Abstract
Assembly of the rotavirus outer capsid is the final step of a complex pathway. In vivo, the later steps include a maturational membrane penetration that is dependent on the scaffolding activity of a viral nonstructural protein. In vitro, simply adding the recombinant outer capsid proteins VP4 and VP7 to authentic double-layered rotavirus subviral particles (DLPs) in the presence of calcium and acidic pH increases infectivity by a factor of up to 107, yielding particles as infectious as authentic purified virions. VP4 must be added before VP7 for high-level infectivity. Steep dependence of infectious recoating on VP4 concentration suggests that VP4-VP4 interactions, probably oligomerization, precede VP4 binding to particles. Trypsin sensitivity analysis identifies two populations of VP4 associated with recoated particles: properly mounted VP4 that can be specifically primed by trypsin, and nonspecifically associated VP4 that is degraded by trypsin. A full complement of properly assembled VP4 is not required for efficient infectivity. Minimal dependence of recoating on VP7 concentration suggests that VP7 binds DLPs with high affinity. The parameters for efficient recoating and the characterization of recoated particles suggest a model in which, after a relatively weak interaction between oligomeric VP4 and DLPs, VP7 binds the particles and locks VP4 in place. Recoating will allow the use of infectious modified rotavirus particles to explore rotavirus assembly and cell entry and could lead to practical applications in novel immunization strategies.
To initiate rotavirus infection, the nonenveloped, icosahedral, triple-layered particle (TLP or virion) must translocate a large transcriptionally active subviral particle, the double-layered particle (DLP), across a membrane and into the cytoplasm of a target cell. The DLP consists of concentric VP2 and VP6 icosahedral protein shells, which encapsidate 11 double-stranded RNA genome segments, the viral polymerase (VP1), and a capping enzyme (VP3). Biochemical and structural studies indicate that conformational rearrangements in VP4 and VP7, the two proteins that make up the outermost shell of the TLP, deliver the DLP into the cytoplasm. The dissociation of trimers of the plate-like protein VP7 in low-calcium environments mediates uncoating in vitro (17, 51). The spike protein VP4 is anchored in the DLP and protrudes through the VP7 layer (53, 58). This protein undergoes a fold-back rearrangement that resembles the fusogenic rearrangements of enveloped virus fusion proteins (18), although the function of the VP4 conformational change has yet to be demonstrated experimentally.
Because VP4 and VP7 are both targets of neutralizing antibodies, understanding the mechanism of cell entry is linked to understanding protection against rotavirus gastroenteritis, which kills approximately 500,000 children each year (43). The development of efficient techniques to incorporate recombinant VP4 and VP7 into infectious rotavirus virions would provide powerful tools to investigate how these proteins mediate translocation of the DLP into the cytoplasm. A recombinant gene encoding VP4 has recently been introduced into the rotavirus genome (30). This first realization of rotavirus reverse genetics relies on selection for recombinant viruses during serial passage and allows the production of replication-competent particles with modified outer capsid proteins.
The ability to engineer virions that are defective for cell entry would greatly expand the possibilities for experiments that probe the rotavirus cell entry pathway. The development of “recoating genetics” for reovirus allows such experiments to be carried out using this virus (4). Reovirus recoating has been used successfully to study assembly and cell entry mechanisms (3, 4, 40, 41). During recoating, minimally infectious authentic reovirus cores (functionally equivalent to rotavirus DLPs) are incubated in vitro with the recombinantly expressed components of the reovirus entry apparatus. Recoating produces particles that are approximately one million times more infectious than cores and half as infectious as authentic reovirus virions (4).
Reovirus particle assembly occurs entirely in the cytoplasm (reviewed in reference 39). In contrast, rotavirus assembly in vivo involves sequential steps in the cytoplasm, the endoplasmic reticulum (ER), and possibly a post-ER compartment (reviewed in reference 20). Rotavirus outer capsid assembly in vivo requires a virally encoded ER-membrane receptor and involves budding of the DLP into the ER followed by a maturational membrane penetration. This complexity raises a potential barrier to developing in vitro recoating for rotavirus.
Transcapsidation of rotavirus particles suggests that it is possible to recoat rotavirus DLPs with virion-derived outer capsid proteins to achieve at least a low level of infectivity (5). In transcapsidation, solubilized outer capsid proteins from virions of one strain are incubated with DLPs from another strain at acidic pH. The resulting transcapsidated particles resemble TLPs by electron microscopy of negatively stained specimens and are 500- to 1,000-fold more infectious than the input DLPs, with a specific infectivity of 106 to 107 particles per PFU (5). The finding that all detectable VP4 in the transcapsidation mixture is degraded by trypsin and, therefore, not properly mounted on the particles led to the hypothesis that the scaffolding activity of NSP4 may be required for proper assembly of VP4 and the production of particles as infectious as authentic virions (5).
The assembly of noninfectious, triple-layered virus-like particles (VLPs) entirely from recombinant rotavirus proteins suggests that DLPs might also be recoated with recombinant VP4 and VP7. VP4-containing, triple-layered VLPs have been produced by coinfecting insect cells with baculoviruses that express recombinant VP2, VP4, VP6, and VP7 (9, 24) and by mixing lysates and supernatants of insect cells infected separately with baculoviruses expressing VP4, VP6, or VP7 (49). Experiments using VLPs that include VP4 and VP7 have provided important insights into rotavirus attachment and membrane interactions (9, 23, 24) and have been evaluated as immunogens (8, 9, 49).
We describe here the efficient recoating of rotavirus with recombinant outer capsid proteins. Recoated rotavirus particles are highly infectious. The use of purified components allows analysis of the in vitro outer capsid assembly pathway. These findings have implications for our understanding of outer capsid assembly in infected cells and the requirements for efficient cell entry.
MATERIALS AND METHODS
Virus infection and cell culture.
Baculoviruses expressing rhesus rotavirus (RRV) VP4 or VP7 have been described previously (22, 34) and were propagated in Sf9 insect cells grown in spinner cultures with Sf900 II serum-free medium (Invitrogen) (16, 17, 57). MA104 cells were grown in M199 (Invitrogen) supplemented with 7.5% fetal bovine serum (HyClone Laboratories, Inc.), 10 mM HEPES, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. For rotavirus inoculation and amplification, fetal bovine serum was omitted and M199 was supplemented to 1 μg/ml with porcine pancreatic trypsin (Sigma-Aldrich).
Infectious focus assay.
For rotavirus titration, confluent MA104 cell monolayers were washed twice with M199 and inoculated with serial dilutions of virus in M199 containing 1 μg/ml trypsin. After incubation for 1 h at 37°C, the inoculum was removed and replaced with serum-supplemented M199 containing 2.5 μg/ml neutralizing monoclonal antibody M159 (27) to inhibit secondary infection. At 14 to 18 h postinfection, cells were washed with phosphate-buffered saline and fixed with methanol. Infectious foci were detected by immunoperoxidase staining. Monoclonal antibody M60, which recognizes VP7 (54), was used as the primary detection antibody.
Rotavirus and recoated particle purification.
Rotavirus, strain RRV (G3, P5B[3]) was purified from the frozen and thawed medium of infected MA104 cells. Cell debris was cleared by low-speed centrifugation. Viral particles were concentrated by pelleting at 158,000 × g for 1 h at 4°C in a 45Ti rotor (Beckman) or by ultrafiltration through a polyethersulfone membrane with a 50-kDa molecular mass cutoff, using a stirred cell (Amicon). The concentrated virus suspension was extracted twice with Freon 113. The aqueous phase was concentrated further, briefly sonicated, and layered over CsCl solutions in TNC (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM CaCl2) for TLPs and recoated particles or TNE (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA) for DLPs. Samples were centrifuged at 256,000 to 286,000 × g in an SW 55Ti rotor at 4°C. Preformed gradients (1.26 to 1.45 g/ml) were centrifuged for 1.5 to 2 h. Self-forming gradients (1.36 g/ml mean density) were centrifuged for 18 h. Densities were determined by refractometry. Particle-containing fractions were identified by mixing 5 μl of each fraction with an equal volume of 2-μg/ml ethidium bromide, boiling for 1 minute, and illuminating at 302 nm. The illuminated samples were imaged using a model 4915-2010 charge-coupled-device camera (Cohu, Inc.) and compared on a relative scale by integrating luminosity in a circle of fixed radius for each drop using Photoshop 7 (Adobe Systems, Inc.). DLPs were banded a second time and dialyzed against TN (20 mM Tris, pH 8.0, 100 mM NaCl) with 0.02% sodium azide. TLPs were dialyzed against TNC with 0.02% sodium azide.
