Abstract
Replication of the plus-stranded RNA genome of hepatitis C virus (HCV) occurs in a membrane-bound replication complex consisting of viral and cellular proteins and viral RNA. NS5B, the RNA polymerase of HCV, is anchored to the membranes via a C-terminal 20-amino-acid-long hydrophobic domain, which is flanked on each side by a highly conserved positively charged arginine. Using a genotype 1b subgenomic replicon (V. Lohmann, F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartensclager, Science 285:110-113, 1999), we determined the effect of mutations of some highly conserved residues in this domain. The replacement of arginine 570 with alanine completely abolished the colony-forming ability by the replicon, while a R591A change was found to be highly detrimental to replication, viability, and membrane binding by the mutant NS5B protein. Mutations of two other highly conserved amino acids (L588A and P589A) reduced but did not eliminate colony formation. It was of interest, if specific amino acid residues play a role in membrane anchoring of NS5B and replication, to determine whether a complete exchange of the NS5B hydrophobic domain with a domain totally unrelated to NS5B would ablate replication. We selected the 22-amino-acid-long hydrophobic domain of poliovirus polypeptide 3A that is known to adopt a transmembrane configuration, thereby anchoring 3A to membranes. Surprisingly, either partial or full replacement of the NS5B hydrophobic domain with the anchor sequences of poliovirus polypeptide 3A resulted in the replication of replicons whose colony-forming abilities were reduced compared to that of the wild-type replicon. Upon continued passage of the replicon in Huh-7 cells in the presence of neomycin, the replication efficiency of the replicon increased. However, the sequence of the poliovirus polypeptide 3A hydrophobic domain, in the context of the subgenomic HCV replicon, was stably maintained throughout 40 passages. Our results suggest that anchoring NS5B to membranes is necessary but that the amino acid sequence of the anchor per se does not require HCV origin. This suggests that specific interactions between the NS5B hydrophobic domain and other membrane-bound factors may not play a decisive role in HCV replication.
Hepatitis C virus (HCV), like other plus-strand RNA viruses, replicates its RNA in membranous replication complexes. These complexes form on the cytosolic surfaces of cellular membranes, and they contain both viral and cellular proteins associated with the viral RNA (10, 11, 16, 36). The exact function of membranes in viral replication is not yet clear but possible functions include (i) giving physical support to the RNA/protein complexes, (ii) concentrating and compartmentalizing the components, (iii) supplying essential lipids that are required for RNA synthesis, and (iv) providing attachment of the viral RNA during unwinding.
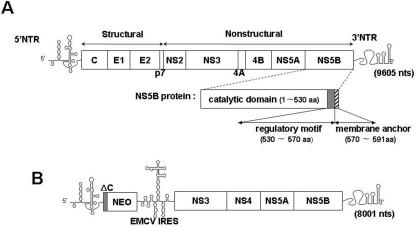
HCV, a member of the Flaviviridae family, contains a positive-sense RNA genome of about 9.6 kb (Fig. 1A). Detailed studies of HCV replication were originally difficult due to the lack of an efficient tissue culture system for the growth of the virus. However, the development of the subgenomic replicon cell culture system enabled studies of HCV RNA replication (4, 30). This system demonstrated that HCV RNA replication requires most of the nonstructural proteins, namely, NS3, NS4A, NS4B, NS5A, and NS5B (Fig. 1B). Although the detailed mechanism of HCV RNA replication has not yet been determined, it is known that replication takes place in two steps. First, a complementary minus strand is synthesized, and it in turn is used as the template for the production of the progeny plus strands. The enzyme primarily responsible is the HCV RNA-dependent RNA polymerase NS5B, an enzyme that has been expressed in both bacterial and insect cells for biochemical characterization (3, 12, 43). In vitro, the enzyme possesses two types of synthetic activities: de novo initiation and the elongation of an oligonucleotide primer on a suitable RNA template (3, 29, 33, 52). In addition, the purified enzyme specifically interacts with an essential cis-replicating RNA element near the C terminus of its own coding sequence (25). The availability of purified enzyme has also facilitated the structural analysis of the NS5B protein. Like other nucleic acid polymerases, NS5B also possesses the structure of a right hand, consisting of finger, thumb, and palm subdomains (6, 27). Oligomerization of the protein was demonstrated both by yeast two-hybrid analyses and by in vitro methods, and two interfaces in the crystal lattice were identified (38, 48). The protein interacts with other viral proteins (NS2, NS3, NS4A, NS5A, and core protein) (8, 20, 41, 42, 47) and cellular proteins (nucleolin, vesicle membrane protein hVAP-33, and eukaryotic initiation factor 4AII) (14, 18, 22). Full-length NS5B consists of 591 amino acids (aa), but the C-terminal 60 residues can be deleted without a loss of enzymatic activity in vitro (Fig. 1A) (6). Amino acids 530 to about 570 comprise a regulatory domain that inhibits both RNA binding and polymerase activity in vitro (Fig. 1A) (1, 28).
FIG. 1.
Genomic organization of HCV and of its subgenomic replicon. (A) Genomic structure of full-length HCV RNA. The single-stranded RNA genome of HCV is divided into the 5′ NTR (single line), the polyprotein coding region (boxed), and the 3′ NTR (single line). The polyprotein coding region is divided into structural and nonstructural regions. The NS5B domain, shown enlarged, contains three functional motifs: the catalytic domain (open box), the regulatory motif (gray box), and the membrane anchor (hatched box). (B) Genomic structure of HCV subgenomic replicon RNA. The single-stranded RNA genome of the subgenomic replicon (30) contains the HCV 5′ NTR (single line), the first 16 codons encoding the HCV core protein (ΔC, gray box), the neomycin phosphotransferase gene (NEO; open box), the EMCV IRES (single line), the nonstructural protein coding regions of HCV from NS3 through NS5B (open box), and the 3′ NTR (single line).
Recently, it was demonstrated that the C-terminal 21 amino acids of NS5B, comprising a highly conserved hydrophobic domain, are sufficient and essential for targeting NS5B to the cytosolic side of endoplasmic reticulum (ER) membranes (39). The NS5B membrane anchor domain, which is required for RNA replication, crosses the lipid bilayer as a transmembrane segment (21, 26, 39). Thus, HCV NS5B belongs to a small class of “tail-anchored” proteins (49). This group of proteins is characterized by (i) posttranslational membrane attachment by a C-terminal hydrophobic domain, (ii) integral membrane association, and (iii) cytosolic orientation of the functional protein domain (49). In addition to NS5B, most of the nonstructural proteins (NS3, NS4A, NS4B, and NS5A) of HCV are also components of the membranous replication complex (5, 10, 32). Protein NS3, which has both proteinase and helicase domains, is membrane bound via binding to polypeptide NS4A. Polypeptide NS4A, a cofactor of NS3 proteinase, contains its hydrophobic membrane anchor segment near its N terminus, while NS4B contains multiple potential membrane-binding domains. NS5A has been called a “tip-anchored” protein (5) in which the membrane anchor domain maps to the N-terminal 30 amino acids. A recent report by Aizaki et al. (2) suggested that during HCV replicon RNA replication, both the nonstructural proteins and the RNA are localized on detergent-insoluble membrane structures characteristic of lipid rafts.
It has been known for some time that the 3′-terminal NS5B coding region has the propensity to form highly conserved and stable stem-loop structures (19, 46). We have recently predicted the presence of four stable stem-loop structures (SL-IV, SL-V, SL-VI, and SL-VII) in the 3′-terminal 249-nucleotide (nt)-long NS5B coding sequence (25). In addition, we demonstrated that two of these hairpins (SL-V and SL-VI) are required for the replication of the subgenomic HCV replicon and one of these (SL-V) specifically binds NS5B (25). You et al. have also reported the existence of a cis-acting RNA element (5BSL3.2) in the C-terminal NS5B coding sequence, which corresponds to SL-V in our experiments (51). Subsequent studies by Friebe et al. demonstrated that that this same stem loop in the NS5B coding sequence (SL-V) is involved in an essential “kissing” interaction with the 3′ nontranslated region (NTR) (13).
The aim of our studies was to determine the effects of single amino acid substitutions in the hydrophobic domain of HCV NS5B on the replication of the subgenomic replicon in Huh-7 cells. The replacement of the positively charged arginine at position 591 of NS5B with alanine, combined with three different assays (colony formation assay, luciferase expression assay, and immunofluorescence analysis of the mutated protein expressed in Huh-7 cells), provided strong evidence that R591 is very important for the replication of the subgenomic replicon. The replacement of R570 with alanine completely ablated replicon replication, but this mutation disrupted a single base pair in a cis-acting replication domain (25, 51), and we cannot be certain what the reason for the lethal effect is. The substitution of two other highly conserved residues in the hydrophobic domain (L588A and P589A) yielded defective replicons, highlighting their importance in replication.
