Abstract
A key genomic characteristic that helps define Hantavirus as a genus of the family Bunyaviridae is the presence of distinctive terminal complementary nucleotides that promote the folding of the viral genomic segments into “panhandle” hairpin structures. The hantavirus nucleocapsid protein (N protein), which is encoded by the smallest of the three negative-sense genomic RNA segments, undergoes in vivo and in vitro trimerization. Trimeric hantavirus N protein specifically recognizes the panhandle structure formed by complementary base sequence of 5′ and 3′ ends of viral genomic RNA. N protein trimers from the Andes, Puumala, Prospect Hill, Seoul, and Sin Nombre viruses recognize their individual homologous panhandles as well as other hantavirus panhandles with high affinity. In contrast, these hantavirus N proteins bind with markedly reduced affinity to the panhandles from the genera Bunyavirus, Tospovirus, and Phlebovirus or Nairovirus. Interactions between most hantavirus N and heterologous hantavirus viral RNA panhandles are mediated by the nine terminal conserved nucleotides of the panhandle, whereas Sin Nombre virus N requires the first 23 nucleotides for high-affinity binding. Trimeric hantavirus N complexes undergo a prominent conformational change while interacting with panhandles from members of the genus Hantavirus but not while interacting with panhandles from viruses of other genera of the family Bunyaviridae. These data indicate that high-affinity interactions between trimeric N and hantavirus panhandles are conserved within the genus Hantavirus.
Hantaviruses are classified as emerging viruses which cause two often fatal diseases that arise by infection of endothelial cells: hemorrhagic fever with renal syndrome and hantavirus cardiopulmonary syndrome (30-33). Each hantavirus is carried by one or a limited number of wild rodent species and transmitted to humans through the aerosol route. The two diseases associated with hantaviruses both cause striking increases in vascular permeability and are elicited by viruses such as Hantaan virus and Sin Nombre virus (SNV), respectively. Hemorrhagic fever with renal syndrome and hantavirus cardiopulmonary syndrome are generally restricted to the Old World and New World, respectively (34). Hantaviruses comprise a genus in the family Bunyaviridae. Members of this virus family have genomes composed of three minus-strand viral RNA (vRNA) segments whose mRNAs encode an RNA-dependent RNA polymerase (RdRp) (L segment), the nucleocapsid protein (N protein; S segment) and G1 and G2 glycoproteins (M segment). The G1 and G2 proteins are posttranslationally processed through the endoplasmic reticulum and Golgi apparatus and ultimately presented on the viral surface. These proteins enable viruses to enter new host cells via their attachment to integrin receptors (7, 8). In the virion the three genomic RNA molecules form a complex with N protein and presumably with RdRp.
During replication, assembly is initiated with the binding of nucleocapsid protein at a unique encapsidation signal on the genomic RNA. The specific recognition of genomic RNA promotes the oligomerization of N protein. The interaction with other viral proteins subsequently results in the formation of virions. For hantaviruses it has been suggested that sequences at the 5′ end of the genomic RNA provide the nucleation point for encapsidation by N protein (25). Studies with Bunyamwera virus indicate that N protein specifically encapsidates vRNA and cRNA but not mRNA or nonviral RNA molecules (13). In vitro studies indicate that nucleocapsid protein preferentially binds vRNA over cRNA and nonviral RNA (9, 23, 27, 35, 37). A cis-acting element that shows a high binding affinity for Hantaan and Bunyamwera N proteins has been mapped to the 5′ end of the viral RNA (23, 35, 37). The RNA binding domain for Hantaan virus N protein has been mapped to the central conserved region corresponding to amino acids 175 to 217 (39). N protein has also been found to undergo trimerization under in vivo (1, 2) and in vitro (14, 15) conditions.
The vRNAs of the Bunyaviridae family each feature a higher-order “panhandle” formed from the hydrogen bonding of the nucleotides that comprise the 5′ and 3′ ends of the RNA. We have recently shown that SNV N protein, in trimeric form, specifically recognizes the panhandle structure of the genomic RNA, and this specific interaction is postulated to have a role in either encapsidation or viral genome replication initiation (19-21). Here we show that trimeric nucleocapsid proteins from other members of the genus Hantavirus, including Andes, Puumala, Prospect Hill, and Seoul viruses, also specifically recognize the panhandle of their viral genome. N proteins from these members of the genus Hantavirus bind viral RNA panhandles with intragenus specificity and affinity. In addition, interaction between N and both homologous and heterologous hantavirus panhandles results in a conformational change in N.
MATERIALS AND METHODS
Oligonucleotides and enzymes.
PCR primers were from Sigma Genosys. All restriction enzymes were from New England Biolabs. Hot Mastertaq polymerase was purchased from Eppendorf. DNase I was from Invitrogen, and T7 transcription reagents were from Fermentase. All other chemicals were purchased from Sigma. RNA purification kits were from QIAGEN.
Cloning of bacterial N expression constructs.