Recombinant protein purification.
RRV VP4 was purified from recombinant baculovirus-infected Sf9 cells by anion exchange and size exclusion chromatography as previously described (16). Recombinant RRV VP7 was purified from the medium of recombinant baculovirus-infected Sf9 cells by lectin affinity, immunoaffinity, and size exclusion chromatography as previously described (17). Purified proteins were quantified by absorbance at 280 nm, using absorption coefficients of 95,100 M−1 cm−1 for VP4 and 62,700 M−1 cm−1 for VP7. Absorption coefficients were calculated based on the sequence using Vector NTI 7 (InforMax, Inc.).
Quantitation of DLPs and TLPs.
Viral and subviral particles were counted by transmission electron microscopy (TEM) of negatively stained DLPs, using an adaptation of a previously described technique (47). Droplets containing purified DLPs mixed with known quantities of 85-nm-diameter latex bead standards (Polysciences, Inc.) were deposited on Formvar grids. The grids were supported on scintered glass so that buffer was drawn through the grids by capillary action, depositing particles on the grid surface. Droplets of 0.25% uranyl acetate and wash solution (TNC) were similarly drawn through the grids to ensure retention of the particles. Images were obtained with a JEOL 1200EX 80-kV electron microscope. This technique gave highly reproducible ratios of DLPs to latex beads. Using these counts as a standard, we determined that 1 A260 unit = 0.213 mg/ml of DLPs, corresponding to 2.2 × 1012 particles/ml. Estimation of TLP quantity by spectrophotometry proved less reliable, possibly due to light scattering by aggregated particles. Therefore, reported TLP quantities were measured by densitometry of VP2 and VP6 bands on Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Bands produced by DLPs quantified by absorbance at 260 nm were used as standards.
Immunoelectron microscopy.
Trypsin-digested particles suspended in TNC were adsorbed to carbon-Formvar-coated grids for 5 minutes, after which excess buffer was blotted away. The grids were blocked with 1% bovine serum albumin in TNC, which was also used as a diluent for antibodies. Primary monoclonal antibodies, 120 μg/ml 7A12 against VP8* (35) or 100 μg/ml anti-six-histidine (QIAGEN, Inc.), were applied to the grids for 15 min. The grids were washed four times with TNC, and 20 μg/ml of a 5-nm colloidal gold-conjugated goat antiserum against mouse immunoglobulin G (IgG; Kirkegaard & Perry Laboratories, Inc.) was applied for 15 min. The grids were washed four times with TNC and negatively stained with 0.75% uranyl formate, pH 4.5 to 5.0.
Recoating reaction.
In the standard recoating protocol, 9.75 μg of purified VP4 (11.7 molecules for each binding site) in 1.0 to 2.5 μl of TNE was added to 3 μg DLPs in 2 to 5 μl of TN with 0.02% sodium azide. The solution was adjusted to pH 5.2 with a 0.1 volume of 0.5 M sodium acetate and incubated for 1 h at room temperature (RT; approximately 25°C). Next, 6 μg of purified VP7 (4.6 molecules for each binding site) in 10 to 20 μl of 0.1× TNC was mixed with 0.5 M sodium acetate, pH 5.2, and a 20 mM Tris-buffered 10 mM CaCl2 solution. The volumes of the sodium acetate and CaCl2 solutions were calculated to yield final concentrations of 50 mM sodium acetate and 1 mM excess calcium (over EDTA) when combined with the DLP and VP4 mixture. After addition of this VP7-containing solution, the recoating reaction mixture was incubated for 1 h at RT. Trypsin activation of the recoated particles is described below (see “Trypsin digestion”). To test the dependence of the recoating reaction on pH, the reaction mixtures were brought to 50 mM in the following buffers during each incubation: sodium acetate (pH 4.6, 5, 5.2, and 5.4), 2-morpholinoethanesulfonic acid (pH 5.8 and 6.2), piperazine-1,4-bis(2-ethanesulfonic acid) (pH 6.6), and 3-(N-morpholino)-propanesulfonic acid (pH 7.2).
For recoating with crude preparations of VP4 and VP7, 40 μl of a clarified recombinant baculovirus-infected cell lysate, containing approximately 30 μg/ml (340 nM) VP4, was added to 1 μg of DLPs, followed by 0.1 volume of 0.5 M sodium acetate, pH 5.2. The mixture was incubated for 1 h at RT. The source of VP7 was a clarified recombinant baculovirus-infected insect cell medium which had been concentrated 10-fold using an Amicon Ultra-4 10-kDa cutoff ultrafiltration device (Millipore Corp.). This concentrated medium contained approximately 0.85 μg/ml (27 nM) VP7. Fifty microliters of the concentrated VP7-containing medium was mixed with acetate buffer and calcium (as described above) and added to the DLPs and VP4 before a second 1-h RT incubation. Variations on these protocols are described in Results and the figure legends.
Trypsin digestion.
Rotavirus particles were primed for infection by digestion with 2.5 to 5 μg/ml tissue culture-grade trypsin (1,000 to 1,500 N-benzoyl-l-arginine ethyl ester (BAEE) units/mg; Sigma-Aldrich) at 37°C for 30 min. For analytical digestion, 1-chloro-3-tosyl-amido-7-amino-2-heptanone-treated trypsin (at least 10,350 BAEE units/mg; Worthington Biochemical Corp.) was freshly diluted from a 5-mg/ml stock in 1 mM HCl and added to 1 μg/ml. After incubation at 37°C for 30 min, analytical digestions were quenched by adding 100 mM ethanolic phenylmethylsulfonyl fluoride to a final concentration of 1 mM and incubating for 5 min on ice before boiling in SDS-PAGE sample buffer.
Western blotting.
Proteins separated by SDS-PAGE were transferred to a polyvinylidene difluoride membrane (Bio-Rad Laboratories) by electroblotting. VP4 was reduced by treatment with approximately 5% β-mercaptoethanol prior to electrophoresis and detected using monoclonal antibody HS2 (42). VP7 was detected using monoclonal antibody M60 (54). To preserve the disulfide-dependent M60 epitope, VP7 was not reduced prior to electrophoresis (55). Peroxidase-conjugated goat anti-mouse IgG was used as a secondary antibody (Kirkegaard & Perry Laboratories, Inc.) and was detected by chemiluminescence (ECL Plus; GE Healthcare).
RESULTS
Reconstitution of infectivity by recoating DLPs with recombinant VP4 and VP7.
Incubation of purified RRV DLPs with purified recombinant RRV VP4 and VP7 in the presence of calcium, followed by trypsin digestion of the mixture, reproducibly increases the infectivity of the particles by a factor of 105 to 107. The purified DLPs added to the reaction mixture are free of authentic VP4 or VP7, as assayed by Western blotting and Coomassie-stained SDS-PAGE (not shown). In the experiment shown in Fig. 1A, the specific infectivity of the DLPs before recoating was 3 × 108 particles per focus-forming unit (FFU); after recoating, the specific activity was 28 particles per FFU.
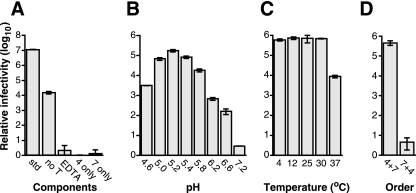
FIG. 1.
Basic recoating parameters. “Relative infectivity” is the infectious titer of each recoating mixture divided by the infectious titer of DLPs alone. The error bars indicate the standard deviations of three titrations of each sample. The standard recoating mixture is described in Materials and Methods. Modifications to this procedure are indicated under each graph. (A) Dependence on the components of the reaction mixture. std, standard procedure; no T, not trypsin digested; EDTA, 5 mM EDTA added before trypsin digestion; 4 only, buffer alone substituted for VP7; 7 only, buffer alone substituted for VP4. (B) Dependence on pH. Recoating reaction mixtures were adjusted to the indicated pH during incubation with VP4 and VP7 (buffers are listed in Materials and Methods). (C) Temperature dependence. Temperatures during incubations with VP4 and VP7 are indicated. (D) Order of addition. VP4 and VP7 were added to the reaction mixtures in the indicated orders: 4 + 7, VP4 first; 7 + 4, VP7 with excess calcium first.