We were curious, if specific amino acid residues play an important role in the membrane anchoring of NS5B and replication, whether a complete exchange of the NS5B hydrophobic domain with one totally unrelated to NS5B would ablate replication. We selected the 22-amino-acid-long hydrophobic domain of poliovirus (PV) polypeptide 3A that is known to adopt a transmembrane configuration, thereby anchoring 3A to membranes. Poliovirus protein 3A, in the context of its precursor, 3AB (40), interacts with the RNA polymerase 3Dpol and anchors it to membranes in replication complexes (34, 50). Moreover, we recently found evidence that 3A can be considered a “tail-anchored” membrane protein (K. Fujita, S. S. Krishnakumar, D. Franco, A. V. Paul, E. London, and E. Wimmer, submitted for publication). We were surprised to discover that the replacement of the hydrophobic domain of NS5B with the membrane anchor domain of poliovirus protein 3A yielded replicating replicons; these replicons, however, had weaker colony-forming abilities than the wild-type (wt) replicon. Immunofluorescence analyses have revealed that the pattern of membrane association of this chimeric protein, expressed in Huh-7 cells, was similar to that of wt NS5B. During the >40 passages of the chimeric replicon in Huh-7 cells, its genotype remained unchanged, suggesting that this foreign membrane anchor sequence in NS5B is functional in the context of the subgenomic replicon.
MATERIALS AND METHODS
Cell cultures.
Monolayers of the human hepatoma cell line (Huh-7) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 units of penicillin, 100 μg streptomycin, and 10% fetal bovine serum. In cell lines carrying HCV replicons, 250 to 500 μg of G418 (Geneticin; Invitrogen Life Technologies) per ml was added to the growth medium.
Plasmids.
The plasmid for the subgenomic replicon (pFK-I389neo/NS3-3′/5.1, abbreviated NK5.1) was generously provided by R. Bartenschlager. It is a dicistronic construct containing the HCV 5′ NTR, the first 16 codons of the core protein coding region, the neo gene, and the encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) for the translation of HCV sequences of NS3 through NS5B, followed by the 3′ NTR (Fig. 1B). The nucleotide positions refer to HCV subtype 1b nucleotide sequence (NCBI accession no. AJ238799). Mutations were introduced into the NS5B C-terminal region of the subgenomic replicon using PCR-based mutagenesis with the oligonucleotides listed in Table 1. Subclone pHCV(Eco-Spe) (EcoRI [nt 6699] to SpeI [nt 9609]) of the HCV replicon in plasmid pFastBac1 was constructed and used as the template for all mutagenesis. The mutated fragments, EcoRI/SpeI cleaved, were transferred back into the original replicon NK5.1. All PCR fragments and final constructs were sequenced using the ABI Prism DNA sequencing kit.
TABLE 1.
Oligonucleotides used for PCR-based mutagenesis
| Oligonucleotide no. | Position (nt) (sense)f | Sequence (5′ to 3′) | Construct |
|---|---|---|---|
| 1 | 8717-8739 (+)a,d | G CAC GAT GCA TCT GGC AAA AGG G | |
| 2 | 9588-9617 (−)a,d,e | TGC ACT AGT AGT ACT TGA TCT GCA GAG AGG | |
| 3 | 9348-9380 (+)a,c | GC ATC TAT CTA GCC CCC AAC CGA TGA ACG GGG | pMT(L588A) |
| 4 | 9348-9380 (−)a,c | CCC CGT TCA TCG GTT GGG GGC TAG ATA GAT GCC | pMT(L588A) |
| 5 | 9348-9380 (+)a,c | GGC ATC TAT CTA CTC GCC AAC CGA TGA ACG GGG | pMT(P589A) |
| 6 | 9348-9380 (−)a,c | CCC CGT TCA TCG GTT GGC GGC TAG ATA GAT GCC | pMT(P589A) |
| 7 | 9351-9383 (+)a,c | ATC TAT CTA CTC CCC AAC GCA TGA ACG GGG AGC | pMT(R591A) |
| 8 | 9348-9380 (−)a,c | GCT CCC CGT TCA TGC GTT GGG GAG TAG ATA GAT | pMT(R591A) |
| 9 | 9363-9380 (+)a,b | GCCAAGCTTC GCC GCA GTG GCT GGA GTT GTC CCC AAC CGA TGA ACG GGG | pMT(3A-I) |
| 10 | 9294-9314 (−)a,b,d | GCCAAGCTTGT CAC CGC TTG TAG AAT TGT CAT GGA CCA GCG GGG TCG GGC ACG | pMT(3A-I) |
| 11 | 5297-5328 (+)b | CTA CAA GCG GTG ACA ACC TTC GCC GCA GTG GC | pMT(3A-I/S68T) |
| 12 | 5297-5328 (−)b | GC CAC TGC GGC GAA GGT TGT CAC CGC TTG TAG | pMT(3A-I/S68T) |
| 13 | 5288-5308 (+)b | CGC GCG ATG ACA ATT CTA CAA GCG GTG | pMT(3A-II) |
| 14 | 9294-9308 (−)a,b | G AAT TGT CAT CGC GCG GGG TCG GGC ACG | pMT(3A-II) |
| 15 | 9372-9390 (+)a,b | TAT GTC ATG TAT AAA TGA ACGGGGAGCTAAACAC | pMT(3A-II) |
| 16 | 5321-5353 (−)b | CA TTT ATA CAT GAC ATA GAC AAC TCC AGC CAC TGC | pMT(3A-II) |
| 17 | 5297-5327 (+)b,c | CTA CAA GCG GTG AAA AAG TTC GCC GCA GTG G | pMT(3A/TT-KK) |
| 18 | 5297-5331 (−)b,c | CC AGC CAC TGC GGC GAA CTT TTT CAC CGC TTG TAG | pMT(3A/TT-KK) |
| 19 | 7602-7621 (+)a,d | GGCGGATCC ATG TCT TAT TCC TGG ACA GG | |
| 20 | 9348-9371 (−)a,d | GGCTCTAGA CTA TCA TCG GTT GGG GAG TAG ATA GAT | pcDNA(NS5B/WT) |
| 21 | 9282-9308 (−)a,d | GGCTCTAGA CTA TCA GCG GGG TCG GGC ACG AGA CAG GCT GTG | pcDNA(NS5B/CΔ21) |
| 22 | 5328-5353 (−)b,d | GGCTCTAGA CTA TCA TTT ATA CAT GAC ATA GAC AAC TCC AGC | pcDNA(NS5B/3A-II) |
| 23 | 5321-5353 (−)b,d | GGCTCTAGA CTA TCA TTT ATA CAT GAC AAC TCC AGC CAC TGC | pcDNA(NS5B/3A-III) |
| 24 | 5321-5338 (−)b,d | GGCTCTAGA CTA TCA TCG GTT GGG GAC AAC TCC AGC CAC TGC | pcDNA(NS5B/3A-IV) |
| 25 | 9348-9371 (−)a,d | GGCTCTAGA CTA TCA TGC GTT GGG GAG TAG ATA GAT | pcDNA(NS5B/R591A) |
Nucleotide positions refer to the HCV subtype 1b sequence (NCBI accession no. AJ238799).
Nucleotide sequences belonging to poliovirus 3A protein are underlined. Nucleotide positions of the poliovirus 3A sequence refer to the poliovirus 1 Mahoney sequence (NCBI accession no. NC002058).
The oligonucleotide contains nucleotide changes, which are in bold.
The oligonucleotide contains an enzyme recognition site, which is shown in italics.
Nucleotide positions after 9605 represent the nucleotide sequence of the vector (pBR322).
+, positive sense; −, negative sense.
(i) Constructs with a substitution of a single amino acid in the hydrophobic domain of NS5B.
For the construction of pMT(L588A), pMT(P589A), and pMT(R591A), PCR mutagenesis was used with complementary primer pairs 3 and 4, 5 and 6, and 7 and 8, respectively, and the pHCV(Eco-Spe) subclone was the template. Primers 1 and 2 were used as the upstream and downstream primers. The mutated fragments, EcoRI/SpeI cleaved, were transferred back into the original NK5.1 replicon.
(a) Construct with a partial replacement of the NS5B hydrophobic domain with the membrane anchor sequence of poliovirus protein 3A.