As reported previously (26, 38), hantavirus N genes were subcloned from the S genomic segment of the P360 strain of Puumala virus (a generous gift of C. Schmaljohn) or directly from viral RNA prepared from the following strains of virus (all grown in Vero E6 cells). Prospect Hill virus strain PHV was provided by R. Yanagihara, Seoul virus strain 80/39 was from H. W. Lee, and Sin Nombre virus strain SN77734 and Andes virus strain CHI-7913 were from H. Galeno. Briefly, in each case, except for that of SNV, primers were designed to enable the in-frame insertion of each N gene in the pET21b (Novagen) backbone so that the initiating ATG immediately follows a HindIII restriction site, and the termination codon was replaced by a site for XhoI. The SNV N gene was cloned in pTri.Ex1.1 vector using the NcoI and HindIII sites. N proteins were expressed as C-terminal histidine-tagged proteins in Escherichia coli. Histidine tags were not removed after purification. Protein expression was carried out as recommended by Novagen. In each case, production of the N proteins was verified by Western analysis. Each N protein was purified using Ni2+ chelating columns under denaturing conditions as recommended by QIAGEN. The bacterial pellet was lysed in lysis buffer (100 mM NaH2PO4, 10 mM Tris-Cl, 8 M urea, pH 8.0) and centrifuged at 3,000 rpm. The resulting supernatant was incubated with N-NTA beads for 15 min. Beads were washed twice with wash buffer A (100 mM NaH2PO4, 10 mM Tris-Cl, 8 M urea, 10 mM imidazole, pH 8.0) followed by two additional washes with wash buffer C (100 mM NaH2PO4, 10 mM Tris-Cl, 8 M urea, 100 mM imidazole, pH 8.0). Bound N protein was eluted with elution buffer (100 mM NaH2PO4, 10 mM Tris-Cl, 8 M urea, 500 mM imidazole, pH 8.0). Proteins were renatured by stepwise dialysis to remove the urea and imidazole from the purified N protein. Protein purity was assessed and confirmed by Coomassie and Western blot analysis. N proteins from these viruses were all assembled into stable trimers at 4°C (Fig. 1). Trimeric N proteins were purified by sucrose density gradient centrifugation as described previously (21).
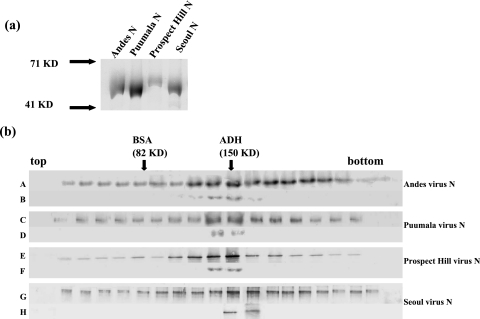
FIG. 1.
(a) N proteins from Andes, Puumala, Prospect Hill, and Seoul viruses were expressed in E. coli with C-terminal histidine fusion proteins. Each protein was purified by using Ni-NTA beads according to the manufacturer's protocol. Proteins were analyzed by SDS-polyacrylamide gel electrophoresis and visualized by Coomassie staining. (b) N proteins from Andes, Puumala, Prospect Hill, and Seoul viruses were sedimented through 10 to 45% sucrose gradients, and N was detected using Western blot analysis with anti-N antibody. Gradients A, C, E, and G correspond to N proteins from Andes, Puumala, Prospect Hill, and Seoul viruses, respectively. Gradients B, D, F, and H are trimeric N proteins that have been resedimented to examine the relative stability of trimeric N. Protein standards (bovine serum albumin [BSA] and alcohol dehydrogenase [ADH]) were run in parallel with the N samples, and arrows indicate the migration of those standards.
Preparation of RNA substrates.
The “minipanhandle” RNAs used in this study contained the 32 nucleotides from both the 5′ and 3′ ends of S segment viral RNA, separated by a loop composed of six uracil residues. This RNA was synthesized by in vitro transcription with T7 RNA polymerase. A single-stranded template sequence with a T7 promoter juxtaposed to the minipanhandle sequences was amplified by PCR using two opposing primers. For the synthesis of mutant panhandles, primers containing the designed mutation or the single-stranded template sequence were used in PCR. All PCR-generated templates were gel purified and used directly in T7 transcription reactions. [α-32P]CTP-radiolabeled transcripts were produced from different templates using a T7 transcription kit (MBI Fermentas). After transcription the reaction mixture was digested with DNase I to remove the DNA template. Purification of the RNA was performed using Trizol (Invitrogen). Purified RNA was stored at −20°C in 20-μl aliquots for up to 2 weeks.
Sucrose density gradient centrifugation.
Sucrose gradients were used to examine the oligomeric forms of N proteins from Andes, Puumala, Seoul, and Prospect Hill viruses using methodology described previously (21). One hundred-microliter samples containing 100 μM purified N protein were layered onto linear gradients containing 10 to 45% (wt/vol) sucrose in RNA binding buffer (21) and centrifuged at 30,000 rpm in an SW40 rotor at 40°C for 22 h. Fractions of 0.6 ml were collected from the bottom of the gradient. The protein molecular mass markers in the form of bovine serum albumin (82 kDa) and alcohol dehydrogenase (150 kDa) were fractionated in parallel. A 100-μl sample of each fraction was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. N proteins were detected by Western blot analysis with anti-SNV N protein antiserum.
RNA secondary structure analysis.