Trypsin digestion primes rotavirus particles for infectivity by cleaving VP4 into two fragments, VP8* and VP5* (21). Trypsin priming increases the infectivity of purified authentic TLPs approximately 20- to 66-fold (10, 21). Trypsin priming increases the infectivity of recoated particles almost 1,000-fold (Fig. 1A). The lesser enhancement of authentic particle infectivity by trypsin priming may reflect adventitious cleavage of some VP4 on authentic particles during growth and purification, resulting in greater baseline infectivity before intentional trypsin cleavage. Calcium chelation destroys the infectivity of TLPs by uncoating virions (7). Correspondingly, recoated particle infectivity is reduced to the level of DLPs by incubation with EDTA (Fig. 1A).
The recoating reaction is strongly pH dependent. It is maximally efficient at pH 5.2 but yields less than a 10-fold increase in infectivity at pH 7.2 (Fig. 1B). Recoating is also temperature dependent, with a broad maximum between 4 and 30°C and a 100-fold-lower yield of infectious particles at 37°C (Fig. 1C). The recoating reaction can be carried out using crude preparations of proteins from recombinant baculovirus-infected insect cell lysates (for VP4) and medium (for VP7, which is secreted from insect cells). We have obtained increases in infectivity by factors of up to 106 using these crude preparations (not shown).
Recoating with limiting amounts of outer capsid proteins.
Incubating DLPs with VP4 alone or with VP7 alone does not significantly increase their infectivity (Fig. 1A). Differences in the dependence of infectivity on VP7 and VP4 concentration and amount (relative to DLPs) reflect differences in the assembly of these proteins and in their function during cell entry. In the descriptions of these experiments, the stoichiometry of outer capsid proteins is calculated relative to the number of available binding sites rather than relative to the number of DLPs. Thus, a stoichiometry of 1 corresponds to 780 VP7 molecules and 180 VP4 molecules per DLP. The numbers of binding sites per DLP are based on evidence from electron cryomicroscopy and X-ray crystallography (18, 44, 58).
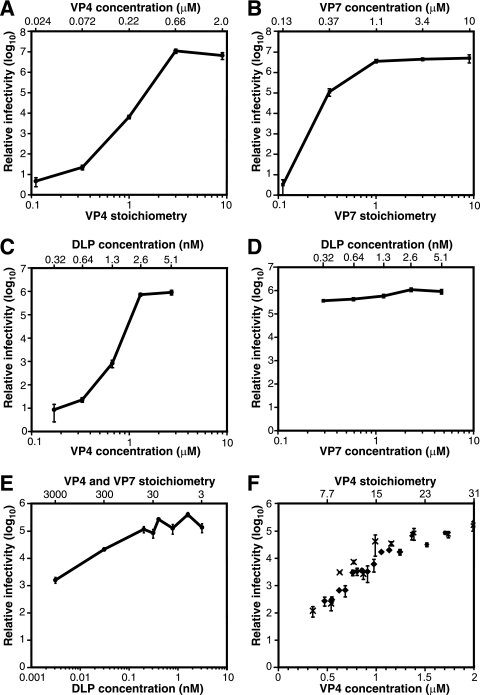
We explored the effect on infectious yield of increasing the amount of each outer capsid protein in the recoating reaction mixture from a stoichiometry of 0 to 9 (Fig. 2A and B). To explore dependence on VP4, the amount and concentration of DLPs and VP7 were held constant (Fig. 2A). The amount and concentration of VP4 varied together. Infectivity rose with each increment of VP4 until it reached a plateau at a VP4 stoichiometry of 3. The increased infectivity (approximately 20-fold greater than with DLPs alone) at a VP4 stoichiometry of 0.3 suggests either that particles with partial VP4 occupancy are infectious or that limiting quantities of VP4 are distributed nonrandomly between particles. The continued rise in infectivity above a VP4 stoichiometry of 1 (and concentration of 0.22 μM) suggests that relatively low-affinity interactions are involved in VP4 addition or that not all added VP4 molecules can assemble and function in entry.
FIG. 2.
Titration of recoating reaction mixture components. Infectivity is reported relative to that of DLPs alone. In panels A to D, 3 × 1010 DLPs were used in each recoating. In panels C to F, saturating amounts of VP4 and VP7 were always present. Error bars indicate the standard deviations of three titrations of each sample. (A) Recoating with limiting amounts of VP4. The VP7 stoichiometry is 3. The VP7 concentration is 3.4 μM. (B) Recoating with limiting amounts of VP7. The VP4 stoichiometry is 9. The VP4 concentration is 2 μM. (C) Dependence of recoating on the concentration of VP4 and DLPs. The VP4 and DLP concentrations were varied in parallel by dilution with VP7 in TNC. The VP7 concentration is 4.7 μM; the VP7 stoichiometry varies from 1 to 16; the VP4 stoichiometry is 3. (D) Dependence of recoating on the concentration of VP7 and DLPs. The VP7 and DLP concentrations were varied in parallel by dilution with VP4 in TNE. The VP4 concentration is 2.8 μM; the VP4 stoichiometry varies from 3 to 48; the VP7 stoichiometry is 1. (E) Titration of DLPs. The VP4 concentration is 1.7 μM, and the VP7 concentration is 8.7 μM. The stoichiometries of VP4 and VP7 vary inversely with the amount of DLPs. VP4 and VP7 are present in the same stoichiometry. (F) Titration of VP4. The VP7 concentration is 0.9 μM; the VP7 stoichiometry is 3.6. The DLP concentration is 0.32 nM; the DLP quantity is 9.4 × 109 particles. The data points marked by diamonds and crosses are taken from separate experiments. Note that the abscissa of this panel is on a linear scale.
To explore the dependence on VP7, the amount and concentration of DLPs and VP4 were held constant (Fig. 2B). The amount and concentration of VP7 varied together. Infectivity plateaued at a VP7 stoichiometry of 1. At a VP7 stoichiometry of 0.3, approximately 3% of the maximum infectious yield was obtained, corresponding to a 105-fold enhancement in infectivity. The large increase in infectivity with a substoichiometric amount of VP7 suggests either that partially coated particles are infectious or that VP7 assembly is cooperative, so that a higher proportion of complete capsids form than can be accounted for by the random distribution of VP7 between particles.
Dependence of recoating on DLP and outer capsid protein concentrations.
In the experiments of Fig. 2A and B, the quantity of one outer capsid protein varied from substoichiometric to saturating amounts. The concentration of that protein varied together with its quantity. Thus, with less recombinant protein added, the lower infectivity could have been due to an insufficient quantity of VP4 or VP7 being available for assembly or due to an insufficient concentration of VP4 or VP7 being available to drive low-affinity interactions. To distinguish these possibilities, we performed experiments in which both outer capsid proteins were always present in excess relative to binding sites on DLPs, and the concentration of each outer capsid protein was varied separately (Fig. 2C and D). In these experiments, the reaction volume was progressively increased, with the concentration of one outer capsid protein being held constant so that its total amount increased. As the volume increased, the concentration of DLPs and of the other outer capsid protein fell together.
To assess the effect of VP4 and DLP concentration on infectious yield, constant amounts of VP4 and DLPs were diluted with varying amounts of buffer and VP7. The final VP4 and DLP concentrations varied inversely with the volume added. VP7 concentration remained constant, and the VP7 stoichiometry was always saturating at 1 or greater (Fig. 2C). The infectious yield was strongly dependent on the concentration of VP4 and DLPs. A twofold decrease in VP4 (1.3 to 0.67 μM) and DLP (2.6 to 1.3 nM) concentration decreased infectivity 869-fold. This steep decrease indicates greater-than-second-order reaction kinetics. Therefore, cooperativity involving VP4 and/or DLPs must drive VP4 assembly.
To assess the effect of VP7 and DLP concentration on the infectious yield of recoating, a constant amount of DLPs was diluted with various amounts of buffer and VP4 (Fig. 2D). VP4 concentration remained constant, and VP4 stoichiometry remained saturating at 3 or greater. After incubation, a constant amount of VP7 was added in a buffer adjusted to maintain a 1 mM final excess of calcium over EDTA. The final VP7 and DLP concentrations varied inversely with volume. Infectious yield was insensitive to the concentration of VP7 and DLPs over the range tested. As the DLP and VP7 concentrations fell 16-fold, infectivity only decreased 3-fold (Fig. 2D). These results indicate that VP7 binds DLPs with high affinity.
Dependence of recoating on DLP concentration and quantity.
