark colors). (D) Genome diagrams of FMDV, teschoviruses, and human enteroviruses drawn to scale and numbered according to the reference sequences NC_011450 for FMDV, PEN011380 for teschoviruses, and POL3L37 for enteroviruses.
The degree of randomness in order was quantified through the use of the segregation scan selectable in the TreeOrder program. This counts the number of serotype label transitions between sequences in the list order of a tree. Expected values for a perfectly segregated tree where sequences grouped by serotype would therefore correspond to one less than the number of serotypes in the data set, while the number in a tree where there was no relationship between serotype and phylogeny (i.e., randomly distributed) would be much larger, depending on the number of assigned groups and numbers of members within each. Actual segregation values from the different data sets can be plotted as segregation values from 1 (random) to 0 (follows group assignment) based on these opposed outcomes (Fig. 1, row B). For the capsid-encoding region, segregation values were 100% or close to 100% for all three virus groups but declined to much lower levels abruptly on either side of this region. HEV-B and teschoviruses showed minor deviations from complete segregation in the capsid region.
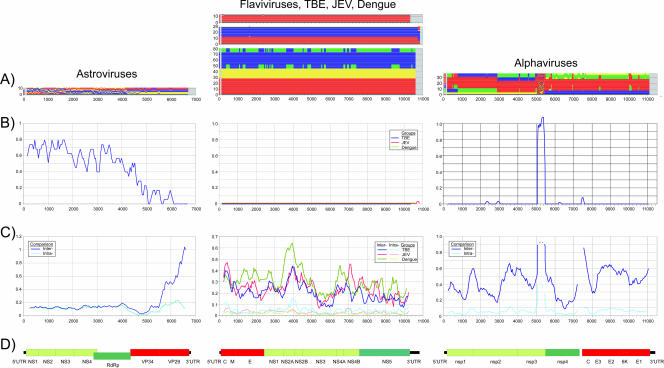
Relationships between serotype/genotype classifications and phylogenies in different regions of the genome were subsequently examined for other mammalian positive-stranded RNA viruses (listed in Table 1). In contrast to the picornaviruses described above, each virus group within the Flaviviridae and alphaviruses retained their segregation into assigned sero- or genogroups/-types throughout the nonstructural regions (Fig. 2), with at most minor incompatibilities in tree order and their group assignments. In the segregation scans for different flavivirus groups, sequences classified into different serotypes (color coded in Fig. 2, middle column of row A) consistently grouped together upon phylogenetic analysis, with changes in branching order restricted to those between whole groups (e.g., between dengue virus serotypes 2 and 4 [blue and green]). Apart from the split in the JEV variants in the last genome fragment (3′ UTR), segregation values were consistently zero throughout the length of the genomes (Fig. 2, middle, row B), distinct from the high values observed for nonstructural regions for picornaviruses (Fig. 1). Similarly, alphaviruses (Fig. 2, right column) showed almost entirely consistent grouping by serotype classifications in both nonstructural and structural regions, except for a 400-nucleotide region at the end of nsp3, corresponding to the hypervariable domain 3 (59), whose extreme amino acid and length variability both between and within serotypes prevented the creation of a coherent alignment.
FIG. 2.
Analysis of sequences for astroviruses, serotypes within three groups of flaviviruses, and alphaviruses (as described for FMDV, teschovirus, and enterovirus sequences shown in Fig. 1; see legend for description). Astroviruses are color labeled as follows: serotype 1 is red, serotype 4 is yellow, and others are blue. TBE-like viruses are labeled as follows: TBE is red, Alkhurma virus is blue, and Powassan virus is yellow. JEV-like viruses are labeled as follows: JEV is red, WNV/Kunjin is blue, and Murray Valley encephalitis virus is yellow. Serotypes 1 to 4 of dengue virus are labeled as red, blue, yellow, and green, respectively. Alphaviruses are labeled as follows: Venezuelan equine encephalitis virus, red; Eastern equine encephalitis virus, blue; Semliki Forest virus, yellow; and Sindbis virus, green. Genome diagrams of astroviruses and alphaviruses were numbered according to the reference sequences NC_001944 and ALSFV42S, respectively. For flaviviruses, a genome diagram of dengue virus (based on the annotation for NC_001477) was included (center, row D). Although not precisely aligned, genome lengths and gene boundaries of the TBE and JEV groups approximated those of NC_001477.
In a calculation of mean values for structural and nonstructural gene regions of each of the virus alignments, mean segregation values for the flavivirus and alphavirus groups were always less than 0.1 and frequently were 0 (Table 3). Among these sequence groups, both genetic (HCV and YFV) or serological/neutralization (JEV/WNV and dengue virus groups, pestiviruses, and alphaviruses) classifications remained congruent with sequence phylogenies (Table 3). Genetic segregation was investigated further through calculation of association index (AI) values for the equivalent genome regions for each of the virus groups (Table 3, right three columns). This independent phylogenetic method scores the degree of segregation of preclassified sequences in bootstrap-replicated phylogenetic trees by calculating an index value representing the ratio of segregation of native sequences to that of replicate sequence sets whose classification has been randomly reassigned (64). Outcomes ranged from 1 (no association between preassigned grouping and phylogeny) to 0 (complete congruence between classification and phylogeny). AI values correlated closely with segregation values, in particular identifying which genome regions were being segregated or not, in a manner almost identical to the segregation scan (Table 3).
TABLE 3.
Segregation and association index values for different genome regions
| Virus | Subset | Segregation value fora:
|
AI value for:
|
||||
|---|---|---|---|---|---|---|---|
| U | S | NS | U | S | NS | ||
| Picornaviridae | |||||||
| FMDV | A-O | 0.824 | 0.007 | 0.473 | 0.193 | 0 | 0.208 |
| SAT | 0.453 | 0.022 | 0.806 | 0.863 | 0.001 | 0.681 | |
| Enterovirus | HEV-A | 0.327 | 0 | 0.301 | 0.012 | 0 | 0.313 |
| HEV-B | 0.649 | 0.032 | 0.717 | 0.700 | 0.022 | 0.849 | |
| HEV-C | 0.456 | 0 | 0.543 | 0.137 | 0.037 | 0.388 | |
| Teschovirus | 0.741 | 0.033 | 0.707 | 0.519 | 0 | 0.440 | |
| Flaviviridae | |||||||
| Flavivirus | JEV group | 0 | 0 | 0 | 0 | ||
| Dengue virus | 0 | 0 | 0 | 0 | |||
| YFV | 0 | 0 | 0 | 0 | |||
| TBE Group | 0 | 0 | 0 | 0 | |||
| HCV | 0.114 | 0.032 | 0.031 | 0.081 | 0.001 | 0 | |
| HGV/GBV-C | 0.046 | 0.091 | 0.054 | 0.063 | 0 | 0.001 | |
| Pestivirus | 0 | 0 | 0.001 | 0 | 0 | 0 | |
| Alphaviridae | 0.024 | 0.098 | 0.007 | 0.063 | 0.002 | ||
| Caliciviridae | Norovirus | 0 | 0 | 0.183 | 0.107 | 0.089 | 0.253 |
| Astroviridae | Human | 0.103 | 0.641 | 0.021 | 0.609 | ||
Boldface indicates values of >0.15. S, structural; NS, nonstructural; U, upstream region from the start of the structural coding region of the genome (including leader protein sequences of FMDV and teschovirus sequences). For noroviruses, U corresponds to open reading frame 3.
While the lack of segregation of serotype classification with sequence phylogenies of nonstructural regions was found predominantly among genera in the picornavirus family, a similar phenomenon was also found among astrovirus sequences (left column of Fig. 2; Table 3), where both segregation and AI values for sequences classified by serotype were low for the NS gene (segregation value, 0.103; AI value, 0.021) but were high (indicating nonsegregation) in the upstream NS region (values of 0.641 and 0.609). Segregation values calculated for sequential 300-base fragments along the genome showed a transition from high to intermediate values at the amino terminus of the first structural protein, VP34, and a subsequent decline to values from zero to near zero for VP29. Breakdown of segregation was also observed to a lesser degree in the nonstructural region of noroviruses.
Segregation and recombination.
The observed loss of segregation between serotypes in phylogenetic trees based on nonstructural region sequences among picornaviruses and astroviruses may have occurred through two mechanisms. As one possibility, it may have arisen through the absence of any phylogenetic signal among nonstructural region sequences, possibly resulting from extensive homoplasy from multiple, saturating substitutions at a restricted number of unconstrained sites. Alternatively, it is possible that nonstructural region phylogenies were informative but that they conflicted with sequence relationships in the structural gene region that determines the serological properties, and therefore the classification, of the virus. Conflicting phylogenies may arise through one or more recombination events between nonstructural regions of different serotypes.