We examined multiple suboptimal secondary structure plots to identify alternative structures that might be nearly as stable as those shown in Fig. 2. In addition, we determined the P-num values for each of the nucleotides in the RNAs. The P-num value of a nucleotide is a representation of the number of potentially stable pairing partners for that nucleotide elsewhere in the same RNA. In particular, P-num is a predictive measure of pairing fidelity at suboptimal free energy values (ΔG), and bases with low values can be used parametrically to identify motifs with the highest probability of assuming similar configurations among a series of energetically related structures. Thus, when viewed from the context of suboptimal folds, the structures composed of low P-num nucleotides still form and are still composed of bases with the fewest alternative pairing partners (low P-num). Examination of suboptimal folds indicated that, with the exception of Rift Valley fever virus, the terminal secondary structures shown in Fig. 2 are much more thermodynamically stable than alternative suboptimal structures. Typically the structures have a ΔG of approximately −30 Kcal/mol, whereas the next most likely structure has a negative free energy that is more than 70% of this low minimum free energy. Moreover, the P-num values for most of the nucleotides that comprise the panhandle are either 1 or 0. Thus, it is highly unlikely that RNAs of alternative secondary structure represent a significant proportion of the RNA population.
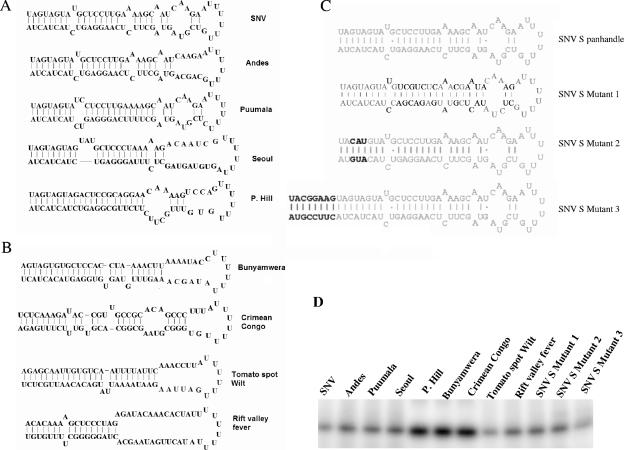
FIG. 2.
S segment genomic minipanhandle RNAs. Minipanhandles corresponding to the panhandles of several bunyaviruses each contained 32 nucleotides from both the 5′ and 3′ ends of viral S segment RNA, separated by six uracil residues. RNAs were synthesized by transcription with T7 polymerase as described in Materials and Methods. (A) Probable secondary structures of S segment minipanhandle RNAs from Sin Nombre, Andes, Puumala, Seoul, and Prospect Hill viruses, belonging to the genus Hantavirus. (B) Probable secondary structures of S segment minipanhandle RNAs from Bunyamwera, Crimean Congo, tomato spot wilt, and Rift Valley fever viruses, which belong to the genera Bunyavirus, Nairovirus, Tospovirus, and Phlebovirus, respectively. (C) Probable secondary structures of the wild-type SNV S segment minipanhandle and three mutants derived from SNV. Bold letters indicate nucleotide substitutions relative to the wild-type sequence. In SNV S mutant 3, extra nucleotides have been added to both the 5′ and 3′ ends of the RNA, such that they undergo additional base pairing and extend the stem structure in the panhandle. The secondary structures of all panhandles, except that of Rift Valley fever virus, are highly probable. Some of the nucleotides in the Rift Valley fever virus panhandle might undergo alternative hydrogen bonding with the middle uracil loop. (D) Twelve-percent acrylamide-8 M urea gel of minipanhandles, from panels A, B, and C. These panhandles were synthesized by in vitro T7 transcription reaction, radiolabeled during synthesis (described in detail in Materials and Methods), run on 12% acrylamide gels containing 8 M urea, and visualized by phosphorimaging. P. Hill, Prospect Hill virus.
Filter binding assay.
RNA molecules were prepared by in vitro T7 transcription reaction and radiolabeled with [α-32P]CTP as described above. Binding reactions were carried out in binding buffer (20, 21) at a constant concentration of RNA with increasing concentrations of N protein trimer. Reaction mixtures were incubated at room temperature for 45 min and filtered through nitrocellulose filters under a vacuum. Filters were washed with 10 ml of binding buffer and dried, and bound RNA was quantified by scintillation counting. Nonspecific retention of RNA was measured by parallel filtering of complete reaction mixtures in the absence of protein. Dissociation constants were calculated by fitting the experimental data points into a hyperbolic equation using the program Origin 6 (Microcal). The apparent dissociation constant (KD) corresponds to the concentration of N protein required to obtain the half saturation, assuming that the complex formation obeys a simple bimolecular equilibrium.
Competition assays.
Competition experiments were performed using filter-binding assays. A constant concentration of N protein (100 nM) from various viruses was incubated with a mixture of [P32]CTP-labeled parental wild-type panhandle RNA and different competitor panhandles (panhandles from other viruses). The concentration of wild-type panhandle was 0.01 nM, and the concentration of competitor panhandles was varied from 0.1 to 10 nM. Reaction mixtures were incubated at room temperature for 45 min and then filtered through nitrocellulose filters under a vacuum as described previously. The percentage of bound parental wild-type panhandle was plotted against the competitor panhandle concentration. Data points were fitted by nonlinear curve fit analysis using the Origin 6 program (Microcal).