To detect potential cooperativity involving DLPs, constant amounts and concentrations of VP4 and VP7 were added to a constant volume of serially diluted DLPs (Fig. 2E). The amount of DLPs varied from 3 × 107 to 3 × 1010 particles, and the DLP concentration varied in parallel. From 3 to 200 pM (3 × 107 to 2 × 109 particles), infectious yield varied linearly with DLP concentration and quantity. At higher concentrations and quantities, the increase in infectivity leveled off. Based on the threefold or greater molar excess of VP4 and VP7 in all reaction mixtures and the results presented in Fig. 2A and B, depletion of outer capsid proteins by an excess of DLPs does not explain the plateau. Thus, there was no indication of cooperativity between DLPs in the recoating reaction. Actually, the results suggest competition between DLPs at higher concentrations. We hypothesize that the plateau in infectivity at higher DLP concentrations and amounts reflects aggregation of recoated particles, which we have observed in larger samples (see “Trypsin priming of VP4 on recoated particles,” below).
Dependence of recoating on VP4 concentration.
The large drop in infectivity when VP4 and DLP concentration decline together (Fig. 2C) and the modest drop when only DLP concentration declines (Fig. 2E) suggest that VP4 concentration is a key determinant of infectious recoating efficiency. To test this hypothesis, we added a constant volume of serially diluted VP4 to a constant amount and concentration of DLPs. The total amount of VP4 was always saturating, with a stoichiometry always greater than 3. After incubation, a constant amount of VP7 was added (Fig. 2F). A 5.7-fold rise in VP4 concentration (from 0.35 to 2.0 μM) increased infectivity approximately 1,000-fold. At higher VP4 concentrations, infectivity fell (not shown). Copelleting of particles and VP4 with low-speed centrifugation (16,000 × g for 10 min) at higher VP4 concentrations suggests that this fall in infectivity is due to coaggregation of VP4 and DLPs (not shown). In the range of concentrations displayed in Fig. 2F, the dependence of infectivity on VP4 concentration is approximately fourth order, although the confounding effect of VP4-mediated aggregation precludes fitting a simple model to the data.
Dependence of recoating on the order of addition of VP4 and VP7.
Adding VP7 to DLPs before VP4 yields minimal (fivefold) enhancement of infectivity; adding VP4 before VP7 yields a full (105- to 106-fold) increase (Fig. 1D). Varying the delay between the addition of VP4 and VP7 does not significantly affect the infectious yield, and simultaneous addition produces an intermediate level of infectivity (not shown). VP4 must also be added before VP7 when crude insect cell lysates containing VP4 are used in place of purified VP4 and unprocessed insect cell medium containing VP7 is used in place of purified VP7 (not shown). Because the recoating mixture contains the same components at the time of trypsin priming, regardless of the order of addition, these results suggest that sequential assembly of VP4, and then VP7, onto DLPs is required to produce infectious particles.
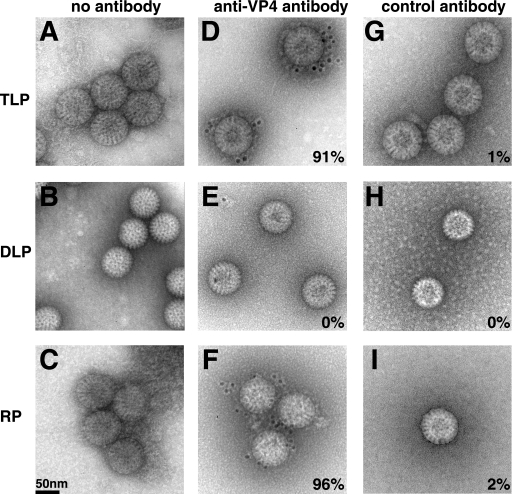
EM and immuno-EM characterization of recoated particles.
Recoated particles resemble authentic rotavirus virions by electron microscopy of negatively stained specimens. Like authentic TLPs (Fig. 3A, D, and G), recoated particles have a smooth outer rim of VP7 (Fig. 3C, F, and I). By contrast, the DLPs used as starting material for recoating have a serrated rim (Fig. 3B, E, and H). VP4 is not detected reliably by electron microscopy of negatively stained authentic TLPs or recoated particles, but it can be detected on particles by immuno-electron microscopy. Monoclonal antibody 7A12 against the VP8* fragment of VP4 decorates authentic TLPs (Fig. 3D) and recoated particles (Fig. 3F), but not DLPs (Fig. 3E). An irrelevant antibody (anti-histidine tag) does not decorate any of the particles. (Fig. 3G, H, and I).
FIG. 3.
TEM and immunogold labeling of negatively stained authentic and recoated rotavirus particles. (A, D, and G) TLPs; (B, E, and H) DLPs; (C, F, and I) DLPs recoated with recombinant VP4 and VP7 under standard conditions. In panels D to F, the grids were incubated with monoclonal antibody 7A12 against VP4 and with a 5-nm colloidal gold-conjugated goat antiserum against mouse IgG. In panels G to I, an anti-six-histidine antibody was used as a primary antibody. Colloidal gold is visible as black dots surrounding the particles in panels D and F. The percentages indicated on panels D to I indicate the proportion of particles in each sample with one or more adjacent black dots. At least 50 particles were counted for each sample.
EM of partially recoated particles.
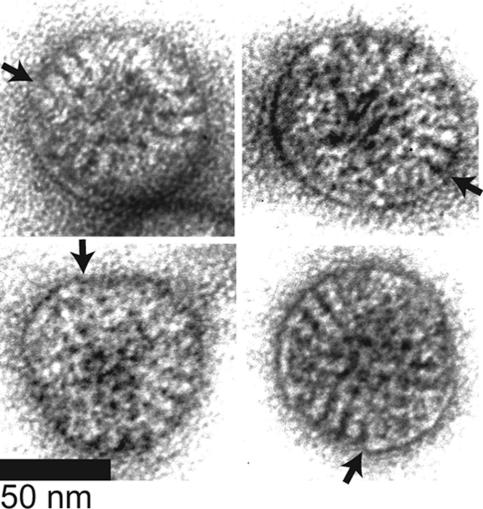
If there were no cooperativity between VP7 trimers during assembly, adding 0.3 molecules of VP7 per binding site would fully recoat approximately 1 out of 10136 particles. If only fully recoated particles were infectious, there would be no measurable increase in infectivity. However, adding 0.3 molecules of VP7 per binding site to DLPs increases infectivity 10,000-fold (Fig. 2B). Therefore, either particles with incomplete VP7 layers are infectious or trimers assemble cooperatively onto each DLP.
EM of negatively stained particles recoated with VP7 at a stoichiometry of 0.3 reveals a mixture of particles with serrated outlines (not shown), particles with complete smooth outlines (not shown), and particles with mixed outlines (Fig. 4). The two-dimensional projections do not reveal whether some particles with serrated outlines bound VP7 that does not contribute to the outline or whether some particles with complete smooth outlines have unobserved defects in their VP7 layer. Therefore, the micrographs do not allow precise quantitation of the proportion of partially recoated particles and do not resolve whether partially recoated particles are infectious. Nevertheless, the presence of many particles with contiguous but incomplete outer capsids indicates that there is cooperativity between VP7 trimers during assembly that is strong enough to create patches of assembled VP7 but not strong enough to produce a mixture of only fully coated and completely uncoated particles.
FIG. 4.
TEM of negatively stained, partially recoated particles. DLPs were recoated with VP7 at a stoichiometry of 0.3. In these examples, VP7 is visible as patches on the particle surface, resulting in segments of smooth contour and segments of serrated contour on the same particle. In each panel, an arrow marks a transition between a region of smooth contour and serrated contour.
Physical characterization of purified recoated particles.
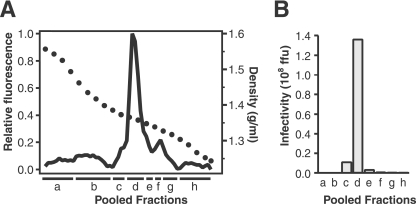
Fractionation of an untrypsinized recoating reaction mixture by isopycnic centrifugation through a CsCl density gradient yields a peak of particles with a density of 1.363 g/ml (Fig. 5A), closely matching the density of authentic TLPs. Infectivity peaks in the pool that contains the peak of physical particles (Fig. 5B). Approximately 17% of the infectivity of the mixture added to the gradient is recovered in the peak fractions. Loss of infectivity during CsCl purification is also observed for authentic rotavirus TLPs (reference 6 and data not shown). The specific infectivity of the recoating mixture before the purification shown in Fig. 5 was 6.1 × 102 particles per FFU. After banding in CsCl, the specific infectivity was 3.8 × 103 particles per FFU, a 6.2-fold decrease. Purified authentic RRV TLPs, cultivated in the absence of trypsin and primed after banding in CsCl, have similar specific infectivity (1.0 × 103 particles per FFU).