In plasmid pMT(3A-I), the C-terminal transmembrane domain of NS5B was replaced with the C-terminal part of the poliovirus protein 3A hydrophobic sequence (see Fig. 3A). Two PCR fragments were made with primer pair 1 and 10 and primer pair 2 and 9 (Table 1), and they were cut with NsiI/HindIII and HindIII/SpeI, respectively. The two fragments were cloned into a NsiI/SpeI-restricted pHCV(Eco-Spe) subclone. Due to the presence of the HindIII site, the resulting clone contained a single nucleotide change (C/G) and an amino acid change (T/S). This was corrected with PCR mutagenesis using primer pairs 11 and 12 (Table 1). The corrected PCR fragment, EcoRI/SpI cleaved, was transferred back into the parental replicon NK5.1.
FIG. 3.
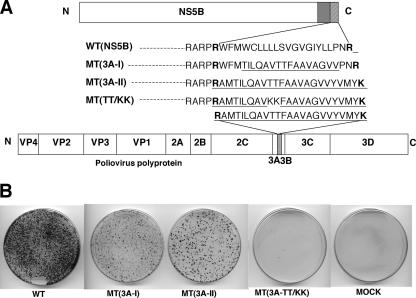
The C-terminal hydrophobic anchor sequence of poliovirus 3A is functional in the HCV NS5B C terminus. (A) Amino acid sequences of the wt and mutant NS5B C-terminal transmembrane domains (hatched box) in the subgenomic replicons. The catalytic domain of NS5B is shown with an open box and the regulatory domain with a gray box. The foreign amino acid sequences from poliovirus 3A are underlined. The entire poliovirus polyprotein and the hydrophobic domain of poliovirus 3A protein are shown with an open box and a dotted box, respectively. The positively charged amino acids (R and K) located at both ends of the hydrophobic domains of HCV NS5B and poliovirus 3A are marked in bold. (B) In vitro RNA transcripts of the wt and mutant replicon constructs were transfected into Huh-7 cells, and the colony-forming efficiencies of the replicons were measured 3 weeks after transfection. The colony-forming efficiencies of the mutant replicons are compared with that of the wt replicon.
(b) Constructs with the complete replacement of the NS5B hydrophobic domain with a wt or mutant membrane anchor sequence of poliovirus protein 3A.
In plasmid pMT(3A-II) (see Fig. 3A), the poliovirus protein 3A sequence of pMT(3A-I) was extended in both the N- and C-terminal directions by using primer pair 13 and 14 and primer pair 15 and 16, respectively (Table 1). The first two codons, one for R (AGG) and one for A (GCA) in the 3A sequence, were changed to CGC and GCG, respectively, to preserve the secondary structure of the stem in the essential RNA element SL-V (25) and to preserve the sequence next to the stem. pMT(3A-I) was used as the template in the PCR mutagenesis, and primers 1 and 2 were used as the upstream and downstream primers (Table 1). The resulting PCR fragment, EcoRI/SpeI cleaved, was ligated back into a similarly restricted plasmid, pNK5.1.
In plasmid pMT(3A-TT/KK) (see Fig. 3A), two threonines of pMT(3A-II) were replaced with lysines. For the construction of this plasmid by PCR mutagenesis, pMT(3A-II) was used as the template with complementary primers 17 and 18 and upstream and downstream primers 1 and 2, respectively.
(iv) Replicon constructs.
To generate the wt luciferase replicon (luc-5.1) for the transient-replication assay, the neo gene of replicon NK5.1 was replaced with the gene encoding the luciferase of the firefly Photinus pyralis by using the AscI and PmeI restriction sites. These sites were introduced at the 5′ and 3′ ends of the luciferase gene by PCR mutagenesis. Replicon luc-R591A, carrying the R591A mutation in NS5B, was constructed by replacing the XhoI-SpeI fragment of luc-5.1 with that of pMT (R591A). The luc-GAA replicon, with an active site mutation (GDD to GAA) in NS5B, was constructed the same way and was used as the negative control.
In vitro transcription, electroporation, and selection of G418-resistant cells.
Wild-type or mutant NK5.1 plasmid DNAs were linearized with ScaI and transcribed into RNA with T7 RNA polymerase. The template DNA was removed by digestion with RNase-free DNase I for 1 h at 37°C. The RNA was purified with an RNeasy mini kit (QIAGEN). Subconfluent monolayers of Huh-7 cells were detached from the culture dish by trypsin treatment, washed three times with phosphate-buffered saline (PBS), and resuspended in Cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM potassium phosphate, pH 7.6, 25 mM HEPES, pH 7.6, 2 mM EGTA, pH 7.6, 5 mM MgCl2) (39). Two to five micrograms of replicon RNA was mixed with the cell suspensions and was transferred into an electroporation cuvette. Electroporation of the RNA was carried out with a Gene Pulser II instrument at 270 V and 960 μF in a cuvette with a 0.4-cm gap width (Bio-Rad). The cells were immediately transferred into 12 ml of complete DMEM (1.25% dimethyl sulfoxide) and were seeded into a 10-cm culture dish. At 24 h, the medium was replaced with complete DMEM supplemented with 500 μg of G418 (Geneticin; Gibco Life Sciences) per ml. The growth medium was changed two or three times a week, and 2 to 3 weeks after transfection, colonies were stained with crystal violet. For each replicon, three to five independent transfections were performed.
Replicon RNA quantitation by real-time RT-PCR.
TRIzol reagent (Gibco-BRL) was used for the purification of total RNA from cells harboring replicons WT(NK5.1), MT(3A-II), and MT(SL-IV) according to the manufacturer's protocol. At each time point, a 500-ng aliquot of total RNA was used for the quantitation of HCV replicon copy number using a LightCycler system (Roche). Real-time reverse transcription-PCR (RT-PCR) amplifications were done with the LightCycler RNA amplification kit with SYBR green I (Roche) with a primer pair specific for HCV NS5B (5′ CCATAGTTACTCTCCAGGTGAGATC 3′ [plus-strand sequence] and 5′ GTGTTTAGCTCCCCGTTCA 3′ [minus-strand sequence]). GAPDH (glyceraldehyde 3′-phosphate dehydrogenase) was used as an internal control. The primer pair 5′ GGAAGGTGAAGGTCGGAGTCAACGG 3′ (plus-strand sequence) and 5′ TCCTGGAAGATGGTGATGGGATTTC 3′ (minus-strand sequence) (25) was used to amplify the mRNA. Reverse transcription was carried out at 50°C for 30 min. The PCR protocol consisted of 40 cycles at 95°C for 10 s, 50°C for 10 s, and 72°C for 15 s. Transcript HCV RNA standards of known concentration were used with each set of reactions and these were used to determine a standard curve. The real-time PCR signals were analyzed using the LightCycler software, version 3.5 (Roche).
Amplification of replicon RNA by RT-PCR and sequencing of amplified DNA fragments.
Total RNA (1 μg) was mixed with 1 μM of reverse transcription primer 5′ CAGGATGGCCTATTGGCCTGGAG 3′ (minus-strand primer, nt 9390 to 9412) in a total volume of 10 μl and denatured for 10 min at 65°C. Reverse transcription was performed with SuperScript first-strand synthesis system for RT-PCR (Invitrogen) in a total volume of 20 μl. Five microliters of the reaction mixture was used for PCR with the Expand Long Template PCR system (Roche Biochemicals). Cycle conditions were 2 min of initial denaturation at 95°C and 10 cycles with 30 s at 95°C, 30 s at 45°C, and 60 s multiplied by the number of kilobase pairs of amplified fragment at 68°C, and 25 cycles with 30 s at 95°C, 30 s at 52°C, and 60 s multiplied by the number of kilobase pairs of amplified fragment at 68°C. The reaction mixtures were incubated for 10 min at 68°C, and the PCR products were purified by preparative agarose gel electrophoresis prior to sequencing.
Transient expression of wt and mutant NS5B proteins in Huh-7 cells and indirect immunofluorescence microscopy.