Fluorescence binding assay.
Values for KD were also calculated by fluorometric titration of N protein trimer with different panhandle RNAs. The fluorescence spectra of N protein trimers in binding buffer, excited at 295 nm, were recorded using a Hitachi spectrofluorometer at excitation and emission band passes of 5 nm and 10 nm, respectively. We observed a decrease in the fluorescence quantum yields of all N protein trimers after association with RNA. The fluorescence signal of N protein trimer at 330 nm was plotted against different input concentrations of panhandle RNA. The plot generated was used to calculate the apparent value for KD as described previously (20, 21).
Fluorescence spectroscopic analysis.
Structural alterations in N protein trimers from different viruses due to their binding with different RNA molecules were monitored by quenching of tryptophan fluorescence of N protein trimers with acrylamide in the presence and absence of RNA according to the Stern-Volmer equation (17): F0/F = 1 + Ksv × [Q], where F0 and F are the fluorescence intensities in the absence and presence of quencher, respectively, [Q] is the concentration of quencher, and Ksv is the Stern-Volmer quenching constant. Fluorescence-quenching measurements of tryptophan in N protein with acrylamide quencher were made by serial addition of small aliquots of concentrated acrylamide solution to 1 ml of sample in a cuvette. If the plot of F0/F versus [Q] is linear, indicating a single class of fluorophores equally accessible to acrylamide, the value of Ksv can be calculated from the slope of a straight line, fitted to the data points.
Calculation of binding stoichiometry.
As described previously (20), the binding stoichiometry (expressed in terms of number of N molecules bound per panhandle) was estimated from the intersection of two straight lines of a least-squares fit plot of the percent increase of bound RNA against the ratio of input concentrations of N protein and RNA. We also used continuous variation plots to verify the binding stoichiometry results (6). This is a reliable method to determine the binding stoichiometry of RNA-protein complexes. At a constant temperature (25°C), the fluorescence signal was recorded for the solutions where the concentrations of both panhandle RNA and N protein were varied, keeping the sum of their concentrations constant at 100 nM. ΔF330 (the difference in the fluorescence intensity of N protein in the absence and presence of panhandle RNA) was plotted as a function of the input mole fraction of N protein. The breakpoint in the resulting plot corresponds to the mole fraction of N in the N-panhandle RNA complex. The binding stoichiometry is obtained in terms of N:panhandle RNA [xligand:(1 − xligand)], where xligand represents the molar concentration of N divided by the total molar concentration of N and panhandle RNA.
RESULTS
Stable formation of trimeric hantavirus N proteins.
Bacterially expressed SNV N forms stable trimers (21). We wanted to determine whether bacterially expressed N proteins from other hantaviruses, including Andes virus, Puumala virus, Prospect Hill virus, and Seoul virus, also form stable trimers. E. coli cells harboring expression plasmids with N genes from these viruses were induced with IPTG (isopropyl-β-d-thiogalactopyranoside) to express the histidine-tagged N protein, and these proteins were isolated and purified as reported previously (21). Figure 1a depicts an SDS-polyacrylamide gel of N proteins from Andes, Puumala, Prospect Hill, and Seoul viruses. The electrophoretic mobility of these histidine-N fusion proteins was consistent with the expected mass of the peptide as evidenced by comigration relative to molecular weight protein markers (Fig. 1a). Moreover, preparation of the protein by denaturation, renaturation, and affinity purification on Ni-NTA beads resulted in N protein preparations that were free of significant detectible heterologous protein.
To assess the subunit composition of the purified N proteins, we carried out sucrose gradient centrifugation. Samples of N protein were run on 10 to 45% sucrose gradients as described in Materials and Methods. To characterize the subunit composition of the N preparation molecular weight standards were centrifuged in parallel with N. Gradient fractions were then collected, and N protein was detected using Western analysis with anti-SNV N polyclonal antibody. All N proteins were typically most abundant in fractions 9 and 10 (Fig. 1b). This yielded a sedimentation profile indicating that N is trimeric compared with the molecular weight standards. To examine the stability of trimeric N, we pooled the fractions corresponding to the trimeric N protein, dialyzed the samples against 1× phosphate-buffered saline, and resedimented N on 10 to 45% sucrose gradients. We observed that the purified N protein trimers from the Andes, Puumala, Prospect Hill, and Seoul viruses retained trimeric form (Fig. 1b). These data are in agreement with previous observations with SNV N, and they indicate that hantavirus N generally forms stable trimers.
Hantavirus N binds specifically to homologous vRNA panhandles.