FIG. 5.
Purification of recoated particles by isopycnic density centrifugation. (A) Particle distribution. The solid line shows the fluorescence of each fraction of a CsCl density gradient after mixing with ethidium bromide and illuminating at 302 nm. Black circles indicate density. The peak of fluorescence, which identifies RNA-containing particles, is at 1.36 g/ml. Fractions were combined into pools a to h. (B) Infectivity of each pool from panel A.
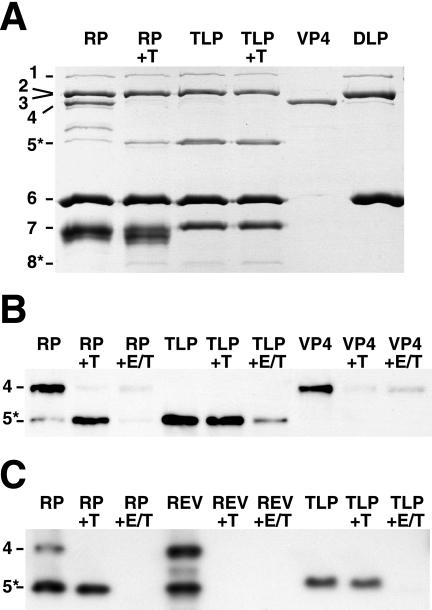
SDS-PAGE of the pooled peak fractions (Fig. 6A, lane RP) shows a recoated particle protein composition that is similar to that of authentic purified TLPs (Fig. 6A, lane TLP). The recombinant VP7 associated with recoated particles migrates slightly more quickly and in a more dispersed band than the authentic VP7 associated with authentic TLPs. The differences in the migration of authentic and recombinant VP7 are probably due to different processing of the proteins' oligosaccharide side chains and possibly due to cleavage of recombinant VP7 by adventitious proteases during purification. Fractions containing recoated particles contain both intact VP4 and fragments that migrate between the positions of VP4 and VP5* by SDS-PAGE. These fragments have blocked N termini and are probably formed by partial cleavage of VP4 (see below). VP5*, but little or no intact VP4, is associated with the TLPs.
FIG. 6.
Trypsin cleavage of free and assembled VP4. In each panel, protein bands are identified by viral protein number on the left. (A) Trypsin cleavage of recoated and authentic particles. A Coomassie-stained SDS-PAGE gel is shown. “RP” indicates purified recoated particles from pool d in Fig. 5. “+T” indicates analytical trypsin digestion (see Materials and Methods). The TLPs were prepared with trypsin in the cell culture medium. Samples of purified recombinant VP4 and DLPs are shown for comparison. (B) Effect of assembly and uncoating on the trypsin cleavage of VP4. A Western blot with monoclonal antibody HS2, which recognizes VP5* and VP4, is shown. Samples are labeled as in panel A. “+E/T” indicates that EDTA was added to 5 mM before trypsin digestion. The less-efficient degradation of VP4 in the presence of EDTA (last lane) reflects mild inhibition of trypsin by EDTA. (C) Effect of order of addition during recoating on trypsin sensitivity of VP4. A Western blot with monoclonal antibody HS2 is shown. “RP” indicates a standard recoating reaction mixture, in which VP4 was added before VP7. “REV” indicates a reverse order of addition, in which VP7 was added before VP4. In this panel, complete recoating mixtures, which also include unassembled VP4 and VP7, were analyzed.
Trypsin priming of VP4 on recoated particles.
To prime virions for infectivity, trypsin specifically cleaves authentic assembled VP4 after three closely spaced arginines between VP8* and VP5* (R231, R241, and R247) (1). Cleavage after R247, which produces the VP5* N terminus at A248, is essential for infectivity and membrane interactions (1, 23). Trypsin cleavage of soluble, unassembled recombinant VP4 is considerably more extensive and does not yield a protease-resistant product that comigrates with authentic VP5* (16) (Fig. 6B, lane VP4+T).
Trypsin digestion of purified recoated particles produces a fragment that comigrates with authentic VP5* by Coomassie-stained SDS-PAGE (Fig. 6A, lanes RP+T and TLP+T), reacts with a monoclonal antibody against VP5* by Western blotting (Fig. 6B, lanes RP+T and TLP+T), and has the authentic VP5* N-terminal sequence AQANE (corresponding to RRV VP4 residues 248 to 252). A slight broadening of the VP7 band upon trypsin digestion of recoated particles (Fig. 6A, lane RP+T) but not upon trypsin digestion of TLPs (Fig. 6A, lane TLP+T) indicates slightly greater protease sensitivity of some of the VP7 on the recoated particles. The difference in protease sensitivity suggests a subtle difference in assembly of VP7 onto authentic and recoated particles. When recoated particles or authentic TLPs are uncoated by calcium chelation, trypsin degrades VP4, yielding little or no VP5* (Fig. 6B and C, lanes RP+E/T and TLP+E/T). Similarly, if VP7 is added to DLPs before VP4, subsequent trypsin digestion degrades VP4 (Fig. 6C, lane REV+T). Thus, recombinant VP4 must be added to DLPs before VP7 to be mounted in a conformation that is specifically susceptible to the priming trypsin cleavage.
Trypsin digestion of purified recoated particles either degrades the proteins that migrate between VP5* and VP4 or converts these proteins to VP5* (Fig. 6A, compare lanes RP and RP+T). Trypsin digestion also has a grossly observable effect on preparations of purified recoated particles. Purified, uncleaved, recoated particles aggregate visibly when CsCl-containing gradient fractions are dialyzed against TNC. Trypsin digestion disperses these aggregates. This finding suggests that, during recoating, some recombinant protein assembles properly onto DLPs and becomes relatively trypsin resistant, while some recombinant protein is bound nonspecifically, possibly as a result of denaturation under the conditions of acidity during recoating and high ionic strength during isopycnic centrifugation. The nonspecifically associated protein (primarily VP4) remains trypsin sensitive and is probably responsible for aggregation.
Densitometry of the Coomassie-stained VP4 band in lane RP and VP5* bands in lanes RP+T and TLP+T of Fig. 6A indicates that trypsin digestion converts approximately 40% of the recoated particle-associated VP4 into VP5* and that trypsin-digested recoated particles have approximately 28% as much VP5* as authentic TLPs. This finding indicates, at most, 28% average spike occupancy on recoated particles. The spike occupancy of authentic purified TLPs is not known precisely and varies among strains (6).
DISCUSSION
Sequentially adding recombinant VP4 and VP7 to DLPs yields recoated rotavirus particles that are more infectious than DLPs by a factor of up to 107 (Fig. 1A). The recoated particles resemble authentic rotavirus TLPs by electron microscopy of negatively stained specimens (Fig. 3). After trypsin priming, freshly recoated particles have a specific infectivity as low as 28 particles per FFU. By contrast, the lowest specific infectivity that we have obtained for purified authentic RRV TLPs is 1 × 103 particles per FFU. The lower infectivity of purified TLPs probably reflects damage to the outer capsid during purification from infected cells. Unlike the outer capsid of purified TLPs, the outer capsid of recoated particles has not been subjected to freezing and thawing, extraction with an organic solvent (Freen), or incubation at high ionic strength (approximately 2.5 M CsCl). Although the DLPs used as starting material for recoating are also subjected to these insults during purification from infected cells, they are hardier than TLPs. Large losses in TLP infectivity during purification have been documented previously (6). Specific infectivity comparable to that of recoated particles has been reported for nonpurified authentic rotavirus-containing preparations (28), although differences in techniques preclude precise comparison to our results.
Calcium and low pH are required for efficient recoating (Fig. 1A and B). Calcium-dependent trimerization of VP7 (15), a prerequisite for VP7 binding to DLPs, explains the calcium requirement. Why is low pH required? Transcapsidation also requires acidic pH (5). The affinity of EDTA for calcium falls steeply at low pH. Because transcapsidation reaction mixtures contain more EDTA than calcium, acidity may be required to free calcium from EDTA. However, recoating is carried out with excess calcium present. Therefore, low pH must also promote outer capsid assembly through another mechanism, possibly facilitating initial VP4-VP4 or VP4-DLP interactions. There is no evidence that outer capsid assembly occurs in an acidic compartment of infected cells.