For the transient expression of the wt and mutant NS5B proteins in Huh-7 cells pcDNA3.1 (Invitrogen), was used. PCR fragments encoding the wild-type NS5B protein, NS5B CΔ21, NS5B 3A-II, NS5B 3A-III, NS5B 3A-IV, the R591A mutant, and 3A TT/KK were made with the following pairs of primers (Table 1): 19 and 20, 19 and 21, 19 and 22, 19 and 23, 19 and 24, 19 and 25, and 19 and 22, respectively. For each PCR, the subgenomic replicon containing the corresponding mutation was used as the template. The PCR fragments were cut with BamHI and XbaI and were ligated into a similarly cut pcDNA3.1 vector. NS5B(CΔ21) lacks the C-terminal 21 amino acids of NS5B. The plasmid DNAs were transfected into Huh-7 cells using Lipofectamine (Invitrogen). Indirect immunofluorescence was performed 72 h posttransfection with NS5B monoclonal antibody 5B-12B7 (35). In brief, cells grown as monolayers on glass coverslips were fixed with 3% paraformaldehyde, and incubated for 1 h at room temperature with primary antibody against NS5B (monoclonal antibody 5B-12B7) in PBS containing 3% bovine serum albumin and 0.1% Triton X-100. Bound primary antibody was detected with a fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin. Coverslips were washed with PBS and mounted to a slide and then examined under a fluorescence microscope.
Western blot analysis.
Huh-7 cells in 10-cm-diameter culture dishes were harvested 4 h after transfection and lysed by sonication (1 min) in denaturing protein buffer (50 mM Tris-HCl, pH 8.8, 100 mM dithiothreitol, 0.1% bromophenol blue, 2% sodium dodecyl sulfate, and 10% glycerol). The cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were transferred to a nitrocellulose membrane (Roche) and were immunoblotted according to standard protocols. The NS5B proteins were specifically detected with polyclonal antibodies, a gift of R. de Francesco (3).
Transient-replication assays.
Huh-7 cells (4 × 106) were transfected by electroporation as described above using 5 μg of luciferase replicon RNA. After the addition of 10 ml of complete DMEM, 2.5-ml aliquots of the cell suspension were seeded in 35-mm- or 60-mm-diameter culture dishes and harvested at 4, 24, 48, and 72 h posttransfection. In order to assay the luciferase activity, cells were washed with phosphate-buffered saline and scraped off the plate into 400 μl lysis buffer (Promega). Then, 20 μl of lysate was mixed with 100 μl of assay buffer (Promega) and the firefly luciferase activity was measured with an Optocomp I luminometer (MGM Instruments, Inc.).
RESULTS
Mutational analyses of conserved amino acids in the HCV NS5B C-terminal transmembrane domain.
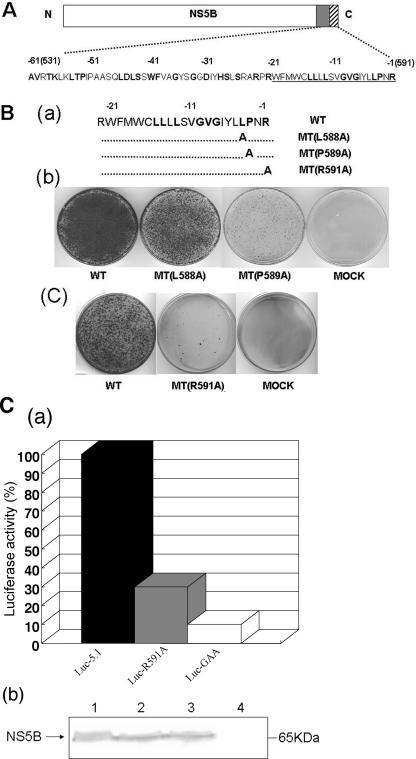
The C-terminal hydrophobic domain of HCV NS5B, encompassing amino acids 570 to 591 (Fig. 2A), is highly conserved among different isolates (Table 2). We selected four amino acids that have been reported by Schmidt-Mende et al. (39) to be fully conserved among 296 different isolates (L588, P589, R570, and R591) (Fig. 2Ba) for mutational analysis. The residues were individually replaced with alanine, and the effects of the changes on the colony-forming abilities of the subgenomic replicons were determined. As shown in Fig. 2Bb, the P589A mutation had a strongly detrimental effect on replication, but the L588A change altered the colony-forming efficiency of the subgenomic HCV replicon to a much lesser extent. These observations support the contention that the identities of the residues in these positions are significant for replicon replication.
FIG. 2.
Mutational analyses of some conserved amino acids in the HCV NS5B C-terminal transmembrane domain. (A) NS5B is divided into three functional motifs. The catalytic domain (open box) of NS5B contains the N-terminal 530 amino acids of the protein, which is followed by the noncatalytic region (aa 530 to 591) at the C terminus. Highly conserved amino acids are shown as boldface letters. Underlined amino acid residues correspond to the transmembrane domain of NS5B (hatched box). Negative numbers above the amino acid sequences represent the amino acid positions counted backward from the C-terminal end of the NS5B protein. (B) Effect of point mutations in the NS5B transmembrane domain on the colony-forming efficiency of the subgenomic replicon. (a) Amino acid sequences of the transmembrane domains of wt and mutant NS5B proteins. (b and c) In vitro RNA transcripts of the wt and mutant replicon constructs were transfected into Huh-7 cells, and the colony-forming efficiency of the replicons was measured as described in Materials and Methods. (C) (a) Effect of the R591A mutation in NS5B on transient replication. Huh-7 cells were transfected with the specified luciferase replicons, and luciferase activities were determined in lysates of cells harvested 4 and 72 h after transfection. The 4-h value (not shown) was used to correct for different transfection efficiencies. The 72-h value of luc-5.1 (7.2 × 103 units) was set as 100%. Data are means of three independent experiments. (b) Western blot analysis of cytoplasmic extracts from naïve and luciferase replicon-transfected Huh-7 cells. Western blot analysis was carried out as described in Materials and Methods with polyclonal antibody to NS5B. Lane 1, luc-5.1-transfected cells; lane 2, luc-R591A-transfected cells; lane 3, luc-GAA-transfected cells; lane 4, naïve Huh-7 cells.
TABLE 2.
Comparison of the hydrophobic domains of HCV and BVDV NS5B and PV 3A
| Strain | Hydrophobic domain of NS5B or 3Aa | EMBL accession no. |
|---|---|---|
| HCV 1a | RWFWFCLLLLAAGVGIYLLPNR | AF009606 |
| HCV 1b | RWFMWCLLLLSVGVGIYLLPNR | AJ238799 |
| HCV 2a | RLLLFGLLLLFVGVGLFLLPAR | AAY24373 |
| HCV 2b | RLLLLCLLLLSVGVGIFLLPAR | AAP55704 |
| HCV 3a | RYLLLCLLLLTVGVGIFLLPAR | AF046866 |
| HCV 3b | RHLLLCLLLLTVGVGIFLLPAR | BAA08372 |
| HCV 4a | RYLLLCLLILTVGCGIFLLPAR | CAA72338 |
| HCV 4b | RWFMWCLLLLSVGVGIYLLPNR | CAA43793 |
| HCV 5a | RNLLLCLLLLSVGVGIFLLPAR | CAA73640 |
| HCV 6a | RFLLLGLLLLTVGVGIFLLPAR | CAA72801 |
| HCV 6b | RMLLLCLLLLSVGVSIFLLPAR | BAA07103 |
| BVDV C | PIVNLLLRRLRVLLMAAVGASS | Q96662 |
| BVDV N | PIVNLLLRRLKILLMTAVGVSS | P19711 |
| PV1 (M) | AMTILQAVTTFAAVAGVVYVMY | NP740474 |
Conserved amino acids are in bold.
Replacement of R570 with an alanine at the N-terminal end of the hydrophobic domain abolished HCV replication (data not shown). We realize, however, that this R570A mutation leads to a disruption of the lower stem of SL-V, an essential cis-acting RNA replication element identified previously by You et al. (51) and by us (25). The effect of the mutation, therefore, could relate to replication rather than to membrane binding. This conjecture is in agreement with previous data by Moradpour et al., who tested the lethal double mutant (R568A/R570A) (37) and reported that the phenotype of this mutant is not due to a lack of membrane binding of the corresponding NS5B protein but rather the result of an RNA replication defect. Moradpour et al. (37) also speculated that the interruption of the structure of SL-V in the R568A/R570A mutant is the culprit of the observed phenotype. The replacement of the arginine at the C terminus of the hydrophobic region (R591A) resulted in a severe reduction of colony formation (Fig. 2Bc). These data differ from results by Moradpour et al. (37), who observed that the R591A mutation altered neither the membrane-binding ability of the double mutant NS5B protein nor the replication efficiency of a dicistronic luciferase replicon. We have repeated the mutational analysis of the R591 residue as described in the legend to Fig. 2A and consistently found that the R591A change had a strongly negative effect on colony formation. It should be noted that our results are similar to those of G. Luo (personal communication), who also observed a severe reduction in the colony-forming efficiency of the NS5B R591A mutant replicon.