To distinguish between these possibilities, frequencies of phylogeny violations were calculated between trees constructed from structural and nonstructural regions of the genome. Detection of frequent phylogeny violations would provide evidence for incompatibilities between phylogeny relationships between different genome regions, consistent with recombination. Alternatively, low frequencies of phylogeny violations would be expected in genome regions that lacked segregation into sero-/genogroups if it arose through a lack of a phylogenetic signal. For the current study, phylogenetic trees were constructed from 300-base sequential fragments incrementing by 50 bases through the structural and nonstructural regions of each virus group listed in Tables 1 and 2. Values for phylogeny violations represent the number of times the phylogeny of one tree had to be violated (i.e., transfer of a sequence or group of sequences between different bootstrap-supported clades) to match tree orders, using a bootstrap value of 70%.
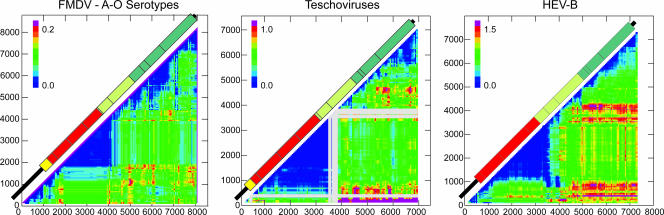
For the three picornavirus genera that lacked phylogenetic segregation into serotypes in nonstructural regions (nonsegregating viruses) (Fig. 1), normalized values for each pair-wise comparison of sequence fragments were plotted as half-diagonal matrices (Fig. 3). Sequence violations invariably occurred at low frequency in or were absent from the region bounded by the capsid-encoding region (dark blue color) and coincided closely with regions that showed segregation by serotype (Fig. 1). In contrast, phylogenetic incongruities between trees generated from structural and nonstructural regions, and between different trees within nonstructural regions for all three virus groups analyzed, were frequently detected.
FIG. 3.
Phylogenetic compatibility between different genome regions of aphthoviruses, teschoviruses, and HEV-B. Matrices show phylogenetic compatibility scores between trees generated from consecutive 300-base fragments of genome alignments of each virus group. Frequencies of phylogeny violations required to order trees generated from each pair-wise comparison of fragments (x and y axes) were recorded in color (legend), using a 60% bootstrap value to define clades. Phylogenetically compatible regions are shown in deep blue; gray-shaded regions indicate regions of the genome lacking any bootstrap-supported clades. Genome diagrams were aligned with the matrix to enable regions of phylogenetic compatibility to be matched to specific genome regions. For other nonsegregating virus groups, FMDV SAT serotypes produced a matrix similar to that for serotypes A to O (data not shown), while matrices for human enteroviruses HEV-A and HEV-C have been presented elsewhere (55). A similar result for a slightly different sequence subset of HEV-B has been presented previously (55) and is included here for comparison with other picornavirus genera.
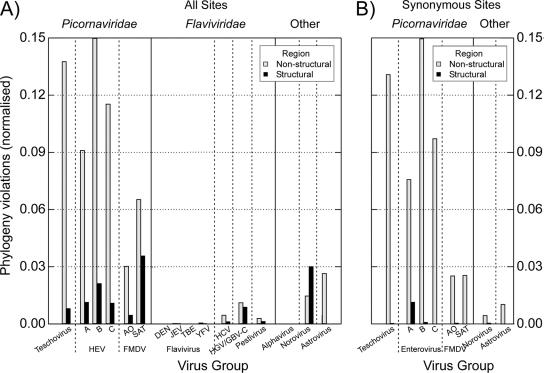
This analysis was extended for the rest of the virus groups, where mean frequencies of phylogeny violations within structural and nonstructural regions were calculated separately (Fig. 4). Because the virus data sets differed in terms of size and the degree of segregation, frequencies of phylogeny violations were normalized for the length of the region analyzed, the number of sequences in the data set, and the number of phylogenetically distinct clades at the selected bootstrap value. As predicted by the analysis above, teschovirus, enterovirus, and FMDV groups showed high degrees of phylogenetic incompatibility between trees constructed from different sequence fragments in the nonstructural region (Fig. 4). In contrast, very low normalized violation values were generally observed in the structural region of each virus genus or group, indicating that trees constructed from different parts of the capsid-encoding region were substantially compatible with each other. Astroviruses also showed high normalized values in the nonstructural region (≈0.03), while noroviruses showed some evidence for phylogenetic incompatibility in both structural and nonstructural regions. In marked contrast to these nonsegregating viruses, parallel analysis of flaviviruses and alphaviruses that showed consistent phylogeny relationships across the genome (segregating viruses) generally revealed no phylogeny violations in either nonstructural or structural genome regions. The only exception was HGV/GBV-C, where phylogeny violations were observed in both structural and nonstructural regions.
FIG. 4.
(A) Normalized frequencies of phylogeny violations in structural and nonstructural regions of picornaviruses, flaviviruses, alphaviruses, noroviruses, and astroviruses. Neighbor-joining phylogenetic trees were created from consecutive pair-wise fragments of alignments of each virus group, and violation scores were calculated for each pair-wise comparison, using a bootstrap value of 70% to define clades. Values (y axis) are expressed as calculated frequencies of phylogeny violations in each data set per thousand bases per phylogenetically defined clade (see Materials and Methods for an explanation of the normalization procedure). (B) Repeated analysis using trees constructed from synonymous distance matrices, representing phylogeny violations between clades defined by synonymous site variation.
The analysis of virus alignments showing high frequencies of intraregional phylogeny violations was repeated using phylogenetic trees reconstructed from synonymous changes (i.e., silent, non-amino changing) only (Fig. 4B). The similarity in results between this and the previous analysis of sequence changes at all sites ruled out the possibility that positive selection mechanisms, such as convergent selection for specific amino acid residues in regions under immune or receptor use selection, were responsible for differences in phylogeny between genome regions.
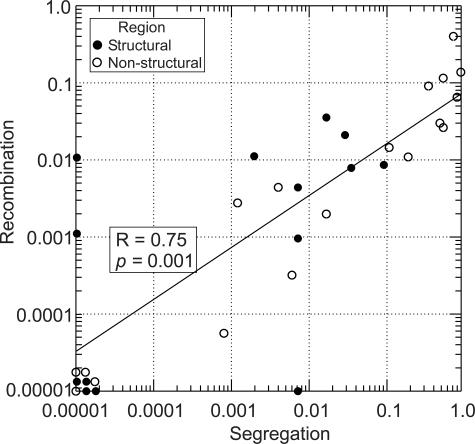
High frequencies of phylogeny violations occurred among the same sequence data sets that showed loss of genetic segregation into sero-/genogroups, with a close correlation between normalized phylogeny violation values and segregation values (Fig. 5). This association provides evidence that conflicting phylogenies, potentially arising through interserotype recombination, rather than lack of phylogenetic differentiation, was largely responsible for the observed abrupt breakdown of segregation into sero-/genogroups in the nonstructural regions of picornaviruses and other nonsegregating viruses (Fig. 1).
FIG. 5.
Association between segregation values for sero-/genotype categories (x axis) and normalized frequencies of phylogeny violations (y axis) for picornaviruses and other virus groups listed in Table 3. The R value for regression was calculated using the nonparametric Spearman rank correlation coefficient test.
Sequence variability.
To gain an insight into the evolutionary mechanisms and selection pressures underlying the frequent disparities in phylogenetic relationships between different genome regions of picornaviruses and astroviruses, nucleotide and inferred amino acid sequence divergence and dN-to-dS substitution frequency ratios were compared between structural and nonstructural regions. Comparisons were carried out separately within and between sero-/genogroups to compare selection processes in recent and more remote periods in the evolutionary history of each virus.