VP4 must be added to DLPs before VP7 for efficient functional recoating (Fig. 1D). In infected cells, VP7 assembles onto DLPs in the ER lumen in a process that is linked to a membrane penetration event. VP7 assembles as the DLP, having acquired a transient lipid envelope by budding into the ER, sheds this envelope. DLP budding and transient envelope loss require the activity of the virally encoded, ER membrane-anchored nonstructural glycoprotein NSP4 (2, 31, 45, 56). VP7 also appears to have a direct role in mediating envelope disruption (31). Nevertheless, in vitro, VP7 assembles onto DLPs efficiently and functionally with no requirement for a chaperone and no link to membrane disruption.
The timing and location of VP4 assembly in infected cells is controversial. Some observations suggest that VP4 binds DLPs in the cytoplasm or the ER, before or during VP7 assembly. These observations include electron cryomicroscopy reconstructions showing a VP4 domain buried beneath the VP7 layer (58), immunoelectron microscopy localizing VP4 in the cytoplasm between viroplasms and the ER membrane (46), colocalization of VP4 with NSP4 by immunofluorescence microscopy and biochemical assays (26), the detection of VP4 in transiently enveloped particles (48), and the accumulation of viral particles in the ER of infected cells depleted of VP4 by small interfering RNA (11). On the other hand, immunofluorescence microscopy using monoclonal antibody 7.7 detects VP4 in lipid rafts, bound to cytoskeletal elements, and exposed on the external face of the apical plasma membrane (14, 38, 52). This antibody does not detect VP4 in the ER lumen. These observations have given rise to the alternative hypothesis that VP4 assembles after VP7, possibly while VP7-coated nascent particles are transported from the ER to the apical cell surface (12, 29, 52). The order of addition required for efficient in vitro recoating is more consistent with the hypothesis that VP4 assembly precedes VP7 assembly in vivo.
VP4 assembly involves low-affinity interactions, as demonstrated by a very steep dependence on VP4 concentration. Between 0.35 and 2.0 μM VP4, reconstituted infectivity varies with VP4 concentration to the fourth power (Fig. 2F). Consistent with a requirement for low-affinity interactions involving VP4, a threefold molar excess of VP4 must be added to maximize infectivity (Fig. 2A). With fixed VP4 and VP7 concentrations, infectivity varies linearly with DLP concentration and quantity (Fig. 2E). Therefore, VP4-VP4 interactions, not VP4-DLP interactions, are primarily responsible for the steep dependence of infectious recoating on VP4 concentration. As VP4 clusters are dispersed on the virion surface, it is likely that the VP4-VP4 interaction occurs between molecules within each cluster rather than between VP4 molecules in different clusters. We hypothesize that this limiting interaction is a low-affinity trimerization of VP4. This oligomerization must be weak, because the distribution of VP4 in an equilibrium analytical ultracentrifugation experiment can be fit well by a nonassociating monomer model (16).
In the lattice of crystallized rotavirus strain RF VP6, residues D286 and N266 from adjacent trimers coordinate a calcium ion (36). Comparison of the VP6 trimer structure to electron cryomicroscopy reconstructions indicates that these residues are in the interface between VP6 and VP4 on trypsin-primed virions (36). Although the growth of the VP6 crystals in 200 mM calcium could lead to the presence of ordered calcium at low-affinity sites, the presence of calcium bound to these residues suggests that calcium could also be coordinated between VP4 and VP6 on primed virions (36). However, after incubation of DLPs with a comparatively high concentration of VP4 (2 μM) in the presence of 1 mM calcium, no VP4 coelutes with DLPs from a gel filtration chromatography column (data not shown). These findings suggest that, like VP4 oligomerization, the VP4-DLP interaction is also weak. There are no published structural data on the interaction of uncleaved VP4 with VP6 or VP7. The mass density for VP4, including the foot domain, is not seen in electron cryomicroscopy reconstructions of uncleaved particles (10).
The calcium-dependent trimerization of VP7 in solution is somewhat less weak. In the presence of calcium, VP7 trimerizes with a KA3 of approximately 2 × 103, and two VP7 peaks by gel filtration chromatography correspond to VP7 trimers and monomers in slow equilibrium (17). VP7 trimers bind DLPs with high affinity, as indicated by the minimal dependence of infectious recoating on VP7 concentration in the range of 0.29 to 4.7 μM VP7 (Fig. 2D). Because of this high affinity, infectious yield from recoating reaches a plateau at a VP7 stoichiometry of 1, and no excess VP7 is required to drive recoating (Fig. 2B). The presence of contiguous patches of VP7 on recoated particles produced in the presence of substoichiometric amounts of VP7 suggests cooperativity between VP7 trimers during assembly (Fig. 4). There is also evidence for cooperativity between dissociating VP7 trimers during disassembly (uncoating). The binding of bivalent antibodies to between 1% and 10% of the VP7 molecules on a virion neutralizes rotavirus infectivity (50). These antibodies neutralize by blocking uncoating (32). The ability of one or a few antibody cross-links to lock the VP7 shell in place suggests that all 780 VP7 molecules on a virion undergo a concerted, cooperative conformational change during uncoating.
These observations suggest a model for the reactions that lead to infectious recoating (Fig. 7). In this model, VP4 transiently oligomerizes. These VP4 oligomers bind DLPs weakly. VP7 undergoes calcium-dependent trimerization and binds the DLPs tightly, locking VP4 in place. VP7 binding does not depend on the prior binding of VP4 (Fig. 4), consistent with the observation that triple-layered particles without VP4 assemble in infected cells depleted of VP4 by small interfering RNA (13). This model does not account for possible facilitation of VP4 or VP7 oligomerization by the transient binding of monomers to adjacent sites on the DLP surface.
FIG. 7.
Recoating reaction scheme. VP4 has a weak tendency to self-associate. Transient VP4 trimers bind weakly to DLPs. VP7 associates into trimers in the presence of calcium. VP7 trimers bind tightly to DLP-VP4 complexes, locking VP4 in place. We cannot rule out the possibility that VP4 trimerization and DLP binding are a cooperative process, with the peripentonal channels of DLPs providing templates that guide VP4 trimerization.
Proper assembly of VP4 sets the stage for triggered conformational changes that lead to cell entry. On authentic particles, newly assembled VP4 is flexible (10) and forms clusters of three (or, less likely, two) molecules in each peripentonal channel of the DLP (18, 44, 58). Trypsin primes rotavirus for membrane interactions and cell entry by specifically cleaving flexible, assembled VP4 between VP8* and VP5* (1, 23). The priming cleavage triggers a rearrangement that rigidifies VP4 into spikes (10). After an unknown second trigger, VP4 folds back on itself, forming a very stable umbrella-shaped trimer (18). This fold-back translocates an exposed hydrophobic region from one end of the molecule to the other.
Trypsin cleaves unassembled VP4 more extensively than assembled VP4 (16). Preventing proper VP4 assembly by adding VP7 first during recoating or solubilizing VP4 by disrupting the outer capsid with a calcium chelator leaves VP4 sensitive to degradation by trypsin (Fig. 6B and C). Thus, specific trypsin cleavage between VP8* and VP5* to produce a fragment with the electrophoretic mobility and N-terminal sequence of authentic VP5* is an indicator of proper VP4 assembly. Serial digestion of recombinant VP4 in solution with chymotrypsin and trypsin triggers the N-terminal half of VP5* to rearrange into a folded-back umbrella-shaped trimer (18). Digestion of VP4 on the virion leads to the formation of the stable protruding spike. We hypothesize that this spike is a metastable intermediate, primed for a second rearrangement to yield the folded-back state. When VP7 locks VP4 into the peripentonal channels of the DLP, it sets up the restraints that allow for specific trypsin cleavage between VP8* and VP5* and the arrest of rearranging VP4 in a metastable conformation.