To confirm the defective replication phenotype of our R591A mutant, we have analyzed HCV RNA replication in Huh-7 cells by using a dicistronic reporter replicon. Although this strategy is similar to that used by Moradpour et al. (37), our construct was very different: translation of the luciferase gene is promoted by the IRES of HCV, while the IRES of EMCV directs translation of the HCV nonstructural proteins. RNA replication was determined by measurements of luciferase activity at 72 h posttransfection in Huh-7 cells. The wt replicon and a replication-defective RNA (GAA) were used as positive and negative controls, respectively. To ascertain that protein expression with the wt and protein expression with the mutant were the same, we monitored the levels of NS5B protein 4 h after transfection by Western blot analysis (Fig. 2Cb). As shown in Fig. 2Ca, the luciferase activity of the R591A RNA was comparable to that of the negative control, indicating the nearly total absence of replication by this mutant. As we show later (see Fig. 4), these data are in agreement with the altered distribution of the NS5B mutant protein (R591A) when expressed in Huh-7 cells.
FIG. 4.
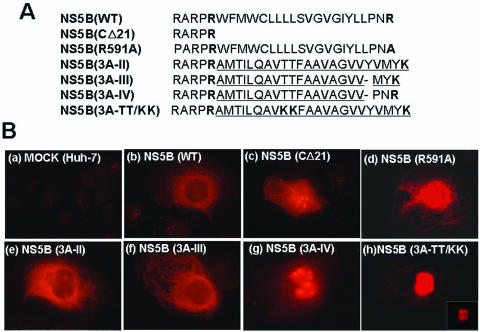
Subcellular localization of wt NS5B and of its variants in Huh-7 cells. (A) Amino acid sequences of the wt and mutant NS5B C-terminal transmembrane domains. The foreign amino acid sequences derived from poliovirus 3A are underlined. The positively charged amino acids located at both ends of the hydrophobic domains of HCV NS5B and poliovirus 3A are marked in bold. NS5B(CΔ21) represents a NS5B protein lacking the C-terminal 21 amino acids. (B) The wt and mutant NS5B proteins were transiently expressed in Huh-7 cells and detected by an indirect immunofluorescence assay with monoclonal antibody 5B-12B7 as described in Materials and Methods.
It is interesting that there is a large discrepancy between the results obtained with the wt and mutant (R591A) replicons in the replication assay using the luciferase replicon (Fig. 2Ca) and the colony formation assay (Fig. 2Bc). While the replication of the mutant is 30% of what is observed with the wt, the colony-forming ability of the mutant is reduced about 1,000-fold. We do not know the exact reason for this observation, but it is most likely related to the use of neo selection when measuring colony formation. It is likely that a 70% decrease in replication of the mutant compared to the wt results in such low levels of neo which are insufficient to promote cell growth and colony formation.
The membrane anchor of HCV NS5B can be functionally exchanged with that of poliovirus polypeptide 3A.
Considering the degree of conservation (39) and the specificity of some amino acids within the hydrophobic domain (37; this work), it was of interest to determine whether the transmembrane domain of HCV NS5B can be functionally replaced by the membrane anchor of another viral protein that is unrelated to HCV. For this purpose, we chose the hydrophobic domain of the poliovirus polypeptide 3A. We designed several chimeric NS5B constructs containing either parts of or the entire C-terminal hydrophobic sequence of poliovirus protein 3A. We selected the hydrophobic domain of poliovirus protein 3A and that of its precursor, 3AB, because (i) the function of the C-terminal hydrophobic domains of both NS5B and 3AB is to anchor their RNA polymerases to membranes (26, 34, 37, 45); (ii) the hydrophobic anchors of both NS5B and PV 3A are transmembrane domains (21; Fujita et al., submitted for publication); (iii) the hydrophobic domains of NS5B and PV 3A are of similar lengths (20 and 22 nonpolar amino acids, respectively); (iv) both hydrophobic domains are flanked on their N and C termini by positively charged amino acids (R and R in NS5B and R and K in 3A); (v) neither hydrophobic domain is interrupted by charged amino acids; and, most importantly for this experiment, (vi) the amino acid sequences of the domains are dramatically different. Although the overall hydrophobicities of the two domains are similar, the shared identity at the amino acid level is only 36%. Moreover, a comparison of the amino acid sequences reveals only one conserved residue, a Gly, seven residues from the C terminus (Fig. 3A).
The first chimeric NS5B construct [MT(3A-I)] contained 16 amino acids of the PV 3A sequence in its central domain flanked by 3 and 2 nonpolar residues of NS5B sequences at the N and C termini, respectively (Fig. 3A). This hydrophobic sequence of residues was located between two positively charged arginines. Transfection of MT(3A-I) RNA into Huh-7 cells yielded a large number of colonies but fewer than that observed with the wt replicon (Fig. 3B). Since the partial replacement of the NS5B transmembrane domain yielded a functional replicon, we subsequently attempted to replace the entire NS5B hydrophobic sequence with the corresponding PV 3A sequence. This new construct [MT(3A-II)] contained an N-terminal arginine followed by 22 hydrophobic residues that were terminated by a positively charged lysine. It is important to note that we introduced four silent mutations into the 3A sequence (for R570, AGG to CGC, and for A571, GCA to GCG) to maintain the authentic structure at the bottom of the stem of the essential cis-acting RNA element SL-V in the NS5B coding sequence (25) and to retain its adjacent RNA sequence (data not shown). Surprisingly, the colony-forming ability of this chimeric replicon, MT(3A-II), was equivalent to that of the construct that contained the partial exchange of NS5B sequences with the PV 3A sequences, MT(3A-I) (Fig. 3B). These results indicate that the heterologous sequence of poliovirus 3A could functionally replace the sequence of the HCV NS5B C terminus in Huh-7 cells, although with reduced efficiency.
To confirm our finding that the replacement of the NS5B hydrophobic sequence with that of PV 3A is functional in the colony-forming ability of the replicon, we reduced the hydrophobicity of the 3A sequence by introducing two positively charged residues. We mutated the two juxtaposed threonine residues (T67 and T68) to lysines in the middle of the hydrophobic domain of poliovirus 3A (Fig. 3A). As expected, the T67K/T68K mutations proved lethal for colony formation (Fig. 3B), an observation supporting our conclusion that the wt 3A sequence can functionally replace the C-terminal hydrophobic sequence in NS5B.
It is important to note that in the poliovirus background, poliovirus 3A did not tolerate the replacement of its hydrophobic domain with the C-terminal membrane insertion sequence of HCV NS5B. That is, the exchange of the hydrophobic region in 3A with that of NS5B, in the context of the poliovirus genome, was lethal (data not shown). The lack of compatibility can be explained by the observation that in poliovirus replication, the hydrophobic domain of polypeptide 3A communicates with other poliovirus proteins. For example, mutations in the 3A hydrophobic domain produce suppressor mutations in polypeptide 2B, a membrane-associated poliovirus protein mapping upstream of 3A (44; Fujita et al., submitted).
Membrane association of wild-type and chimeric NS5B proteins.
To compare the subcellular localization of wt NS5B with that of the chimeric NS5B polypeptides, we used indirect immunofluorescence analyses. The wt and chimeric polypeptides (Fig. 4A) were transiently expressed in Huh-7 cells and then probed with NS5B monoclonal antibodies. As shown in Fig. 4Bb, wt NS5B was detected in the ER membrane network extending from the nuclear membrane through the cytoplasm, confirming previous results (10). In contrast, NS5B from which the C-terminal 21 amino acids were deleted (Fig. 4A) showed a diffuse staining pattern in Huh-7 cells with accumulation of the polypeptides in the nucleus and nucleoli (Fig. 4Bc). These patterns are very similar to those previously observed by Moradpour et al. (37). The NS5B proteins of the defective R591A and lethal TT/KK mutants displayed staining primarily in the nucleus (Fig. 4Bd and h). The chimeric NS5B MT(3A-II) polypeptide (Fig. 4A) revealed a distribution in the cytoplasm reminiscent of that of wt NS5B, although there was also some deposition in the nucleoli (Fig. 4Be). In an attempt to improve the membrane-binding ability of the PV 3A hydrophobic sequence in the NS5B polypeptide, we have made two additional chimeric constructs and determined the cellular localization of the resulting NS5B proteins. In the first construct, NS5B(3A-III), two amino acid residues (Y77 and V78) of the hydrophobic domain of PV 3A were deleted, and its replicon exhibited colony-forming efficiency that was about the same as that of MT(3A-II) (data not shown). This change reduced the number of the hydrophobic amino acids in the membrane insertion sequence of 3A from 22 to 20 amino acids, the same number as that in wt NS5B. Interestingly, this modification of the 3A insertion sequence did not significantly alter the membrane localization of the chimeric polypeptide. NS5B(3A-III) was deposited in the cytoplasm in a pattern similar to that observed with NS5B(3A-II) (Fig. 4Bf). The result that the membrane localizations of NS5B(3A-II) and NS5B(3A-III) are similar to that of wt NS5B is in full agreement with our observation reported here that the hydrophobic PV 3A sequence is functional as a membrane anchor of NS5B in the production of drug-resistant colonies.