Mean pair-wise distances were calculated for sequential 300-base fragments generated from each virus alignment. For viruses shown to be nonsegregating in the nonstructural region (FMDV, teschoviruses, human enteroviruses, and astroviruses), regions of the genome encoding the capsid proteins showed amino acid sequence variability consistently greater than that for nonstructural and untranslated regions, with sharp transitions in variability at the boundaries of the capsid regions (row C in Fig. 1; also left column of Fig. 2). In contrast, there was little or no consistent difference in amino acid sequence variability between regions among flaviviruses and alphaviruses in which recombination was not detected (row C in Fig. 1; also middle and right columns of Fig. 2). Segregating and nonsegregating viruses also differed in terms of the pattern of sequence diversity between and within sero-/genogroups. For the latter, structural region diversity was much greater between serotypes than between members of the same serotype. In contrast, intra- and intergroup sequence divergences were indistinguishable in nonstructural regions, where the two graph lines generally coincided exactly, consistent with the evidence for the loss of phylogenetic segregation into serogroups in regions outside the capsid. In contrast, the much greater sequence divergence between serotypes of flaviviruses and alphaviruses was maintained throughout the genome (Fig. 2, middle and right columns), with no evidence for greater genetic differentiation of serotypes in structural gene regions observed in nonsegregating viruses.
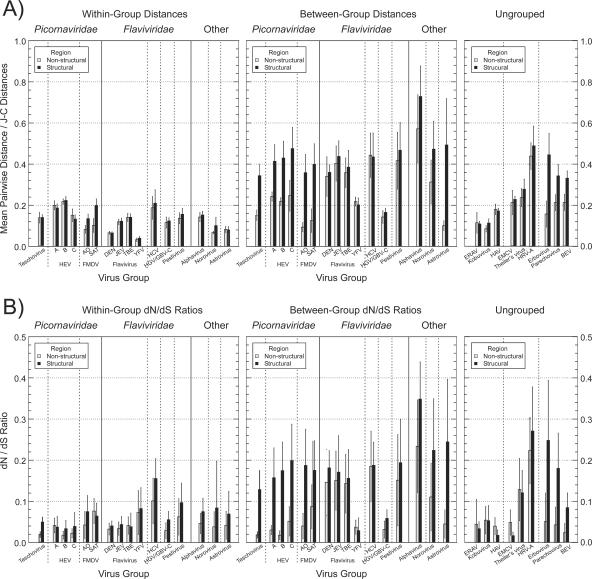
To analyze this phenomenon more systematically for all virus groups, mean pair-wise nucleotide distances and the variance between different fragments within each region were calculated between classification groups and within classification groups (Fig. 6, upper row). Nonsegregating picornaviruses (enteroviruses, FMDV, and teschoviruses) showed consistent differences in their patterns of variability from segregating viruses in flavivirus and alphavirus families. For the former viruses, nucleotide sequence variability was substantially greater in the structural region than in the nonstructural region (ratios of 1.7 to 3.8) (Fig. 6), whereas for segregating viruses, sequence divergence was approximately evenly distributed in the two parts of the genome (range, 0.9 to 1.3). Also, in contrast to the examples shown in Fig. 1 and 2, there was consistently very high sequence divergence in the nonstructural region between groups compared to sequence divergence within groups among the set of segregating viruses (Fig. 6, right panel), consistent with the previous evidence for genetic segregation throughout the viral genome.
FIG. 6.
Mean pair-wise distances (A) and dN/dS ratios (B) of nonsegregating picornaviruses and other virus groups between and within assigned geno-/serogroups (left and middle panels). Corresponding within-group values for other picornavirus genera not analyzed for recombination are shown in the right panels. Bar heights represent mean pair-wise distances and dN/dS ratios of consecutive 300-base fragment sequences within structural and nonstructural regions of each virus group alignment. Error bars show standard deviations between sequence fragments within each region.
Differences in sequence divergence between structural and nonstructural regions were even more marked at the amino acid level (Table 4), with ratios varying from three- to sevenfold among the nonsegregating picornavirus groups (values shown in boldface type). These ratios formed an even greater contrast with the segregating flaviviruses and alphaviruses, where ratios between regions averaged 1.1 (range, 0.7 to 1.4). There was a similar contrast between nonsegregating and segregating viruses in ratios of synonymous to nonsynonymous substitutions in different parts of the genome between groups (Fig. 6; Table 4). Comparing sero-/genogroups, the nonsegregating picornaviruses showed markedly greater dN/dS ratios (2- to 10-fold) in the structural region than in the nonstructural region, whereas for the segregating viruses, ratios between genome regions were invariably similar (ratio range, 0.8 to 1.6). In contrast to the between-group comparisons, there was no correspondingly increased dN/dS ratio for within-serotype comparisons among nonsegregating viruses; variability within each serotype was characterized by low ratios in both genome regions (Fig. 6B).
TABLE 4.
Distance, selection, and codon usage among picornaviruses and other virus families
| Virus | Value for:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid divergence
|
dN/dS
|
ENc
|
||||||||||||||||
| S (%) | NS (%) | Ratioa | S | NS | Ratiob | S | NS | Ratioc | ||||||||||
| Picornaviruses | ||||||||||||||||||
| Nonsegregating | ||||||||||||||||||
| HEV-A | 20.9 | 5.9 | 3.571 | 0.157 | 0.030 | 5.192 | 0.986 | 0.920 | 0.933 | |||||||||
| HEV-B | 24.0 | 3.4 | 7.101 | 0.175 | 0.018 | 9.904 | 0.971 | 0.906 | 0.933 | |||||||||
| HEV-C | 26.9 | 8.8 | 3.041 | 0.205 | 0.051 | 3.991 | 0.958 | 0.874 | 0.912 | |||||||||
| FMDV A-O | 23.8 | 3.1 | 7.577 | 0.187 | 0.040 | 4.675 | 0.996 | 0.904 | 0.908 | |||||||||
| FMDV SAT | 25.9 | 6.3 | 4.108 | 0.175 | 0.087 | 2.013 | 0.970 | 0.896 | 0.924 | |||||||||
| Teschovirus | 22.4 | 3.7 | 6.054 | 0.128 | 0.018 | 7.111 | 0.916 | 0.888 | 0.969 | |||||||||
| Ungrouped | ||||||||||||||||||
| ERAV | 3.4 | 3.7 | 0.911 | 0.033 | 0.059 | 0.565 | 0.926 | 0.843 | 0.910 | |||||||||
| HAV | 1.8 | 5.7 | 0.320 | 0.017 | 0.039 | 0.432 | 0.761 | 0.758 | 0.996 | |||||||||
| Kobuvirus | 4.1 | 3.0 | 1.349 | 0.053 | 0.054 | 0.974 | 0.814 | 0.766 | 0.941 | |||||||||
| EMCV | 2.3 | 6.4 | 0.365 | 0.014 | 0.044 | 0.320 | 0.912 | 0.892 | 0.978 | |||||||||
| Theiler's virus | 12.9 | 12.6 | 1.028 | 0.111 | 0.111 | 0.998 | 0.951 | 0.917 | 0.964 | |||||||||
| BEV | 13.1 | 3.4 | 3.833 | 0.081 | 0.021 | 3.782 | 0.991 | 0.944 | 0.953 | |||||||||
| Erbovirus | 27.5 | 3.1 | 8.906 | 0.475 | 0.051 | 9.352 | 0.899 | 0.880 | 0.979 | |||||||||
| Parechovirus | 21.5 | 5.9 | 3.624 | 0.228 | 0.043 | 5.334 | 0.910 | 0.854 | 0.938 | |||||||||
| HRV-A | 32.8 | 28.4 | 1.153 | 0.363 | 0.231 | 1.573 | 0.899 | 0.901 | 1.002 | |||||||||
| Other virus groups | ||||||||||||||||||
| Nonsegregating | ||||||||||||||||||
| Astrovirus | 31.7 | 3.9 | 8.191 | 0.318 | 0.045 | 7.080 | 0.966 | 0.930 | 0.963 | |||||||||
| Norovirus | 30.6 | 15.7 | 1.951 | 0.233 | 0.122 | 1.916 | 0.969 | 0.874 | 0.902 | |||||||||
| HGV/GBV-C | 6.0 | 3.0 | 1.979 | 0.058 | 0.031 | 1.860 | 1.038 | 0.982 | 0.946 | |||||||||
| Segregating | ||||||||||||||||||
| Alphavirus | 51.9 | 35.7 | 1.452 | 0.460 | 0.281 | 1.634 | 0.972 | 0.977 | 1.005 | |||||||||
| HCV | 28.1 | 25.8 | 1.090 | 0.188 | 0.193 | 0.971 | 1.019 | 0.981 | 0.963 | |||||||||
| Pestivirus | 30.8 | 25.7 | 1.201 | 0.208 | 0.160 | 1.300 | 0.893 | 0.861 | 0.964 | |||||||||
| Flavivirus
|
||||||||||||||||||
| Dengue virus | 33.5 | 29.6 | 1.131 | 0.181 | 0.156 | 1.163 | 0.870 | 0.826 | 0.949 | |||||||||
| JEV | 27.1 | 23.2 | 1.166 | 0.171 | 0.150 | 1.137 | 0.933 | 0.921 | 0.987 | |||||||||
| TBE | 23.4 | 19.8 | 1.183 | 0.156 | 0.143 | 1.090 | 0.950 | 0.940 | 0.989 | |||||||||
| YFV | 4.4 | 6.1 | 0.714 | 0.029 | 0.037 | 0.772 | 0.905 | 0.885 | 0.978 | |||||||||
Ratio of intraregion distances of structural (S) and nonstructural (NS) regions; values above 1.5 are shown in bold.