In vitro assembly of VP4 is relatively inefficient. Recoated particles have a decreased total complement of VP4, and protease sensitivity analysis shows that only 40% of the associated VP4 is properly assembled. Although the association of the remaining 60% is stable to CsCl gradient purification, its protease sensitivity indicates that it is not mounted properly. This improper association may reflect aggregation of VP4 with DLPs at the low pH of recoating and/or the high ionic strength of purification. Relatively low occupancy and the confounding presence of improperly mounted VP4 are barriers to using recoated particles to image spike morphology. However, because high occupancy is not required for efficient infectivity, recoated particles are highly suitable for functional studies. Consistent with the finding that a relatively small complement of functional spikes suffices for efficient infectivity, most, if not all, of the VP4 on a virion must be bound by antibody to neutralize infectivity (50).
Previous studies of transcapsidated particles and recombinant VLPs have examined VP4 association and cleavage. The level of specifically cleaved VP4 that mediates the low-level infectivity of transcapsidated particles appears to be below the detection limit of silver-stained SDS-PAGE (5). Western blot evidence of digestion of VP4 to VP5* suggests that some VP4 is properly mounted on VLPs formed by coinfecting insect cells with baculoviruses expressing VP2, VP4, VP6, and VP7 (9, 23, 24). Baculovirus-expressed VP7 is probably cotranslationally translocated into the ER and appears to be secreted from insect cells (17); the other proteins probably accumulate in the cytoplasm. Therefore, VP4 probably binds VP2/6 particles in the insect cell cytoplasm, with VP7 released from the ER or secreted into the cell culture medium binding afterwards. Thus, compartmentalization in insect cells sets up an order of addition conducive to proper VP4 assembly.
Indeed, we hypothesize that, during authentic rotavirus infection of mammalian cells, segregation of VP7 in the ER from DLPs in the cytoplasm might allow VP4 to bind DLPs before VP7. Scaffolding by NSP4 (33) or chaperoning by cellular factors may facilitate efficient VP4 assembly. Although the activity of NSP4 or cellular factors could potentially permit a reverse order of assembly, VP4 also must be added before VP7 when recoating is carried out in the presence of an insect cell lysate (not shown). Therefore, any factors that allow the late addition of VP4 are absent or inactive in the insect cell extract.
Recoating in vitro has allowed the determination of key parameters underlying rotavirus assembly, such as differences in the binding affinities of VP4 and VP7 and a required order of addition. Recoating also demonstrates the ability of relatively few functional VP4 spikes to mediate efficient infectivity. Recoating with engineered outer capsid proteins will allow rigorous experiments to test whether observed associations between mutations in outer capsid protein genes and viral phenotypes reflect causal relationships. The high particle-to-infectious unit ratio that is typical of rotavirus virions could be explained, in part, by many particles entering cells through “dead-end” pathways. The ability to use infection as an end point to verify productive entry is a key advantage of using infectious recoated particles rather than noninfectious VLPs for assembly and entry studies. The ability to engineer particles with blocks to productive entry will expand the range of potential experiments beyond those that can be carried out with virus strains produced by reverse genetics.
We have obtained high-resolution structures for the head and body domains of VP4 (18, 19, 37, 59) and anticipate a high-resolution structure for VP7 (17). Structure-based engineering of the influenza virus hemagglutinin has produced substantial insights into membrane fusion (25). Studies using reovirus have pioneered the use of recoating to study the assembly and entry of this nonenveloped virus (3, 4, 40, 41). We will combine structure-based engineering of VP4 and VP7 with recoating to determine how the rearrangements of rotavirus outer capsid proteins lead to membrane penetration by rotavirus during cell entry. Finally, recoating could provide a novel “sheep in wolf's clothing” approach to designing highly attenuated rotavirus preparations capable of a single round of efficient entry and replication in the host.
Acknowledgments
We thank Marina Babyonyshev for skillful technical assistance; Stephen C. Harrison, Max L. Nibert, David M. Knipe, and Sean P. J. Whelan for scientific advice; Harry B. Greenberg for recombinant baculoviruses and monoclonal antibodies; Joshua D. Yoder and Ethan C. Settembre for helpful discussions; Michael Berne and the Tufts Protein Microchemistry Facility for N-terminal sequencing; and Maria Ericsson and the Harvard Medical School Electron Microscopy Facility for assistance and technical advice.
This work was supported by Public Health Service grant RO1 AI 053174 from the National Institute of Allergy and Infectious Diseases and by an Ellison Medical Foundation New Scholars in Global Infectious Diseases Award to P.R.D.
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Arias, C. F., P. Romero, V. Alvarez, and S. Lopez. 1996. Trypsin activation pathway of rotavirus infectivity. J. Virol. 70:5832-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au, K. S., W. K. Chan, J. W. Burns, and M. K. Estes. 1989. Receptor activity of rotavirus nonstructural glycoprotein NS28. J. Virol. 63:4553-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandran, K., J. S. Parker, M. Ehrlich, T. Kirchhausen, and M. L. Nibert. 2003. The δ region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. 77:13361-13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandran, K., X. Zhang, N. H. Olson, S. B. Walker, J. D. Chappell, T. S. Dermody, T. S. Baker, and M. L. Nibert. 2001. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. J. Virol. 75:5335-5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, D., and R. F. Ramig. 1993. Rescue of infectivity by in vitro transcapsidation of rotavirus single-shelled particles. Virology 192:422-429. [DOI] [PubMed] [Google Scholar]
- 6.Chen, D. Y., and R. F. Ramig. 1992. Determinants of rotavirus stability and density during CsCl purification. Virology 186:228-237. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, J., J. Laporte, A. Charpilienne, and R. Scherrer. 1979. Activation of rotavirus RNA polymerase by calcium chelation. Arch. Virol. 60:177-186. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, S. E., M. K. Estes, M. Ciarlet, C. Barone, C. M. O'Neal, J. Cohen, and M. E. Conner. 1999. Heterotypic protection and induction of a broad heterotypic neutralization response by rotavirus-like particles. J. Virol. 73:4813-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford, S. E., M. Labbe, J. Cohen, M. H. Burroughs, Y. J. Zhou, and M. K. Estes. 1994. Characterization of virus-like particles produced by the expression of rotavirus capsid proteins in insect cells. J. Virol. 68:5945-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford, S. E., S. K. Mukherjee, M. K. Estes, J. A. Lawton, A. L. Shaw, R. F. Ramig, and B. V. Prasad. 2001. Trypsin cleavage stabilizes the rotavirus VP4 spike. J. Virol. 75:6052-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuadras, M. A., B. B. Bordier, J. L. Zambrano, J. E. Ludert, and H. B. Greenberg. 2006. Dissecting rotavirus particle-raft interaction with small interfering RNAs: insights into rotavirus transit through the secretory pathway. J. Virol. 80:3935-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuadras, M. A., and H. B. Greenberg. 2003. Rotavirus infectious particles use lipid rafts during replication for transport to the cell surface in vitro and in vivo. Virology 313:308-321. [DOI] [PubMed] [Google Scholar]
- 13.Dector, M. A., P. Romero, S. Lopez, and C. F. Arias. 2002. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 3:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delmas, O., A. M. Durand-Schneider, J. Cohen, O. Colard, and G. Trugnan. 2004. Spike protein VP4 assembly with maturing rotavirus requires a postendoplasmic reticulum event in polarized Caco-2 cells. J. Virol. 78:10987-10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dormitzer, P. R., and H. B. Greenberg. 1992. Calcium chelation induces a conformational change in recombinant herpes simplex virus-1-expressed rotavirus VP7. Virology 189:828-832. [DOI] [PubMed] [Google Scholar]
- 16.Dormitzer, P. R., H. B. Greenberg, and S. C. Harrison. 2001. Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core. J. Virol. 75:7339-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dormitzer, P. R., H. B. Greenberg, and S. C. Harrison. 2000. Purified recombinant rotavirus VP7 forms soluble, calcium-dependent trimers. Virology 277:420-428. [DOI] [PubMed] [Google Scholar]
- 18.Dormitzer, P. R., E. B. Nason, B. V. Prasad, and S. C. Harrison. 2004. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature 430:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dormitzer, P. R., Z.-Y. J. Sun, G. Wagner, and S. C. Harrison. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott, Williams, and Wilkins, Philadelphia, Pa. [Google Scholar]