For the second chimeric construct, NS5B(3A-IV), in addition to deleting Y77 and V78, we changed the three C-terminal residues (MYK) of the poliovirus 3A-specific sequence to PNR, which is highly conserved in the NS5B proteins of different HCV isolates (39). Surprisingly, NS5B(3A-IV) totally abolished the ER staining patterns typical of wt NS5B. Just like the NS5B(CΔ21) polypeptide, it was deposited predominantly in the nucleus (Fig. 4Bg). These results suggest that the highly conserved PNR sequence of the NS5B hydrophobic domain is not functional in the context of the 3A anchor sequence for replication of the chimeric replicon. One possible explanation of this observation is that the presence of the proline within the 3A sequence alters the structure of the hydrophobic domain in such a way that it can no longer be correctly inserted into the membranes.
Analyses of cell lines containing MT(3A-II) replicon RNA.
To determine whether the replication of the chimeric MT(3A-II) replicon RNA required adaptive mutations for initial replication in clonal cell lines, we selected drug-resistant Huh-7 cells following transfection with the corresponding transcript RNAs and drug selection (250 μg/ml of G418). Surviving cells were then plated again under drug selection and passaged two or three times per week. After each passage, a fraction of the cells was harvested and the replicon RNA therein was sequenced following RT-PCR. Finally, several cell lines harboring the replicon with the chimeric NS5B polypeptide, MT(3A-II), were established following long-term passages at a high drug concentration.
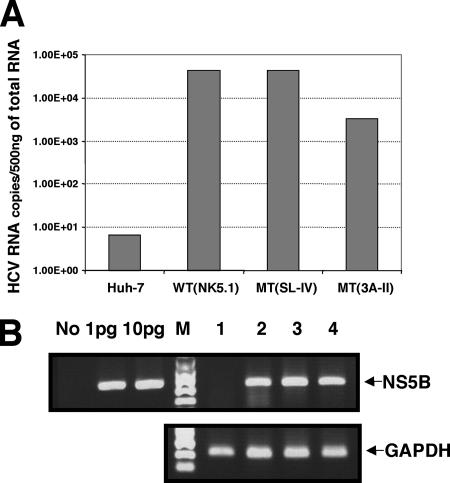
The original colony-forming ability of the MT(3A-II) (Fig. 3B) replicon RNA was less than that of the wt replicon. However, upon passage for four months with 500 μg/ml of G418, individual cell lines harboring the chimera grew as well as cell lines carrying the wt replicon. To determine whether the improved growth properties of the cell lines harboring MT(3A-II) resulted from an increase of replication efficiency of the replicon RNA through genotypic adaptation, by selection of cells more permissive to RNA replication, or by selection of drug-resistant host cells, real-time RT-PCR was performed. For comparison, a cell line containing the parental replicon RNA (NK5.1), which had been maintained for more than 1 year in the presence of a drug (G418, 500 μg/ml), was included in the study. The quantity of HCV RNA was normalized with that of GAPDH RNA. As shown in Fig. 5, the Huh-7 cell lines contained high levels of MT(3A-II) replicon RNA but somewhat less than the cells containing the parental NK5.1 replicon had (compare lanes 2 and 4).
FIG. 5.
Efficient replication of HCV mutant replicons MT(3A-II) and MT(SL-IV) in the established cell lines. (A) Measurement of HCV RNA levels by real-time PCR. Cells from three established cell lines were harvested, and the total RNAs were purified and used for real-time RT-PCR analyses. The quantity of HCV RNA was normalized with that of GAPDH RNA. (B) Agarose gel electrophoresis of the final PCR products obtained by real-time RT-PCR. The PCR products were analyzed by 2% native agarose gel electrophoresis. Lane No, RT-PCR with no RNA; lanes 1pg and 10pg, RT-PCRs with 1 pg and 10 pg of in vitro-transcribed HCV replicon RNA, respectively; lane M, DNA molecular weight marker; lane 1, naïve Huh-7 cells; lane 2, the parental HCV subgenomic replicon (NK5.1); lane 3, MT(3A-II); and lane 4, MT(SL-IV).
To confirm our previous finding that the predicted RNA structure formed by the sequence encoding the hydrophobic domain of NS5B(SL-IV) has no function in replication (25), we have included in our real-time PCR analysis a replicon [MT(SL-IV)] that contained mutations disrupting this stem loop. Cell lines derived from MT(SL-IV) were passaged more than 40 times (G418, 500 μg/ml), and the quantity of replicon RNA in the cells was determined. As expected, the number of HCV RNA copies per 500 ng of total RNA from MT(SL-IV) was essentially the same as that derived from the wt replicon RNA (NK5.1) (Fig. 5, compare lanes 2 and 3). It is interesting that not only SL-IV but also a segment of the 3AB sequence is predicted to form a hairpin structure (7), but its role in PV replication, if any, is not known.
It was of interest to us to determine whether the MT(3A-II) replicon had undergone genotypic changes upon long-term passage. For this reason, we sequenced the NS5B coding regions of the replicons, which were isolated at intervals, and found no nucleotide substitutions in the RNAs. Thus, the replacement of the hydrophobic region in NS5B with that of poliovirus 3A in MT(3A-II) was genetically stable under the conditions of the experiment. We then sequenced the 3′ NTR and the coding region of the MT(3A-II) replicon from NS3 through NS5B by using total RNA isolated from long-term cell lines (>4 months). No nucleotide changes were found in MT(3A-II) and the parental NK5.1 RNA analyzed in parallel. Taken together, our results suggest that the improved growth properties of the replicon in the cell lines are likely to be due to a selection of Huh-7 cells, which we speculate are more permissive to HCV replication than naïve Huh-7 cells (31).
DISCUSSION
Although the importance of a membranous replication complex during HCV replication has been well established, the mechanism of membrane association and the protein-protein interactions involved in this process are not well understood. It has been previously shown that the C-terminal 21-amino-acid-long hydrophobic domain of NS5B is necessary and sufficient for targeting the RNA polymerase to membranes (39). This domain, containing 20 hydrophobic amino acids, is both preceded and terminated by positively charged arginines (R570 and R591, respectively).
A mutation of the arginine at position 591 to alanine strongly reduced the colony formation of the replicon (Fig. 2Bc) and replication of a luciferase-expressing replicon (Fig. 2C). Moreover, the R591A mutant polymerase NS5B, expressed in Huh-7 cells, displayed an aberrant distribution pattern compared to wt NS5B (Fig. 4Bd). Thus, R591 is a very important component of hydrophobic domain function in NS5B. This is in disagreement with the published results of Moradpour et al. (37), who concluded that R591 was not required for replicon replication. The reason for this discrepancy is not clear. Moradpour et al. based their conclusion on luciferase reporter assays in a chimeric construct containing the genotype 1a sequence (aa 467 to 591) at the C terminus of NS5B. Moreover, the poliovirus IRES rather than the HCV IRES was used to promote translation of the luciferase gene. Since detailed assays of colony-forming assays were not included in the report (37), the results cannot be directly compared.
The replacement of the arginine at the N terminus of the hydrophobic domain (R570), which ablated colony formation of the replicon, involved the mutation of two nucleotides (C9306G/C9307G). These two nucleotide substitutions impair formation of the lower stem of the essential cis-acting replication element SL-V (25, 51) and, thus, the lethal phenotype of R570A may be related to RNA synthesis rather than membrane binding. This is likely, since Moradpour et al. (37) have shown that replication in the R568A/R570A double mutant was inhibited, whereas the membrane localization pattern of the corresponding mutant NS5B protein was not significantly altered. The double mutation also altered the structure of the cis-replicating RNA element (37).
To investigate further the role of the NS5B hydrophobic domain in RNA replication, we converted two highly conserved hydrophobic residues into alanines (L588 and P589). Transfection of these mutant RNAs into Huh-7 cells resulted in replicons with reduced colony-forming abilities compared to that of the wt. The replication defect was more evident with the P589A mutant than with the L588A mutant, in which the amino acid substitution was more conservative. These results suggest that these conserved amino acids have a role in but are not essential for the formation of the HCV replication complex.