Ratio of dN/dS values for structural and nonstructural regions; values above 1.75 are shown in bold.
Ratio of ENc values for nonstructural and structural regions; values below 0.96 shown in bold.
The three other virus groups showing evidence for recombination (astroviruses, HGV/GBV-C, and noroviruses) showed sequence divergence characteristics similar (although less marked in two cases) to those of the nonsegregating picornaviruses. Astroviruses showed fourfold increases in amino acid sequence variability in the structural region and in dN/dS ratio over those for the nonstructural region, well within the range of values observed among the nonsegregating picornavirus groups. Noroviruses and HGV/GBV-C showed twofold-greater amino acid sequence variability and dN/dS ratios in the structural region, less extreme than the other nonsegregating virus groups but still distinct from segregating virus groups (mean values of 1.1 and 1.2, respectively).
Although there were too few published sequences of the remaining picornavirus genera (HAV, cardioviruses, parechoviruses, kobuviruses, the ERAV and erbovirus genera, and human rhinoviruses) to be included in the recombination analysis, it was possible to carry out an analysis of sequence diversity restricted to the available complete genome sequences (Table 4; also Fig. 6, right panels). Capsid-encoding regions of HAV, kobuviruses, EMCV, and ERAV showed amino acid sequence diversity ranging from 1.8% to 4.1% (Table 4), comparable to that observed between members of the same serotypes of other picornavirus genera. In contrast, sequence diversity in the capsid-encoding regions of erboviruses, parechoviruses, BEV, and HRV-A (13% to 33%) was comparable to that observed between different serotypes of FMDV, human enteroviruses, and teschoviruses (21% to 27%).
The parechoviruses, erboviruses, and bovine enteroviruses showed sequence divergence profiles comparable to those observed for nonsegregating viruses, with much higher sequence variability in the structural region of the genome (ratios to values for the nonstructural region of 3.6- to 8.9-fold; indicated in bold in Table 4) and higher dN/dS values (ratios from 3.8 to 9.4). In contrast, HRV-A showed approximately equal levels of amino acid sequence divergence in the two genome regions (32% and 28%) and similar dN/dS ratios (0.36 and 0.23). These values are comparable to those for segregating control viruses (flaviviruses and alphaviruses).
Codon usage of structural and nonstructural regions.
Further information on possible differences in the evolutionary processes operating in different parts of the genome was obtained through analysis of compositional constraints and biases between structural and nonstructural gene sequences of the different virus groups. A large number of mononucleotide, dinucleotide, and coding usage parameters were compared between structural and nonstructural regions. These included frequencies of each base, base combinations (i.e., S, R, and K), and normalized dinucleotide frequencies based on mononucleotide base frequencies at each codon position (see Materials and Methods). For analysis of codon usage, RSCU, bias, and variance values for each amino acid, base, and base-pair combination and ENc value were determined for each genome region (see Materials and Methods). ENc values were compared to values expected from the sequences based on the G+C content of third-codon positions.
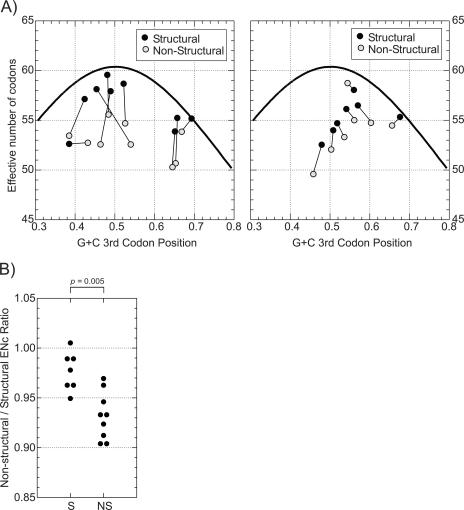
Direct comparison of many of these composition measurements between structural and nonstructural regions was complicated by frequent natural amino acid composition differences between proteins with different functions in different parts of the genome. Differences in amino acid usage in turn influence base and particularly dinucleotide composition between regions, which was the reason for developing methods for dinucleotide frequency normalization that accommodates amino acid usage. For the current analysis, viruses were considered as either nonsegregating (FMDV, enteroviruses, teschoviruses, astroviruses, noroviruses, and HGV/GBV-C; nine groups) or segregating (four groups of flavivirus, HCV, and pestivirus genera within the Flaviviridae, and alphaviruses; seven groups). Ratios of each composition variable (mean value for each virus group) between structural and nonstructural regions were compared between these two sets of viruses. By linear regression and separate nonparametric analysis of categories, the only composition value ratio consistently different between the nonsegregating and segregating virus groups was the ENc (Fig. 7A, left panel; also Table 4). Among nonsegregating viruses, ENc values for structural regions were consistently closer to expected values based on G+C content (i.e., closer to the bell-shaped curve) than values for nonstructural regions or either structural and nonstructural regions of the segregating viruses (Fig. 7A, right panel). For the latter group, ENc values for nonstructural and structural regions both showed consistent nonrandom codon usage (Table 4). Combining the data, nonstructural regions of nonsegregating viruses showed ENc values 7% (range, 3 to 10%) lower than those structural regions, compared with only 2% (range, −1% to 5%) for segregating viruses (P = 0.005; Kruskal-Wallace nonparametric test) (Fig. 7B).
FIG. 7.
(A) Codon usage (ENc values; y axis) and G+C content at third codon positions (x axis) of structural and nonstructural regions of nonsegregating (left panel) and segregating (right panel) viruses. Nonsegregating viruses were FMDV, HEV-A, HEV-B, HEV-C, teschoviruses, astroviruses, noroviruses, and HGV/GBV-C; segregating viruses were HCV, flavivirus groups, alphaviruses, and pestiviruses. The solid line shows the expected Enc value for random codon usage in sequences for G+C contents ranging from 30% to 80% (x axis). (B) Comparison of ratios of codon usage between nonstructural and structural regions of segregating (S) and nonsegregating (NS) virus groups.
The remaining picornavirus genera also showed considerable variation between genomic regions in codon usage. Parechoviruses, BEV, kobuviruses, and ERAV showed low ratios between nonstructural and structural regions (91 to 95%), while HRV-A, erboviruses, both cardiovirus groups, and HAV showed ratios similar to those of the segregating control virus groups. Codon usage correlated with sequence diversity profiles for these virus groups. With one exception (erboviruses), picornavirus groups showing greater codon usage in capsid-encoding regions (ENc ratios of <0.95) also showed greater amino acid sequence variability and dN/dS ratios in the structural region. The possible mechanistic basis for the observed correlation between recombination, sequence divergence profiles, and codon usage in these and the designated nonsegregating and segregating virus groups is discussed below.
DISCUSSION
Genetic segregation in structural and nonstructural regions.
In this study, the occurrence and evolutionary basis for the complete and frequently abrupt breakdown in the relationship between sequence phylogenies of nonstructural regions with serotype in a variety of picornaviruses, astroviruses, and other nonenveloped viruses were investigated. Among various hypotheses for the observed loss of segregation, one possibility that could be discounted was that the lack of segregation arose through sequence variation in the nonstructural region being phylogenetically uninformative, perhaps as a result of the low degree of sequence variability compared to that observed for capsid-encoding regions. Lack of segregation might have arisen through homoplasy arising from multiple, saturating substitutions at a restricted number of sites that obscured the evolutionary history of the variants. The finding of high frequencies of phylogeny violations between structural and nonstructural regions, and within nonstructural regions of nonsegregating viruses, demonstrated that the nonstructural regions contained robust phylogenetic information but that it conflicted with that of structural gene regions.