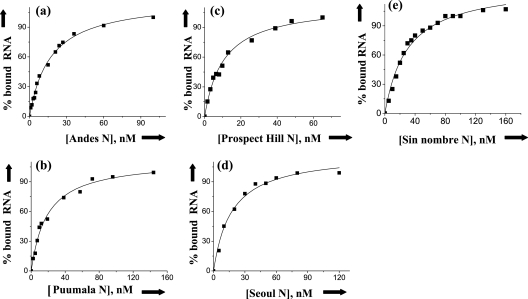
We synthesized a set of minipanhandle RNAs containing 32 nucleotides from both the 5′ and 3′ ends of the S segment from several hantaviruses (Fig. 2A) and carried out RNA filter binding for each of the appropriate homologous N protein trimers. Binding profiles that depict the interaction of N from the Andes, Puumala, Prospect Hill, and Seoul viruses with their S segment genomic RNA minipanhandles are shown in Fig. 3. The binding profiles are hyperbolic, suggesting that RNA-protein interaction follows first-order kinetics indicative of interaction between independent N trimers and RNA. The dissociation constants derived from these and additional binding profiles are presented in Table 1. As indicated in the table, N trimers from the Andes, Puumala, Prospect Hill, Seoul and Sin Nombre viruses specifically bind to their S segment viral RNA panhandles. Binding studies were carried out at three different salt concentrations. There were no significant changes in the dissociation constants at higher salt concentrations consistent with specific binding (Table 1).
FIG. 3.
Filter binding analysis. Binding profile for trimeric N protein from Andes virus (a), Puumala virus (b), Prospect Hill virus (c), Seoul virus (d), and SNV (e) with their parental S segment-derived minipanhandle RNAs, shown in Fig. 2. Data points plotted in each figure were obtained from filter binding analysis (see Materials and Methods). Similar data were obtained from three independent experiments.
TABLE 1.
Dissociation constants for the purified trimeric N proteins from Andes, Puumala, Prospect Hill, Seoul, and Sin Nombre viruses with their negative-sense S segment minipanhandles in binding buffer at 25°Ca
| Protein | Panhandle |
KD (nM) at indicated NaCl concn
|
||
|---|---|---|---|---|
| 80 mM | 160 mM | 220 mM | ||
| Andes virus | Andes virus | 13 ± 2.1 | 21 ± 3.0 | 23 ± 2.2 |
| Puumala virus | Puumala virus | 17 ± 3.2 | 25 ± 2.2 | 27 ± 3.1 |
| PHV | PHV | 22 ± 4.1 | 21 ± 2.1 | 26 ± 3.1 |
| Seoul virus | Seoul virus | 11 ± 1.0 | 17 ± 1.5 | 19 ± 2.3 |
| SNV | SNV | 39 ± 2.2 | 22 ± 3.0 | 38 ± 3.0 |
PHV, Prospect Hill virus.
Hantavirus N protein binds to heterologous hantavirus panhandles.
We carried out experiments to see whether N protein from Andes, Puumala, Prospect Hill, Seoul, and Sin Nombre viruses can recognize heterologous hantavirus panhandles. We also wanted to determine whether N protein from these hantaviruses could bind to the S segment viral RNA panhandles from viruses of different genera from the family Bunyaviridae. Thus, we also synthesized the S segment viral RNA minipanhandles from Bunyamwera virus, tomato spotted wilt virus, Rift Valley fever virus, and Crimean Congo virus, which belong to the genera Bunyavirus, Tospovirus, Phlebovirus, and Nairovirus, respectively (Fig. 2B).
We calculated the dissociation constants for each N protein trimer with each heterologous panhandle (Table 2) by using a fluorescence binding assay (see Materials and Methods). All N protein trimers, except that of SNV, interacted with heterologous hantavirus panhandles with high affinity. In contrast, all hantavirus N protein trimers bound the panhandles from Bunyamwera virus (Bunyavirus genus), tomato spotted wilt virus (Tospovirus genus), Rift Valley fever virus (Phlebovirus genus), and Crimean Congo virus (Nairovirus genus) with significantly lower affinity. SNV N protein trimer bound with high affinity to the parental and closely related Andes virus panhandles. However, the binding affinity was reduced fourfold with the panhandles from Prospect Hill, Puumala, and Seoul viruses (Table 2). Taken together, these binding studies indicate that N-panhandle interaction is generally genus specific.
TABLE 2.
Dissociation constants for the interaction of N protein trimers, from Sin Nombre, Andes, Puumala, Prospect Hill, and Seoul viruses with wild-type S segment panhandles from different viruses in binding buffer containing 160 mM NaCl at 25°Ca
| Panhandle |
KD (nM) for interaction with indicated N protein
|
||||
|---|---|---|---|---|---|
| SNV | Andes virus | Puumala virus | PHV | Seoul virus | |
| Andes virus | 27 ± 3 | 17 ± 2 | 18 ± 2 | 15 ± 1 | 15 ± 1 |
| PHV | 114 ± 3 | 15 ± 2 | 21 ± 2 | 13 ± 1 | 13 ± 1 |
| Puumala virus | 103 ± 4 | 18 ± 1 | 17 ± 1 | 14 ± 1 | 31 ± 2 |
| Seoul virus | 97 ± 4 | 21 ± 2 | 18 ± 1 | 17 ± 1 | 10 ± 1 |
| SNV | 29 ± 2 | 19 ± 2 | 19 ± 1 | 19 ± 1 | 11 ± 1 |
| Bunyamwera virus | 228 ± 6 | 101 ± 5 | 113 ± 5 | 109 ± 4 | 103 ± 3 |
| Tomato spotted wilt virus | 185 ± 5 | 17 ± 4 | 115 ± 6 | 111 ± 3 | 97 ± 4 |
| Rift Valley fever virus | 201 ± 5 | 125 ± 4 | 119 ± 4 | 103 ± 4 | 102 ± 2 |
| CCHFV | 229 ± 7 | 119 ± 2 | 117 ± 5 | 117 ± 4 | 91 ± 3 |
PHV, Prospect Hill virus; CCHFV, Crimean Congo hemorrhagic fever virus.