Based on the mutational analyses reported here and in a previous study (37), it seemed unlikely that the hydrophobic domain of NS5B could be replaced with a hydrophobic domain of different origin and a different sequence. The functional replacement of the NS5B domain with the hydrophobic domain of poliovirus polypeptide 3A was therefore surprising, and it is likely to alter our thinking about the role of the NS5B domain in viral replication.
The 3A protein consists of 87 amino acids, of which the N-terminal 59 residues form the soluble part of the protein. This is followed by a 22-amino-acid-long hydrophobic domain and by 7 additional residues at the C terminus. The hydrophobic domain of PV 3A (amino acids 59 to 81) has been the subject of numerous studies and was found to be essential for PV RNA replication (15, 44, 45). The segment has been subdivided into subdomains I (aa 64 to 72) and II (aa 73 to 80) (45). Based on in vitro membrane-binding studies, it has been determined that the most critical amino acids for membrane association in 3A are located in subdomain II (45; Fujita et al., submitted). In addition to being the membrane anchor of the RNA polymerase, in the context of its precursor 3AB (34, 50), protein 3A is also involved in viral RNA replication, and it is the target of the antiviral drug enviroxime, which blocks viral RNA synthesis (17). In poliovirus-infected HeLa cells, protein 3A alters host cell membrane permeability and inhibits cellular protein secretion (9, 24).
In spite of significant sequence differences, the hydrophobic domains of NS5B and PV 3A are similar in a number of respects. The C-terminal hydrophobic anchor of 3A is nearly equal in size to the corresponding hydrophobic segment in NS5B, and these domains are flanked by positively charged amino acids at both the N and C termini. In addition, both the NS5B and the PV 3A membrane anchors are transmembrane domains that are not interrupted by any charged residues (21; Fujita et al., submitted).
We have analyzed the colony-forming abilities of HCV replicons in which the NS5B hydrophobic domain was either partially or fully replaced with that of poliovirus protein 3A. Several lines of evidence indicate that, in the context of the subgenomic chimeric replicon, the hydrophobic PV 3A sequence is functional in promoting HCV RNA replication, albeit with reduced efficiency. First, the replacement of the central segment (15 amino acids long) of the NS5B hydrophobic domain with 16 residues from PV 3A results in MT(3A-I) replicons, whose colony-forming efficiency is about 20% of that of the wt. Similar results are obtained with a replicon in which the NS5B 21-residue-long hydrophobic domain is fully replaced with the 22-amino-acid-long hydrophobic segment of PV 3A [MT(3A-II)]. This exchange also included the replacement of the C-terminal arginine with a lysine. The observation that the replacement of two threonines with lysine in MT(3A-II) NS5B abolished membrane binding by the mutant protein and the colony-forming ability of the chimeric replicon confirms the importance of the PV 3A sequence for NS5B function. Second, the cellular localization of the mutant MT(3A-II) NS5B is similar to that observed with the wt protein. Third, passaging of the MT(3A-II) replicon in Huh-7 cells did not lead to the emergence of dominant adaptive mutations. An analysis of the total RNA by real-time RT-PCR indicated a high HCV replicon copy number in the MT(3A-II) mutant cell line but a number somewhat less than that in the wt cell line.
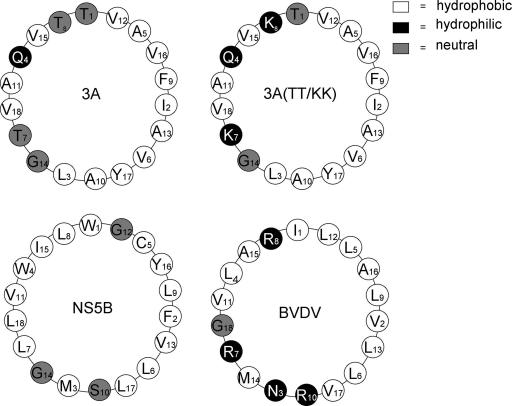
It is interesting that the hydrophobic domain of HCV cannot be replaced by the corresponding domain of bovine viral diarrhea virus (BVDV), another member of the Flaviviridae family, in the context of the HCV subgenomic replicon (26). The hydrophobic domain of BVDV NS5B is 24 amino acids long (PIVNLLLRRLRVLLMAAVGASS) and has been shown to confer membrane binding to BVDV NS5B (23). A comparison of the helical wheels of the BVDV hydrophobic domain with those of HCV NS5B and PV 3A (Fig. 6) reveals one major difference among them. One face of the BVDV helical wheel is highly positively charged, with three positively charged arginines, which are absent in the HCV NS5B and PV 3A helical wheels. This might explain, at least in part, why the NS5B transmembrane domain of BVDV is incompetent to replace the NS5B domain of HCV (26). On the other hand, the distributions of hydrophobic residues in the NS5B and 3A domains, but not the overall sequences, are similar (Fig. 6). Fittingly, the introduction of two basic amino acids into the center of the 3A hydrophobic domain, which makes one face of the helix positively charged (PV 3A TT/KK) (Fig. 6), ablated the function of the membrane binding element.
FIG. 6.
α-Helix projections of HCV NS5B, wt and mutant PV 3A, and BVDV NS5B sequences. The following amino acid sequences were used drawing the helical wheels: HCV NS5B (aa 571 to 588), PV 3A (aa 61 to 78), the PV 3A TT/KK mutant (aa 61 to 78), and BVDV NS5B (aa 699 to 716). The circles are colored according to the hydrophobic character of the residues (39).
The finding that the hydrophobic domain of 3A is functional in HCV NS5B suggests that the precise length and sequence in the primary structure of the anchor domain are not prerequisites of membrane binding by NS5B and RNA replication. The hydrophobic anchors of 3A and NS5B consist of 22 and 20 uncharged amino acids, respectively, flanked by two positively charged arginines (NS5B) or one arginine and one lysine (3A). Shortening the hydrophobic domain of 3A from 22 to 20 amino acids caused no significant change in the cellular localization of the mutant NS5B protein. The lengths of these transmembrane segments are consistent with the typical lengths of transmembrane α-helices. For NS5B, amino acids 571 to 588 were predicted to form a transmembrane α-helix (39). We have recently shown that the hydrophobic domain of 3A can form a transmembrane topology (Fujita et al., submitted).
As we have pointed out above, there is essentially no amino acid sequence similarity between the two hydrophobic segments except for a G seven positions from the C-terminal end. However, there is a similarity in the types of amino acids that are contained within the two segments and in their locations within the segments. Both hydrophobic anchors contain 12 residues with nonpolar aliphatic side chains, most with bulky side chains, and these are found in the central portion of the segments. These stretches of residues are interrupted by one or more G residues (GVG in NS5B and GVV in 3A), which are known to act as helix breakers. In NS5B, this flexible segment was also predicted to adopt an α-helical fold (39). The hydrophobic domains of NS5B and 3A are also similar, in that 2 to 4 residues, located between the primary hydrophobic stretch of 12 amino acids and the positively charged N- and C-terminal residues, consist of amino acids with mostly aromatic R groups (F, Y, or W) or polar, uncharged R groups (M, T, C, S, or N). These residues are most likely located at the membrane interface. We suggest that the numbers of amino acids, the types of amino acids, and their locations within the hydrophobic domains of HCV NS5B and PV 3A are sufficiently similar to permit the functional exchange of these segments in the context of membrane binding by the NS5B protein and the replication of the subgenomic replicon. It will be interesting to analyze other hydrophobic domains with similar amino acid signatures. However, we consider it unlikely that these two different domains per se engage in identical and virus-specific complex formations. Our observation that not only the 3A domain but the entire replicon sequence was genetically stable over many passages seems to indicate that the only essential function of the HCV domain is to anchor the polymerase to the membrane and that the amino acid sequence per se is not involved in the formation of the membrane-associated HCV replication complexes. This is different from the function of the 3A hydrophobic domain in poliovirus replication. Single mutations within the poliovirus 3A hydrophobic domain (15; Fujita et al., submitted) or the exchange of the 3A domain with that of human rhinovirus 14 (44) show severe replication phenotypes that are, upon passage, rapidly changed by direct reversion, by massive amino acid changes within 3A, or by second site suppressor mutations in polypeptide 2B. 2B is a viral membrane-associated protein mapping upstream of 3A. This suggests that the function of the 3A hydrophobic domain is not simply that of a membrane anchor. This also explains why the NS5B hydrophobic domain, when it replaced the 3A domain in the poliovirus genome, yielded a lethal phenotype and no replicating, adapted viruses were ever isolated.