The possibility that phylogeny violations between regions arose from positive selection mechanisms leading to convergence at certain amino acid residues that distorted overall phylogenetic relationships (perhaps in response to immune-mediated selection pressures) was discounted by phylogeny comparisons of trees based on synonymous distances, which showed equivalent frequencies of phylogeny violations between genome regions to those based on all sites (Fig. 4B).
The occurrence of widespread phylogenetic incompatibilities between genome regions of picornaviruses and other nonenveloped viruses at synonymous sites can therefore most consistently be interpreted as resulting from extensive recombination between compatible serotypes in nonstructural regions, as documented between serotypes within HEV-A, HEV-B, and HEV-C (2, 11, 14, 18, 31, 38, 41, 43, 43, 50, 51, 55). The finding of comparable frequencies of phylogeny violations and loss of segregation in nonstructural regions, as well as a spectrum of sequence divergence and compositional characteristics among nonsegregating viruses that match those of human enteroviruses, provides clear evidence for similar underlying evolutionary processes among a variety of nonenveloped viruses.
Location of recombination sites.
As found previously for human enteroviruses (14, 42, 50, 51, 55), there was striking restriction of recombination sites between serotypes to nonstructural regions of the genomes of FMDV and teschoviruses (Fig. 1 and 3), with sharp boundaries to recombination events across the VP1-VP2A boundary, as well as a more diffuse change in recombination frequency and segregation between the L protein and VP4. The main exception to the block-like nature of the capsid region was among HEV-B sequences, which, in contrast to what was found in previous analysis of HEV-A and HEV-C (55), showed evidence of a lesser degree of segregation and reduced phylogenetic incompatibility of VP4 sequences with the rest of the capsid region. This is consistent with previous observations for individual sequences showing phylogenetic incongruence between these regions in this species (40). Both FMDV and teschoviruses showed phylogenetic incompatibility not only between nonstructural and structural regions but also between nonstructural regions, such as between P2 and P3 and with the 5′ UTR/L sequences (Fig. 3). These observations are consistent with multiple, complex recombination events in the evolutionary history of these regions of the genomes of all three picornavirus genera.
In this study, sequence variants were assigned to groups on the basis of serotype, and recombination and segregation analysis was restricted to the detection of recombination events between these groups. The data obtained for a low frequency or an absence of recombination between serotypes within the capsid-encoding region (Fig. 3 and 4) were therefore not incompatible with other data demonstrating recombination in the capsid region between members of the same serotype (13, 22, 42, 60, 67), likely a biologically distinct phenomenon. For example, within-serotype phylogenetic incongruities between FMDV serotype A variants in the structural gene region spanning the 1B/1C junction have been observed (60). SAT serotypes of FMDV have shown linkage disequilibrium across VP1, attributable to frequent recombination events in the evolutionary history of these serotypes (22). Analogously, Yang et al. (67) obtained evidence for recombination between different lineages of poliovirus type 1 (PV1) evolving within a chronically infected, immunodeficient individual. Intraserotype recombination between VP4/2 and VP1 was observed for the HEV-B serotype coxsackie B1 virus (CBV-1) (42). Similarly, there is evidence for a recombination event between a HAV variant at the start of VP1 and other HAV genotypes (13). Although HAV genotypes are frequently referred to as (genotypically defined) counterparts of serotypes of other picornavirus genera, they show much less sequence divergence from each other and are serologically monotypic (29); mean pair-wise distances in the capsid region between genotypes of HAV are indeed comparable to intraserotype diversity of other genera (Table 4; Fig. 6). The recently reported evidence for a recombination event (13) is therefore again equivalent to an intraserotype recombination event rather than an interserotype one.
Biological compatibility is likely to play an important role in the varied patterns of recombination observed for these genera. Although the degree of amino acid sequence similarity between protein regions required to retain functionality has not been experimentally determined for picornaviruses and is, in any case, likely to differ between proteins with different functions, current and previously published observations are consistent with a “compatibility barrier” that apparently limits recombination. As described, within-serotype recombination has been documented to occur both in structural and nonstructural regions of picornavirus genomes among variants differing from each other by 7% (range, 4 to 14%) in the structural regions and 4% (range, 3 to 5%) in the nonstructural regions in the three genera analyzed for recombination in the current study. At the opposite extreme, members of different species within a genus differ substantially in both structural and nonstructural regions (e.g., 53% and 38% mean amino acid sequence divergence between structural and nonstructural gene regions of HEV-A and those of HEV-D), a level at which recombination events have not been demonstrated to occur (31, 40, 51). This limitation may also underlie the absence of observed recombination between variants that vary at the species level in other genera, such as the Theiler's virus and EMCV groups in the cardioviruses (38% and 49%) and human rhinovirus A and B groups (49% and 50%). Between these extremes at the serotype level, the marked difference in amino sequence variability between structural and nonstructural gene regions (Fig. 1 and 6; Table 4) may account for the different patterns of recombination observed. Specifically, the interserotype 21 to 27% diversity in amino acid sequences between structural gene regions of enterovirus, teschoviruses, and FMDV may be too great to allow viable recombinants to be generated, while the much more conserved nonstructural region sequences (3 to 9% between serotypes of these viruses) may be biologically compatible and allow them to be frequently interchangeable. Indeed, the explanation for the apparently anomalous occurrence of recombination between VP4 and the rest of capsid-encoding region in enteroviruses and FMDV may be because this is the most conserved region in the capsid (approximately 10% amino acid sequence divergence) (Fig. 1) and therefore may be below the compatibility barrier.
The limited experimental data on compatibility that have been obtained are consistent with the barrier hypothesis. Chimeric viruses assembled from anything but the shortest exchange of capsid segments of different poliovirus serotypes replicate extremely poorly if at all in vitro (7, 36, 58). Similarly, the viable poliovirus 2/3 recombinant isolated from a vaccinee contained a breakpoint at the conserved C terminus of VP1, leading to only four amino acid changes in VP1, none of which were serotype specific (6). In contrast, interserotype recombinants between two HEV-B serotypes constructed with breakpoints within nonstructural regions were almost invariably fully replication competent both in vitro and in vivo, albeit with alterations in biological properties such as tropism and pathogenicity in mice (19, 20).
Evolution and potential recombination among other picornavirus genera.
Human enteroviruses, FMDV, and teschoviruses showing evidence for recombination and greater capsid-encoding region diversity also showed consistently elevated proportions of nonsynonymous-to-synonymous nucleotide substitutions (dN/dS ratios) compared to those of other genome regions (Table 4). This phenomenon was observed for other picornavirus genera; parechoviruses, erboviruses, and bovine enteroviruses showed comparable levels of amino acid sequence divergence in the capsid-encoding region and elevated dN/dS ratios in the structural gene region. These virus groups therefore demonstrate the same differential sequence diversity barriers as found in the nonsegregating viruses; further analysis when more-complete genome sequences become available will be of importance in determining whether recombination occurs in these groups as well.
Detection of interserotype recombination among ERAV, HAV, kobuvirus, and cardiovirus genera was precluded by the absence among published sequences of variants differing from each other at the serotype level. Because comparisons of sequences within the same serotype do not show the characteristic differences in sequence divergence or dN/dS ratios between genomic regions, even among nonsegregating viruses (Fig. 6, left panels), their absence among ERAV, kobuviruses, cardioviruses, and HAV provides no evidence for or against recombination in these groups. However, ERAV and kobuviruses, but not HAV or cardioviruses, show the same pattern of differential codon usage between structural and nonstructural regions that is characteristic of the nonsegregating virus groups analyzed in this study (Fig. 7; Table 4). This observation provides some evidence, therefore, that ERAV and kobuviruses may have encountered selection pressure of the same type as that encountered by nonsegregating viruses in their evolutionary history (see next section).
Finally, the distance profile data for HRV-A serotypes were substantially different from those for other picornavirus genera with serotypes. Structural and nonstructural regions showed comparable levels of sequence divergence, with the nonstructural region showing amino acid sequence divergence (28%) far greater than that between serotypes of nonsegregating picornaviruses (3 to 9%). Furthermore, dN/dS ratios were similar between regions, in contrast to the nonsegregating virus groups. There is currently a lack of complete genome sequences of HRV-A or -B groups with which to carry out recombination analysis. However, a published comparison of subgenomic sequences from the VP2-VP4 junction and a region within 3D Pol revealed phylogenetic relationships within and between serotypes of both HRV-A and -B groups that were almost entirely congruent (52). Although these authors stressed the difference in phylogeny between the two regions for the 62 sequences analyzed, inspection of the trees revealed almost complete concordance (even in relative branch lengths) between the two trees, with only one difference in branching order supported at a bootstrap level of 70% or greater being identified.