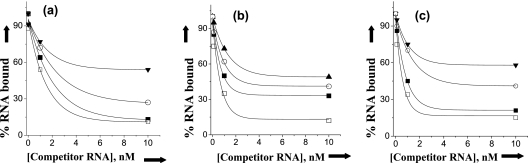
We extended these studies by carrying out competition binding analysis. In this approach, N protein trimers from each hantavirus were incubated with the corresponding homologous radiolabeled S segment minipanhandle RNA in the presence of increasing concentrations of cold competitor panhandle RNA from the same or alternative viruses. The percentage of bound radiolabeled RNA in the presence of increasing input concentrations of competitor RNA was then determined. Representative competition curves are shown in Fig. 4 and summarized in Table 3. In competition assays such as these, effective competition significantly reduces binding of the radiolabeled RNA where as competitor RNA's that are recognized at lower affinity compete only marginally. The competition data are in good correspondence with the observed dissociation constants. It is evident that, aside from SNV, panhandles from Andes, Puumala, Prospect Hill, and Seoul viruses effectively compete for N binding with N from other viruses of the genus Hantavirus. However, competition by panhandles from viruses belonging to other genera from the family Bunyaviridae is significantly weaker. Interestingly, the Andes virus panhandle can compete with high efficiency for SNV N, but competition by hantavirus panhandles from Prospect Hill, Puumala, and Seoul viruses is comparatively weaker. The data of Table 3 also indicate that Puumala N binds with high affinity to panhandles of both the Puumala and Seoul viruses but that interaction between Seoul virus N and the Puumala virus S segment panhandle is less robust.
FIG. 4.
Analysis of RNA binding using competition analysis. (a) A constant amount of Andes virus trimeric N protein was incubated with a constant amount of radiolabeled Andes virus S segment panhandle and increasing concentrations of unlabeled minipanhandles from Andes, Seoul, Bunyamwera, and tomato spotted wilt viruses. The quantitative reduction in the binding of trimeric Andes virus N protein with radiolabeled Andes virus panhandle as a function of increasing concentrations of unlabeled minipanhandles was measured for each competitor panhandle RNA. The figure depicts the competition curves for panhandles from Andes virus (□), Seoul virus (▪), Bunyamwera virus (○), and tomato spotted wilt virus (▾). (b) A similar analysis was carried out with Seoul virus N protein, in which competition curves for panhandles from Seoul virus (□), Puumala virus (▪), Rift valley fever virus (○), and Bunyamwera virus (▾) are presented. (C) A similar analysis was carried out with Puumala virus N protein, in which competition curves for panhandles from Puumala virus (□), Prospect Hill virus (▪), Bunyamwera virus (○), and Rift valley fever virus (▾) are presented. The maximum competition at a competitor concentration of 10 nM in presence of other panhandles and also for other proteins with different panhandles is shown in Table 3. The data shown here and in Table 3 are the average of three separate experiments. The standard deviations for the measured values are shown in Table 3.
TABLE 3.
Competition analysis for the interaction of N proteins with their wild-type S segment panhandles in the presence of competitor RNA panhandles from different viruses in binding buffer containing 80 mM NaCl at 25°Ca
| Competitor panhandle | % binding of wild-type S segment panhandle with indicated N protein
|
||||
|---|---|---|---|---|---|
| SNV | Andes virus | Puumala virus | PHV | Seoul virus | |
| Andes virus | 12 ± 1 | 13 ± 2 | 19 ± 1 | 27 ± 1 | 13 ± 2 |
| PHV | 40 ± 2 | 14 ± 1 | 21 ± 1 | 22 ± 2 | 14 ± 1 |
| Puumala virus | 42 ± 3 | 15 ± 1 | 15 ± 1 | 21 ± 2 | 33 ± 2 |
| Seoul virus | 38 ± 3 | 11 ± 1 | 16 ± 2 | 23 ± 2 | 12 ± 1 |
| SNV | 10 ± 1 | 12 ± 1 | 20 ± 1 | 19 ± 2 | 18 ± 1 |
| Bunyamwera virus | 62 ± 3 | 27 ± 2 | 41 ± 3 | 37 ± 4 | 49 ± 4 |
| Tomato spotted wilt virus | 53 ± 4 | 54 ± 3 | 54 ± 4 | 48 ± 3 | 38 ± 3 |
| Rift Valley fever virus | 55 ± 4 | 48 ± 4 | 58 ± 4 | 35 ± 3 | 41 ± 3 |
| CCHFV | 54 ± 3 | 52 ± 4 | 58 ± 4 | 43 ± 2 | 37 ± 2 |
N protein (100 nM) was incubated with parental radiolabeled panhandle (0.01 nM) with increasing concentrations of competitor RNA panhandle (0.1 nM to 10 nM). PHV, Prospect Hill virus; CCHFV, Crimean Congo hemorrhagic fever virus.