- 21.Estes, M. K., D. Y. Graham, and B. B. Mason. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiore, L., S. J. Dunn, B. Ridolfi, F. M. Ruggeri, E. R. Mackow, and H. B. Greenberg. 1995. Antigenicity, immunogenicity and passive protection induced by immunization of mice with baculovirus-expressed VP7 protein from rhesus rotavirus. J. Gen. Virol. 76:1981-1988. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert, J. M., and H. B. Greenberg. 1998. Cleavage of rhesus rotavirus VP4 after arginine 247 is essential for rotavirus-like particle-induced fusion from without. J. Virol. 72:5323-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert, J. M., and H. B. Greenberg. 1997. Virus-like particle-induced fusion from without in tissue culture cells: role of outer-layer proteins VP4 and VP7. J. Virol. 71:4555-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godley, L., J. Pfeifer, D. Steinhauer, B. Ely, G. Shaw, R. Kaufmann, E. Suchanek, C. Pabo, J. J. Skehel, and D. C. Wiley. 1992. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 68:635-645. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez, R. A., R. Espinosa, P. Romero, S. Lopez, and C. F. Arias. 2000. Relative localization of viroplasmic and endoplasmic reticulum-resident rotavirus proteins in infected cells. Arch. Virol. 145:1963-1973. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg, H. B., J. Valdesuso, K. van Wyke, K. Midthun, M. Walsh, V. McAuliffe, R. G. Wyatt, A. R. Kalica, J. Flores, and Y. Hoshino. 1983. Production and preliminary characterization of monoclonal antibodies directed at two surface proteins of rhesus rotavirus. J. Virol. 47:267-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hundley, F., B. Biryahwaho, M. Gow, and U. Desselberger. 1985. Genome rearrangements of bovine rotavirus after serial passage at high multiplicity of infection. Virology 143:88-103. [DOI] [PubMed] [Google Scholar]
- 29.Jourdan, N., M. Maurice, D. Delautier, A. M. Quero, A. L. Servin, and G. Trugnan. 1997. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J. Virol. 71:8268-8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komoto, S., J. Sasaki, and K. Taniguchi. 2006. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc. Natl. Acad. Sci. USA 103:4646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez, T., M. Camacho, M. Zayas, R. Najera, R. Sanchez, C. F. Arias, and S. Lopez. 2005. Silencing the morphogenesis of rotavirus. J. Virol. 79:184-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludert, J. E., M. C. Ruiz, C. Hidalgo, and F. Liprandi. 2002. Antibodies to rotavirus outer capsid glycoprotein VP7 neutralize infectivity by inhibiting virion decapsidation. J. Virol. 76:6643-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maass, D. R., and P. H. Atkinson. 1990. Rotavirus proteins VP7, NS28, and VP4 form oligomeric structures. J. Virol. 64:2632-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackow, E. R., J. W. Barnett, H. Chan, and H. B. Greenberg. 1989. The rhesus rotavirus outer capsid protein VP4 functions as a hemagglutinin and is antigenically conserved when expressed by a baculovirus recombinant. J. Virol. 63:1661-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackow, E. R., R. D. Shaw, S. M. Matsui, P. T. Vo, M. N. Dang, and H. B. Greenberg. 1988. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc. Natl. Acad. Sci. USA 85:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathieu, M., I. Petitpas, J. Navaza, J. Lepault, E. Kohli, P. Pothier, B. V. Prasad, J. Cohen, and F. A. Rey. 2001. Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J. 20:1485-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monnier, N., K. Higo-Moriguchi, Z. Y. Sun, B. V. Prasad, K. Taniguchi, and P. R. Dormitzer. 2006. High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain. J. Virol. 80:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nejmeddine, M., G. Trugnan, C. Sapin, E. Kohli, L. Svensson, S. Lopez, and J. Cohen. 2000. Rotavirus spike protein VP4 is present at the plasma membrane and is associated with microtubules in infected cells. J. Virol. 74:3313-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott, Williams, and Wilkins, Philadelphia, Pa. [Google Scholar]
- 40.Odegard, A. L., K. Chandran, S. Liemann, S. C. Harrison, and M. L. Nibert. 2003. Disulfide bonding among μ1 trimers in mammalian reovirus outer capsid: a late and reversible step in virion morphogenesis. J. Virol. 77:5389-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Odegard, A. L., K. Chandran, X. Zhang, J. S. Parker, T. S. Baker, and M. L. Nibert. 2004. Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J. Virol. 78:8732-8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Padilla-Noriega, L., R. Werner-Eckert, E. R. Mackow, M. Gorziglia, G. Larralde, K. Taniguchi, and H. B. Greenberg. 1993. Serologic analysis of human rotavirus serotypes P1A and P2 by using monoclonal antibodies. J. Clin. Microbiol. 31:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parashar, U. D., E. G. Hummelman, J. S. Bresee, M. A. Miller, and R. I. Glass. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9:565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pesavento, J. B., S. E. Crawford, E. Roberts, M. K. Estes, and B. V. Prasad. 2005. pH-induced conformational change of the rotavirus VP4 spike: implications for cell entry and antibody neutralization. J. Virol. 79:8572-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrie, B. L., M. K. Estes, and D. Y. Graham. 1983. Effects of tunicamycin on rotavirus morphogenesis and infectivity. J. Virol. 46:270-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrie, B. L., H. B. Greenberg, D. Y. Graham, and M. K. Estes. 1984. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1:133-152. [DOI] [PubMed] [Google Scholar]
- 47.Pinteric, L., and J. Taylor. 1962. The lowered drop method for the preparation of specimens of partially purified virus lysates for quantitative electron micrographic analysis. Virology 18:359-371. [Google Scholar]
- 48.Poruchynsky, M. S., and P. H. Atkinson. 1991. Rotavirus protein rearrangements in purified membrane-enveloped intermediate particles. J. Virol. 65:4720-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redmond, M. J., M. K. Ijaz, M. D. Parker, M. I. Sabara, D. Dent, E. Gibbons, and L. A. Babiuk. 1993. Assembly of recombinant rotavirus proteins into virus-like particles and assessment of vaccine potential. Vaccine 11:273-281. [DOI] [PubMed] [Google Scholar]
- 50.Ruggeri, F. M., and H. B. Greenberg. 1991. Antibodies to the trypsin cleavage peptide VP8 neutralize rotavirus by inhibiting binding of virions to target cells in culture. J. Virol. 65:2211-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz, M. C., A. Charpilienne, F. Liprandi, R. Gajardo, F. Michelangeli, and J. Cohen. 1996. The concentration of Ca2+ that solubilizes outer capsid proteins from rotavirus particles is dependent on the strain. J. Virol. 70:4877-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sapin, C., O. Colard, O. Delmas, C. Tessier, M. Breton, V. Enouf, S. Chwetzoff, J. Ouanich, J. Cohen, C. Wolf, and G. Trugnan. 2002. Rafts promote assembly and atypical targeting of a nonenveloped virus, rotavirus, in Caco-2 cells. J. Virol. 76:4591-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw, A. L., R. Rothnagel, D. Chen, R. F. Ramig, W. Chiu, and B. V. Prasad. 1993. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell 74:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaw, R. D., P. T. Vo, P. A. Offit, B. S. Coulson, and H. B. Greenberg. 1986. Antigenic mapping of the surface proteins of rhesus rotavirus. Virology 155:434-451. [DOI] [PubMed] [Google Scholar]
- 55.Svensson, L., P. R. Dormitzer, C. H. von Bonsdorff, L. Maunula, and H. B. Greenberg. 1994. Intracellular manipulation of disulfide bond formation in rotavirus proteins during assembly. J. Virol. 68:5204-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor, J. A., J. A. O'Brien, V. J. Lord, J. C. Meyer, and A. R. Bellamy. 1993. The RER-localized rotavirus intracellular receptor: a truncated purified soluble form is multivalent and binds virus particles. Virology 194:807-814. [DOI] [PubMed] [Google Scholar]
- 57.Willis, S. H., C. Peng, M. Ponce de Leon, A. V. Nicola, A. H. Rux, G. H. Cohen, and R. J. Eisenberg. 1998. Expression and purification of secreted forms of HSV glycoproteins from baculovirus-infected insect cells, p. 131-156. In S. M. Brown and A. R. McLean (ed.), Methods in molecular medicine: herpes simplex virus protocols, vol. 10. Humana Press Inc., Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 58.Yeager, M., J. A. Berriman, T. S. Baker, and A. R. Bellamy. 1994. Three-dimensional structure of the rotavirus haemagglutinin VP4 by cryo-electron microscopy and difference map analysis. EMBO J. 13:1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoder, J. D., and P. R. Dormitzer. 2006. Alternative intermolecular contacts underlie the rotavirus VP5* two- to threefold rearrangement. EMBO J. 25:1559-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]