This lack of detectable recombination between rhinovirus serotypes appears initially at odds with the high frequency observed for their closest relatives, the human enteroviruses, but is potentially explained by a different pattern of sequence diversity among sequenced human rhinovirus isolates. Although classified as “serotypes,” almost all isolates actually show sequence divergence in the nonstructural region at levels approaching that observed between different species of human enteroviruses (≈35%). For the latter, the lack of interspecies recombination is attributable to biological compatibility barriers that prevent generation of viable chimeric sequences (31, 40, 51). Recombination between the similarly diverse human rhinoviruses may thus be similarly constrained. In the future, the availability of multiple complete genome sequences within the same rhinovirus “species,” equivalent to serotypes of HEVs, may reveal a pattern of recombination and sequence variation equivalent to that observed for human enteroviruses in the current study.
Selection pressures operating in picornavirus evolution.
Data obtained on the pattern of sequence divergence and codon usage reveal a number of shared features of the evolution and constraints operating among the various genera of picornaviruses and a number of differences from other (enveloped) positive-stranded RNA virus families. Examination of relative frequencies of synonymous and nonsynonymous substitution in different genomic regions of picornaviruses revealed a striking discrepancy in dN/dS ratios between structural and nonstructural regions. The higher ratios coincide with the marked increase in amino acid sequence divergence in the capsid-encoding regions (Fig. 1).
The nature of the selection processes implicit in these regional differences varies substantially depending on whether comparisons are made between or within serotypes. Remarkably, comparisons of sequences of the same serotype (including genera without serologically defined groups, such as HAV, ERAV, and kobuviruses) revealed invariably low (0.1 or less) dN/dS ratios in the structural regions, which were comparable to those in the nonstructural regions (Fig. 6). It thus appears that the recent evolution within each of the picornavirus genera that led to intraserotype sequence variation operated under generally negative or purifying selection (49). In this respect, the evidence for positive selection at certain amino acid sites in FMDV during short-term (intraserotypic) drift likely mediated through immune selection (16, 17, 21, 23, 33, 35, 61) clearly occurs on a scale insufficient to significantly modify the dN/dS ratio of the structural region as a whole. Even for FMDV, the general evolutionary trend is for synonymous substitution to dominate the process of recent sequence diversification.
The evolutionary process underlying the emergence of serotypes of picornaviruses was quite distinct. As well as being marked by its much greater magnitude, diversity between serotypes was characterized its pervasive effect on the whole of the capsid-encoding region rather than being restricted to individual amino acid sites or protein domains (Fig. 1). It is as if the differentiation process remodeled the entire virus capsid into novel structural forms during the genesis of a new serotype. The consistent differences in levels of codon usage between structural and nonstructural regions of nonsegregating picornaviruses (Fig. 7) indicate that the evolutionary process underlying the differentiation of serotypes may have also driven out the codon optimization evident in the nonstructural regions. In general, codon use is restricted throughout the genomes of the various genera of flaviviruses and other segregating virus groups, with ENc values consistently below those expected from their G+C content at third codon positions (Fig. 7; Table 4). Restricted codon usage is also found in the nonstructural regions of the nonsegregating virus groups. The basis for this codon optimization strategy in RNA viruses is unclear, but the strategy contrasts with what is seen for host mammalian genes, in which codon use is generally unrestricted (53). Irrespective of the underlying mechanism, the observation of greater ENc values in structural coding regions of nonsegregating viruses suggests that whatever selection process that operated to reduce codon usage was substantially overridden in the evolution of the capsid region during serotype differentiation.
Serotypes and receptor use.
What selection pressure could so profoundly modify the structural region of picornaviruses? One explanation is that the differentiation process reflects selection of different receptor usages. Receptor use by picornaviruses is characterized by extreme diversity even among closely related viruses. For example, serotypes within the four human enterovirus species use at least seven different receptors, including the structurally diverse cell surface molecules ICAM-1 (intracellular adhesion molecule-1), sialic acid, integrins, CD155 (PVR [poliovirus receptor]), CAR (coxsackievirus/adenovirus receptor), and DAF (decay-accelerating factor) (reviewed in references 15, 25, 26, 48, and 65). In some examples, changes in receptor use have arisen through major reorganization of the capsid proteins, such as the insertion of a host-derived sequence from tumor necrosis factor alpha containing the RGD motif, which allows integrin binding. The observation that the three serotypes of poliovirus remain monophyletic in the structural gene region, distinct from what is seen for other HEV-C serotypes, provides evidence that use of PVR by polioviruses is a relatively recent innovation within HEV-C (46).
A unifying theory that accounts for the pattern of highly diverse receptor use in picornaviruses and other virus families proposes that changes in receptor binding are driven by antibody-mediated selection (4). Because viruses are typically neutralized though antibody binding close to sites in the capsid that interact with its receptors, changes in receptor use may render a virus neutralization resistant. By escaping from preexisting immunity in its host, the opportunity to reinfect a newly susceptible population might be the strong selection pressure that drives the evolution of structural change in viral capsids. The gradient of amino sequence variability across the capsid region between serotypes of nonsegregating viruses (Fig. 1C), in which VP1 shows the greatest variability while VP4 is the least divergent, fits this model of selection driving the differentiation of serotypes. Selection would be predicted to act most strongly on the region of the capsid responsible for receptor interactions, while acting much more weakly on the internal capsid protein VP4, which is shielded from direct antibody interactions.
Evolution in other virus groups.
Variants of other nonenveloped virus groups analyzed in this study showed evidence for evolutionary dynamics and constraints remarkably similar to those found for picornaviruses. Both astroviruses and noroviruses are known to have undergone recombination, as evident from comparison of complete genome sequences and findings of phylogeny incongruities of subgenomic sequence fragments from different regions of the genome (1, 5, 28, 44, 45, 47, 63). In the current study, segregation analysis (Fig. 2; Table 3) and quantitation of phylogeny violations (Fig. 4) demonstrated that astroviruses were indeed similar to picornaviruses both in the degree of recombination detected and also in terms of the restriction of recombination to the nonstructural genome region (Fig. 2). Astroviruses, and to a lesser extent, the human noroviruses, also showed the characteristically greater amino sequence divergence and increased dN/dS ratios in the structural gene region, as well as differences in codon usage between genomic regions (higher ENc values were found for the structural gene region) (Table 4). There is therefore a clear commonality in many aspects of the past evolution and selective pressures in the evolutionary history of these viruses and potentially in the nature of the process driving the evolution of different serotypes.
The evolution of these nonenveloped viruses differed from that of the enveloped virus groups analyzed as controls in the current study. With the exception of HGV/GBV-C, none of the flavivirus genera or alphaviruses showed evidence for consistent differences in sequence variability between structural regions, and they showed no differences in dN/dS ratios between regions and similar codon usages (Fig. 6; Table 4). Furthermore, there was no evidence for large-scale phylogenetic incongruities between genome regions (Fig. 4). While it could be argued that compatibility barriers may limit recombination in some virus groups (such as alphaviruses with >35% similarity in amino acid sequences between serotypes in the nonstructural region), the absence of recombination among genotypes of YFV (6.1% divergence in the nonstructural region) indicates this cannot be the only explanation. Biological barriers similarly do not impede the creation of viable intergenotype recombinants of HCV in vitro, and large-scale epidemiological screening has identified at least one naturally occurring intergenotype recombinant of HCV in human populations (27). Furthermore, as described for HAV and FMDV, the existence of intraserotype recombinants of dengue virus and members of the JEV group (62) demonstrates that epidemiological opportunities and the mechanism for recombination within the vector-borne flaviviruses do exist. Broadly, however, the evolution of flaviviruses does not appear to be driven by the selection process driving the evolution of picornavirus serotypes. Possession of a virus envelope with its greater scope for shielding interaction domains with cell receptors through glycosylation may be a major factor that protects them from the intense immune pressure to which nonenveloped viruses are exposed. The exception is HGV/GBV-C, which shows evidence for recombination between genotypes (Fig. 4) (66). Intriguingly, unlike those of all other flavivirus groups, HGV/GBV-C structural region sequences also showed amino acid sequence divergence and dN/dS ratios greater than those for the nonstructural regions (ratios of approximately twofold) as well as the same skewed codon usage characteristic of the nonsegregating picornavirus groups (Table 4). The unusual configuration of the HGV/GBV-C virion, which is formed apparently without a virus nucleocapsid and an almost complete lack of N-linked glycosylation sites in the E1 and E2 proteins, may indeed be more susceptible to antibody-mediated immune pressure, analogous to that directed against nonenveloped viruses.