N protein trimers undergo a conformational change coincident with binding to the vRNA panhandle.
To further characterize the interaction between hantavirus N and the various panhandles, we carried out fluorescence spectroscopic analysis to monitor possible conformational changes in N protein trimers that arise from their association with RNA. The spectroscopic analysis characterizes the disposition of tryptophan residues. All N proteins except that of Seoul virus have four tryptophan residues, and the position of tryptophan residues in all five N proteins is identical. Seoul virus N protein contains an additional tryptophan residue at position 239 from the N terminus. N proteins were excited at 290 nm, and emission spectra from 300 to 500 nm were recorded. The neutral quencher acrylamide was used to quench the tryptophan fluorescence of N trimers in the presence and absence of panhandle RNA. Tryptophan fluorescence at different input concentrations of acrylamide was determined to generate Stern-Volmer plots (Fig. 5), and the associated Stern-Volmer quenching constants were determined (Table 4). In the absence of panhandle RNA, the Ksv value for each N trimer varies in a virus-specific manner, suggesting that the accessibility of one or more tryptophan residues for the quencher also varies. This indicates that the environment surrounding each tryptophan is slightly different. For all N molecules there is a remarkable decrease in Ksv value after association with the RNA panhandles. This indicates that after association with panhandle RNA, N trimers undergo a conformational change, whereby tryptophan residues become less accessible to quencher molecules. In accordance with the binding data, SNV N exhibits a marked conformational change with only the homologous SNV panhandles, whereas the other hantavirus N proteins underwent a conformational change with any of the tested hantavirus panhandles. In contrast, the Ksv values were not altered by association of hantavirus N trimers with panhandle RNA molecules from viruses in other genera of the Bunyaviridae family. This is consistent with the idea that specific and correct interaction between hantavirus N and a hantavirus RNA panhandle is necessary for the observed induction of a conformational change in N.
FIG. 5.
Stern-Volmer analysis of N-associated tryptophan quenching. Plots depicting tryptophan fluorescence quenching of trimeric N protein in the presence of neutral quencher acrylamide are shown. A solution containing 25 nM trimeric N was treated with an excitation wavelength of 292 nm, and emission was recorded at 330 nm. The fluorescence intensity at 330 nm for Prospect Hill virus (PHV; a), Puumala virus (b), Seoul virus (c), SNV (d), and Andes virus (e) was determined by using different input concentrations of acrylamide. Plots of F0/F as a function of acrylamide concentration (Stern-Volmer plots) for free N protein trimer in the absence of any panhandle (▪) and in the presence of minipanhandle from the same (○) or a different virus (□). The panhandle RNA concentration was three times the dissociation constant for the panhandles used in the experiment. The Stern-Volmer plots of N protein trimers in the presence of different panhandles is shown by arrows, and the corresponding Ksv values are also presented. The same method was used to calculate the Ksv values for the interaction of other N protein trimers with different panhandles. These data are presented in Table 4. TSW, tomato spotted wilt virus; RVF, Rift Valley fever virus.
TABLE 4.
Stern-Volmer quenching constants for the association of N proteins from Sin Nombre, Andes, Puumala, Prospect Hill, and Seoul viruses with wild-type S segment panhandles from different viruses in binding buffer containing 80 mM NaCl at room temperaturea
| Panhandle | Quenching constant for interaction with indicated N protein
|
||||
|---|---|---|---|---|---|
| SNV | Andes virus | Puumala virus | PHV | Seoul virus | |
| None | 6.4 ± 0.1 | 7.2 ± 0.3 | 9.7 ± 0.4 | 7.9 ± 0.1 | 6.3 ± 0.1 |
| Andes virus | 2.8 ± 0.05 | 2.4 ± 0.03 | 4.2 ± 0.1 | 5.2 ± 0.3 | 3.3 ± 0.2 |
| PHV | 5.2 ± 0.1 | 2.7 ± 0.1 | 4.8 ± 0.3 | 4.9 ± 0.2 | 3.1 ± 0.2 |
| Puumala virus | 5.4 ± 0.1 | 2.7 ± 0.02 | 3.5 ± 0.1 | 5.4 ± 0.3 | 3.6 ± 0.1 |
| Seoul virus | 4.0 ± 0.3 | 3.0 ± 0.2 | 4.5 ± 0.2 | 5.1 ± 0.2 | 3.0 ± 0.3 |
| SNV | 2.6 ± 0.06 | 2.5 ± 0.1 | 4.3 ± 0.1 | 5.3 ± 0.2 | 3.3 ± 0.3 |
| Bunyamwera virus | 5.0 ± 0.3 | 5.9 ± 0.3 | 8.1 ± 0.4 | 7.1 ± 0.2 | 5.2 ± 0.1 |
| Tomato spotted wilt virus | 5.4 ± 0.2 | 6.2 ± 0.1 | 8.0 ± 0.2 | 7.3 ± 0.1 | 5.6 ± 0.1 |
| Rift Valley fever virus | 5.0 ± 0.3 | 6.3 ± 0.2 | 8.6 ± 0.3 | 7.7 ± 0.2 | 5.3 ± 0.2 |
| CCHFV | 4.9 ± 0.2 | 6.8 ± 0.3 | 8.8 ± 0.4 | 7.2 ± 0.2 | 5.9 ± 0.1 |
Tryptophan fluorescence of N protein (25 nM) was quenched with acrylamide in the presence or absence panhandle RNA to generate the Stern-Volmer plots. The panhandle RNA concentration was three times the dissociation constant for the panhandles used in each experiment. PHV, Prospect Hill virus; CCHFV, Crimean Congo hemorrhagic fever virus.