The chimeric replicon replicated with higher efficiency without apparent changes of the genotype (no adaptive mutations) after numerous passages in Huh-7 cells. We suggest that this is related to a selection of Huh-7 cells with a phenotype better accommodating the chimeric replicon. Changes in the phenotypes of Huh-7 cells that increased proliferation of HCV replicons after the “curing” of Huh-7 cells from HCV replicons with interferon have been described previously (4).
Acknowledgments
The subgenomic replicon construct was a generous gift of R. Bartenschlager. NS5B monoclonal antibody (5B-12B7) was kindly provided by D. Moradpour and NS5B polyclonal antibody was a gift of R. de Francesco.
This work was supported by a grant from the NIH NIAID (no. 5R37AI15122).
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta 1601:38-48. [DOI] [PubMed] [Google Scholar]
- 2.Aizaki, H., K. J. Lee, V. M. H. Sung, H. Ishiko, and M. M. C. Lai. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450-461. [DOI] [PubMed] [Google Scholar]
- 3.Behrens, S.-E., L. Tomei, and R. de Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic α-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Torre, J. C., C. Giachetti, B. L. Semler, and J. J. Holland. 1992. High frequency of single base transitions and extreme frequency of precise multiple-base reversion mutations in poliovirus. Proc. Natl. Acad. Sci. USA 89:2531-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimitrova, M., I. Imbert, M. P. Kieny, and C. Schuster. 2003. Protein-protein interactions between hepatitis C virus nonstructural proteins. J. Virol. 77:5401-5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubuisson, J., F. Penin, and D. Moradpour. 2002. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 12:517-523. [DOI] [PubMed] [Google Scholar]
- 11.El-Hage, N., and G. Luo. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761-2769. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari, E., J. Wright-Minogue, J. W. S. Fang, B. M. Baroudy, J. Y. N. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friebe, P., J. Boudet, J.-P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, L., H. Aizaki, J.-W. He, and M. M. C. Lai. 2004. Interactions between nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78:3480-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giachetti, C., S.-S. Hwang, and B. L. Semler. 1992. cis-acting lesions targeted to the hydrophobic domain of a poliovirus membrane protein involved in RNA replication. J. Virol. 66:6045-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinz, B. A., and L. M. Vance. 1996. Sequence determinants of 3A-mediated resistance to enviroxime in rhinoviruses and enteroviruses. J. Virol. 70:4854-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirano, M., S. Kaneko, T. Yamashita, H. Luo, W. Qin, Y. Shirota, T. Nomura, K. Kobayashi, and S. Murakami. 2003. Direct interaction between nucleolin and hepatitis C virus NS5B. J. Biol. Chem. 278:5109-5115. [DOI] [PubMed] [Google Scholar]
- 19.Hofacker, I. L., M. Fekete, C. Flamm, M. A. Huynen, S. Rauscher, P. E. Stolorz, and P. F. Stadler. 1998. Automatic detection of conserved RNA structure elements in complete RNA virus genomes. Nucleic Acids Res. 26:3825-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishido, S., T. Fujita, and H. Hotta. 1998. Complex formation of NS5B with NS3 and NS4A proteins of hepatitis C virus. Biochem. Biophys. Res. Commun. 244:35-40. [DOI] [PubMed] [Google Scholar]
- 21.Ivashkina, N., B. Wolk, V. Lohmann, R. Bartenschlager, H. E. Blum, F. Penin, and D. Moradpour. 2002. The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J. Virol. 76:13088-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyono, K., M. Miyashiro, and I. Taguchi. 2002. Human eukaryotic initiation factor 4AII associates with hepatitis C virus NS5B protein in vitro. Biochem. Biophys. Res. Commun. 292:659-666. [DOI] [PubMed] [Google Scholar]
- 23.Lai, V. C. H., C. C. Kao, E. Ferrari, J. Park, A. S. Uss, J. Wright-Minogue, Z. Hong, and J. Y. N. Lau. 1999. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J. Virol. 73:10129-10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lama, J., and L. Carrasco. 1992. Expression of poliovirus nonstructural proteins in Escherichia coli cells. Modification of membrane permeability induced by 2B and 3A. J. Biol. Chem. 267:15932-15937. [PubMed] [Google Scholar]
- 25.Lee, H., H. Shin, E. Wimmer, and A. V. Paul. 2004. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J. Virol. 78:10865-10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, K. J., J. Choi, J.-H. Ou, and M. M. M. Lai. 2004. The C-terminal transmembrane domain of hepatitis C virus (HCV) RNA polymerase is essential for HCV replication in vivo. J. Virol. 78:3797-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 28.Leveque, V. J.-P., R. B. Johnson, S. Parsons, J. Ren, C. Xie, F. Zhang, and Q. M. Wang. 2003. Identification of a C-terminal regulatory motif in hepatitis C virus RNA-dependent RNA polymerase: structural and biochemical analysis. J. Virol. 77:9020-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmann, V., F. Korner, J.-O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 31.Lohmann, V., S. Hoffman, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundin, M., M. Monne, A. Widell, G. von Heijne, and M. A. A. Persson. 2003. Topology of the membrane-associated hepatitis C virus protein NS4B. J. Virol. 77:5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyle, J. M., A. Clewell, K. Richmond, O. C. Richards, D. A. Hope, S. C. Schultz, and K. Kirkegaard. 2002. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J. Biol. Chem. 277:16324-16331. [DOI] [PubMed] [Google Scholar]
- 35.Moradpour, D., E. Bieck, T. Hugle, W. Wels, J. Z. Wu, Z. Hong, H. E. Blum, and R. Bartenschlager. 2002. Functional properties of a monoclonal antibody inhibiting the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:593-601. [DOI] [PubMed] [Google Scholar]
- 36.Moradpour, D., R. Gosert, D. Egger, F. Penin, H. E. Blum, and K. Bienz. 2003. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antivir. Res. 60:103-109. [DOI] [PubMed] [Google Scholar]
- 37.Moradpour, D., V. Brass, E. Bieck, P. Friebe, R. Gosert, H. E. Blum, R. Bartenschlager, F. Penin, and V. Lohmann. 2004. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J. Virol. 78:13278-13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin, W., H. Luo, T. Nomura, N. Hayashi, T. Yamashita, and S. Murakami. 2002. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J. Biol. Chem. 277:2132-2137. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 40.Semler, B. L., C. W. Anderson, R. Hanecak, L. F. Dorner, and E. Wimmer. 1982. A membrane-associated precursor to poliovirus VPg identified by immunoprecipitation with antibodies directed against a synthetic heptapeptide. Cell 28:405-412. [DOI] [PubMed] [Google Scholar]
- 41.Shimakami, T., M. Hijikata, H. Luo, Y. Y. Ma, S. Kaneko, K. Shimotohno, and S. Murakami. 2004. Effect of interaction between hepatitis C virus NS5A and NS5B on hepatitis C virus RNA replication with the hepatitis C virus replicon. J. Virol. 78:2738-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirota, Y., H. Luo, W. Qin, S. Kaneko, T. Yamashita, K. Kobayashi, and S. Murakami. 2002. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRP) NS5B and modulates RNA-dependent RNA polymerase activity. J. Biol. Chem. 277:11149-11155. [DOI] [PubMed] [Google Scholar]
- 43.Tomei, L., R. L. Vitale, I. Incitti, S. Serafini, S. Altamura, A. Vitelli, and R. de Francesco. 2000. Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J. Gen. Virol. 81:759-767. [DOI] [PubMed] [Google Scholar]
- 44.Towner, J. S., D. M. Brown, J. H. C. Nguyen, and B. L. Semler. 2003. Functional conservation of the hydrophobic domain of polypeptide 3AB between human rhinovirus and poliovirus. Virology 314:432-442. [DOI] [PubMed] [Google Scholar]
- 45.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 46.Tuplin, A., J. Wood, D. J. Evans, A. H. Patel, and P. Simmonds. 2002. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA 8:824-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uchida, M., N. Hino, T. Yamanaka, H. Fukushima, T. Imanishi, Y. Uchiyama, T. Kodama, and T. Doi. 2002. Hepatitis C virus core protein binds to a C-terminal region of NS5B RNA polymerase. Hepatol. Res. 22:297-306. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wattenberg, B., and T. Lithgow. 2001. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2:66-71. [DOI] [PubMed] [Google Scholar]
- 50.Xiang, W., A. Cuconati, D. Hope, K. Kirkegaard, and E. Wimmer. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and genetic variants of 3AB. J. Virol. 72:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong, W., A. S. Uss, E. Ferrari, J. Y. N. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]