In summary, this study brings together a large amount of disparate data on the nature of sequence variability among the different picornavirus genera and identifies a number of unifying features in the evolution of picornaviruses and other nonenveloped viruses that distinguishes their evolution from that of other positive-stranded RNA virus families. Biological compatibility barriers that could account for the markedly different frequencies of recombination between structural and nonstructural regions of picornaviruses, and other nonenveloped virus groups can indeed be experimentally investigated. For example, virus chimeras can be constructed from naturally occurring HEV-B or HEV-C variants with differing degrees of amino acid sequence divergence by use of recently developed reverse genetic systems, enabling the viabilities of recombinant viruses with breakpoints in structural and nonstructural regions to be compared. Understanding the tempo and mode of picornavirus evolution and the restrictions on replication exerted by the host immune response are clearly of major importance in the vaccine control of many of the virus groups examined in this study.
Acknowledgments
I am grateful to David Evans and Heli Harvala for their careful reading of the manuscript and their many suggestions for improvement.
Footnotes
Published ahead of print on 6 September 2006.
REFERENCES
- 1.Ambert-Balay, K., F. Bon, F. Le Guyader, P. Pothier, and E. Kohli. 2005. Characterization of new recombinant noroviruses. J. Clin. Microbiol. 43:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, P., K. Edman, and A. M. Lindberg. 2002. Molecular analysis of the echovirus 18 prototype: evidence of interserotypic recombination with echovirus 9. Virus Res. 85:71-83. [DOI] [PubMed] [Google Scholar]
- 3.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. [DOI] [PubMed] [Google Scholar]
- 4.Baranowski, E., C. M. Ruiz-Jarabo, and E. Domingo. 2001. Evolution of cell recognition by viruses. Science 292:1102-1105. [DOI] [PubMed] [Google Scholar]
- 5.Belliot, G., H. Laveran, and S. S. Monroe. 1997. Detection and genetic differentiation of human astroviruses: phylogenetic grouping varies by coding region. Arch. Virol. 142:1323-1334. [DOI] [PubMed] [Google Scholar]
- 6.Blomqvist, S., A. L. Bruu, M. Stenvik, and T. Hovi. 2003. Characterization of a recombinant type 3/type 2 poliovirus isolated from a healthy vaccinee and containing a chimeric capsid protein VP1. J. Gen. Virol. 84:573-580. [DOI] [PubMed] [Google Scholar]
- 7.Burke, K. L., G. Dunn, M. Ferguson, P. D. Minor, and J. W. Almond. 1988. Antigen chimaeras of poliovirus as potential new vaccines. Nature 332:81-82. [DOI] [PubMed] [Google Scholar]
- 8.Burns, C. C., J. Shaw, R. Campagnoli, J. Jorba, A. Vincent, J. Quay, and O. Kew. 2006. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J. Virol. 80:3259-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caro, V., S. Guillot, F. Delpeyroux, and R. Crainic. 2001. Molecular strategy for ‘serotyping’ of human enteroviruses. J. Gen. Virol. 82:79-91. [DOI] [PubMed] [Google Scholar]
- 10.Carrillo, C., E. R. Tulman, G. Delhon, Z. Lu, A. Carreno, A. Vagnozzi, G. F. Kutish, and D. L. Rock. 2005. Comparative genomics of foot-and-mouth disease virus. J. Virol. 79:6487-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chevaliez, S., A. Szendroi, V. Caro, J. Balanant, S. Guillot, G. Berencsi, and F. Delpeyroux. 2004. Molecular comparison of echovirus 11 strains circulating in Europe during an epidemic of multisystem hemorrhagic disease of infants indicates that evolution generally occurs by recombination. Virology 325:56-70. [DOI] [PubMed] [Google Scholar]
- 12.Copper, P. D., A. Steiner-Pryor, P. D. Scotti, and D. Delong. 1974. On the nature of poliovirus genetic recombinants. J. Gen. Virol. 23:41-49. [DOI] [PubMed] [Google Scholar]
- 13.Costa-Mattioli, M., V. Ferre, D. Casane, R. Perez-Bercoff, M. Coste-Burel, B. M. Imbert-Marcille, E. C. Andre, C. Bressollette-Bodin, S. Billaudel, and J. Cristina. 2003. Evidence of recombination in natural populations of hepatitis A virus. Virology 311:51-59. [DOI] [PubMed] [Google Scholar]
- 14.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, D. J., and J. W. Almond. 1998. Cell receptors for picornaviruses as determinants of cell tropism and pathogenesis. Trends Microbiol. 6:198-202. [DOI] [PubMed] [Google Scholar]
- 16.Fares, M. A., S. F. Elena, J. Ortiz, A. Moya, and E. Barrio. 2002. A sliding window-based method to detect selective constraints in protein-coding genes and its application to RNA viruses. J. Mol. Evol. 55:509-521. [DOI] [PubMed] [Google Scholar]
- 17.Fares, M. A., A. Moya, C. Escarmis, E. Baranowski, E. Domingo, and E. Barrio. 2001. Evidence for positive selection in the capsid protein-coding region of the foot-and-mouth disease virus (FMDV) subjected to experimental passage regimens. Mol. Biol. Evol. 18:10-21. [DOI] [PubMed] [Google Scholar]
- 18.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvala, H., H. Kalimo, J. Bergelson, G. Stanway, and T. Hyypia. 2005. Tissue tropism of recombinant coxsackieviruses in an adult mouse model. J. Gen. Virol. 86:1897-1907. [DOI] [PubMed] [Google Scholar]
- 20.Harvala, H., H. Kalimo, L. Dahllund, J. Santti, P. Hughes, T. Hyypia, and G. Stanway. 2002. Mapping of tissue tropism determinants in coxsackievirus genomes. J. Gen. Virol. 83:1697-1706. [DOI] [PubMed] [Google Scholar]
- 21.Haydon, D., N. Knowles, and J. McCauley. 1998. Methods for the detection of non-random base substitution in virus genes: models of synonymous nucleotide substitution in picornavirus genes. Virus Genes 16:253-266. [DOI] [PubMed] [Google Scholar]
- 22.Haydon, D. T., A. D. Bastos, and P. Awadalla. 2004. Low linkage disequilibrium indicative of recombination in foot-and-mouth disease virus gene sequence alignments. J. Gen. Virol. 85:1095-1100. [DOI] [PubMed] [Google Scholar]
- 23.Haydon, D. T., A. D. Bastos, N. J. Knowles, and A. R. Samuel. 2001. Evidence for positive selection in foot-and-mouth disease virus capsid genes from field isolates. Genetics 157:7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillis, D. M., and J. J. Bull. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182-192. [Google Scholar]
- 25.Hogle, J. M. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 56:677-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson, T., A. M. King, D. I. Stuart, and E. Fry. 2003. Structure and receptor binding. Virus Res. 91:33-46. [DOI] [PubMed] [Google Scholar]
- 27.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishna, N. K. 2005. Identification of structural domains involved in astrovirus capsid biology. Viral Immunol. 18:17-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemon, S. M., R. W. Jansen, and E. A. Brown. 1992. Genetic, antigenic and biological differences between strains of hepatitis A virus. Vaccine 10(Suppl. 1):S40-S44. [DOI] [PubMed] [Google Scholar]
- 30.Lindberg, A. M., P. Andersson, C. Savolainen, M. N. Mulders, and T. Hovi. 2003. Evolution of the genome of human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 84:1223-1235. [DOI] [PubMed] [Google Scholar]
- 31.Lukashev, A. N. 2005. Role of recombination in evolution of enteroviruses. Rev. Med. Virol. 15:157-167. [DOI] [PubMed] [Google Scholar]
- 32.Lukashev, A. N., V. A. Lashkevich, G. A. Koroleva, J. Ilonen, and A. E. Hinkkanen. 2004. Recombination in uveitis-causing enterovirus strains. J. Gen. Virol. 85:463-470. [DOI] [PubMed] [Google Scholar]
- 33.Martin, M. J., J. I. Nunez, F. Sobrino, and J. Dopazo. 1998. A procedure for detecting selection in highly variable viral genomes: evidence of positive selection in antigenic regions of capsid protein VP1 of foot-and-mouth disease virus. J. Virol. Methods 74:215-221. [DOI] [PubMed] [Google Scholar]
- 34.Melnick, J. L., V. Rennick, B. Hampil, N. J. Schmidt, and H. H. Ho. 1973. Lyophilized combination pools of enterovirus equine antisera: preparation and test procedures for the identification of field strains of 42 enteroviruses. Bull. W. H. O. 48:263-268. [PMC free article] [PubMed] [Google Scholar]
- 35.Mittal, M., C. Tosh, D. Hemadri, A. Sanyal, and S. K. Bandyopadhyay. 2005. Phylogeny, genome evolution, and antigenic variability among endemic foot-and-mouth disease virus type A isolates from India. Arch. Virol. 150:911-928. [DOI] [PubMed] [Google Scholar]
- 36.Murdin, A. D., H. H. Lu, M. G. Murray, and E. Wimmer. 1992. Poliovirus antigenic hybrids simultaneously expressing antigenic determinants from all three serotypes. J. Gen. Virol. 73:607-611. [DOI] [PubMed] [Google Scholar]
- 37.Noel, J. S., T. W. Lee, J. B. Kurtz, R. I. Glass, and S. S. Monroe. 1995. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 33:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norder, H., L. Bjerregaard, and L. O. Magnius. 2002. Open reading frame sequence of an Asian enterovirus 73 strain reveals that the prototype from California is recombinant. J. Gen. Virol. 83:1721-1728. [DOI] [PubMed] [Google Scholar]
- 39.Oberste, M. S., K. Maher, D. R. Kilpatrick, M. R. Flemister, B. A. Brown, and M. A. Pallansch. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oberste, M. S., K. Maher, and M. A. Pallansch. 2004. Evidence for frequent recombination within species human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 78:855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oberste, M. S., S. Penaranda, K. Maher, and M. A. Pallansch. 2004. Complete genome sequences of all members of the species human enterovirus A. J. Gen. Virol. 85:1597-1607. [DOI] [PubMed] [Google Scholar]
- 42.Oberste, M. S., S. Penaranda, and M. A. Pallansch. 2004. RNA recombination plays a major role in genomic change during circulation of coxsackie B viruses. J. Virol. 78:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oprisan, G., M. Combiescu, S. Guillot, V. Caro, A. Combiescu, F. Delpeyroux, and R. Crainic. 2002. Natural genetic recombination between co-circulating heterotypic enteroviruses. J. Gen. Virol. 83:2193-2200. [DOI] [PubMed] [Google Scholar]
- 44.Pantin-Jackwood, M. J., E. Spackman, and P. R. Woolcock. 2006. Phylogenetic analysis of Turkey astroviruses reveals evidence of recombination. Virus Genes 32:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reuter, G., H. Vennema, M. Koopmans, and G. Szucs. 2006. Epidemic spread of recombinant noroviruses with four capsid types in Hungary. J. Clin. Virol. 35:84-88. [DOI] [PubMed] [Google Scholar]
- 46.Rieder, E., A. E. Gorbalenya, C. Xiao, Y. He, T. S. Baker, R. J. Kuhn, M. G. Rossmann, and E. Wimmer. 2001. Will the polio niche remain vacant? Dev. Biol. (Basel) 105:111-122, 149-150. [PubMed] [Google Scholar]
- 47.Rohayem, J., J. Munch, and A. Rethwilm. 2005. Evidence of recombination in the norovirus capsid gene. J. Virol. 79:4977-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossmann, M. G., Y. He, and R. J. Kuhn. 2002. Picornavirus-receptor interactions. Trends Microbiol. 10:324-331. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez, G., A. Bosch, and R. M. Pinto. 2003. Genome variability and capsid structural constraints of hepatitis A virus. J. Virol. 77:452-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santti, J., H. Harvala, L. Kinnunen, and T. Hyypia. 2000. Molecular epidemiology and evolution of coxsackievirus A9. J. Gen. Virol. 81:1361-1372. [DOI] [PubMed] [Google Scholar]
- 51.Santti, J., T. Hyypia, L. Kinnunen, and M. Salminen. 1999. Evidence of recombination among enteroviruses. J. Virol. 73:8741-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savolainen, C., P. Laine, M. N. Mulders, and T. Hovi. 2004. Sequence analysis of human rhinoviruses in the RNA-dependent RNA polymerase coding region reveals large within-species variation. J. Gen. Virol. 85:2271-2277. [DOI] [PubMed] [Google Scholar]
- 53.Sharp, P. M., and W. H. Li. 1986. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 24:28-38. [DOI] [PubMed] [Google Scholar]
- 54.Simmonds, P., and S. Midgley. 2005. Recombination in the genesis and evolution of hepatitis B virus genotypes. J. Virol. 79:15467-15476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simmonds, P., and J. Welch. 2006. Frequency and dynamics of recombination within different species of human enteroviruses. J. Virol. 80:483-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith, D. B., M. Basaras, S. Frost, D. Haydon, N. Cuceanu, L. Prescott, C. Kamenka, D. Millband, M. A. Sathar, and P. Simmonds. 2000. Phylogenetic analysis of GBV-C/hepatitis G virus. J. Gen. Virol. 81:769-780. [DOI] [PubMed] [Google Scholar]
- 57.Stanway, G., F. Brown, P. Christian, T. Hovi, T. Hyypia, A. M. Q. King, N. J. Knowles, S. M. Lemon, P. D. Minor, M. A. Pallansch, A. C. Palmenberg, and T. Skern. 2005. Family Picornaviridae, p. 757-778. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom.
- 58.Stanway, G., P. J. Hughes, G. D. Westrop, D. M. Evans, G. Dunn, P. D. Minor, G. C. Schild, and J. W. Almond. 1986. Construction of poliovirus intertypic recombinants by use of cDNA. J. Virol. 57:1187-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tosh, C., D. Hemadri, and A. Sanyal. 2002. Evidence of recombination in the capsid-coding region of type A foot-and-mouth disease virus. J. Gen. Virol. 83:2455-2460. [DOI] [PubMed] [Google Scholar]
- 61.Tosh, C., D. Hemadri, A. Sanyal, and S. K. Bandyopadhyay. 2003. Genetic and antigenic analysis of two recently circulating genotypes of type A foot-and-mouth disease virus in India: evidence for positive selection in the capsid-coding genes. Arch. Virol. 148:853-869. [DOI] [PubMed] [Google Scholar]
- 62.Twiddy, S. S., and E. C. Holmes. 2003. The extent of homologous recombination in members of the genus Flavivirus. J. Gen. Virol. 84:429-440. [DOI] [PubMed] [Google Scholar]
- 63.Walter, J. E., J. Briggs, M. L. Guerrero, D. O. Matson, L. K. Pickering, G. Ruiz-Palacios, T. Berke, and D. K. Mitchell. 2001. Molecular characterization of a novel recombinant strain of human astrovirus associated with gastroenteritis in children. Arch. Virol. 146:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, T. H., Y. K. Donaldson, R. P. Brettle, J. E. Bell, and P. Simmonds. 2001. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J. Virol. 75:11686-11699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitton, J. L., C. T. Cornell, and R. Feuer. 2005. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 3:765-776. [DOI] [PubMed] [Google Scholar]
- 66.Worobey, M., and E. C. Holmes. 2001. Homologous recombination in GB virus C/hepatitis G virus. Mol. Biol. Evol. 18:254-261. [DOI] [PubMed] [Google Scholar]
- 66a.Wright, F. 1990. The ‘effective number of codons’ used in a gene. Gene 87:23-29. [DOI] [PubMed] [Google Scholar]
- 67.Yang, C. F., H. Y. Chen, J. Jorba, H. C. Sun, S. J. Yang, H. C. Lee, Y. C. Huang, T. Y. Lin, P. J. Chen, H. Shimizu, Y. Nishimura, A. Utama, M. Pallansch, T. Miyamura, O. Kew, and J. Y. Yang. 2005. Intratypic recombination among lineages of type 1 vaccine-derived poliovirus emerging during chronic infection of an immunodeficient patient. J. Virol. 79:12623-12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zell, R., M. Dauber, A. Krumbholz, A. Henke, E. Birch-Hirschfeld, A. Stelzner, D. Prager, and R. Wurm. 2001. Porcine teschoviruses comprise at least eleven distinct serotypes: molecular and evolutionary aspects. J. Virol. 75:1620-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zell, R., A. Krumbholz, M. Dauber, E. Hoey, and P. Wutzler. 2006. Molecular-based reclassification of the bovine enteroviruses. J. Gen. Virol. 87:375-385. [DOI] [PubMed] [Google Scholar]