The data suggest that the requirements for high-affinity binding of SNV N to the panhandle are more stringent than those for binding of N molecules from other hantaviruses. This observation is also in agreement with the previously described filter binding data, which indicate that association of N with the RNA panhandle may be somewhat more stringent than that of N from other hantaviruses.
The terminal nucleotides in the panhandle are necessary for binding.
Since, with the exception of SNV, trimeric hantavirus N protein binds to panhandles derived from other viruses in the genus Hantavirus, it appears that N recognizes a common structure or sequence in these panhandle RNA molecules. All of the panhandles have the same first eight nucleotides (Fig. 2A). We synthesized a panhandle (SNV S, mutant 1) in which nucleotide pairs internal to the first nine bases were randomly shuffled (Fig. 2C). We observed that all N trimers, except that of SNV N, bound this mutant panhandle with affinity and specificity (Table 5). We synthesized another similar mutant panhandle (SNV S, mutant 2) in which the nucleotide pairs at the third, fourth, and fifth positions were changed but the secondary structure of the panhandle would be maintained. We observed that none of the trimeric N molecules could bind mutant 2 with high affinity, indicating that these nucleotides are necessary for binding. In another mutant (SNV S, mutant 3), extra nucleotides were added to both the 5′ and 3′ ends of the RNA such that there would be extended hydrogen bonding and maintenance of the continuous stem structure. This mutant was recognized by all the N trimers with specificity. Based on these observations it appears that all N protein trimers require the first 9 nucleotides for specific binding to the panhandle, whereas the SNV N trimer requires the first 20 nucleotides of the panhandle for binding.
TABLE 5.
Dissociation constants for purified trimeric N proteins from Andes, Puumala, Prospect Hill, Seoul, and Sin Nombre viruses with different panhandles in binding buffer at 25°C
| Proteina | Panhandle |
KD (nM) at indicated NaCl concn
|
|
|---|---|---|---|
| 80 mM | 220 mM | ||
| Andes virus | SNV S mutant 1 | 27 ± 3.1 | 31 ± 3.2 |
| Puumala virus | 25 ± 3.5 | 29 ± 2.0 | |
| PHV | 22 ± 4.1 | 27 ± 3.3 | |
| Seoul virus | 21 ± 2.0 | 27 ± 2.6 | |
| SNV | 212 ± 4.0 | ||
| Andes virus | SNV S mutant 2 | 257 ± 3.1 | |
| Puumala virus | 227 ± 5.2 | ||
| PHV | 262 ± 4.1 | ||
| Seoul virus | 229 ± 3.0 | ||
| SNV | 319 ± 4.0 | ||
| Andes virus | SNV S mutant 3 | 26 ± 2.1 | 31 ± 3.0 |
| Puumala virus | 19 ± 3.2 | 29 ± 2.2 | |
| PHV | 27 ± 4.1 | 31 ± 2.1 | |
| Seoul virus | 21 ± 1.0 | 27 ± 1.5 | |
| SNV | 22 ± 1.0 | 25 ± 2.1 | |
PHV, Prospect Hill virus.
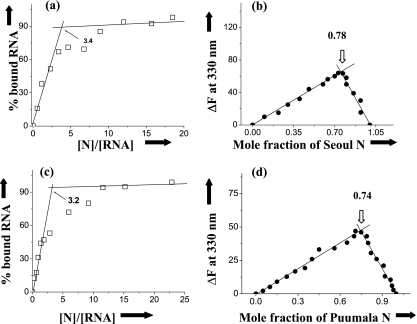
Binding stoichiometry.
As reported previously, the SNV N trimer binds to the panhandle with 1:1 binding stoichiometry, and the minimum size required for the association was 23 nucleotide pairs (20, 21). The binding data from all N protein trimers were replotted to calculate the binding stoichiometry. We used two different methods to verify our results. The binding stoichiometry, expressed in terms of the number of N molecules bound per panhandle, was estimated from the intersection of two straight lines from a least-squares fit plot of the percent increase of bound RNA against the ratio of input concentrations of N protein and RNA (Fig. 6a). We also used continuous variation plots to verify this binding stoichiometry calculation (Fig. 6b). The change in the fluorescence signal of N trimers upon binding to vRNA panhandles was plotted as a function of the input mole fraction of N protein. The breakpoint in the resulting plot corresponds to the mole fraction of N in the N-panhandle RNA complex (see Materials and Methods for details). Figure 6 depicts these plots for Seoul and Puumala viruses, and similar plots were obtained for other N protein trimers (data not shown). These data are consistent with the idea that all N protein trimers bind to the panhandle RNA with a stoichiometry of 1:1.
FIG. 6.
Calculation of binding stoichiometry. Data from binding experiments were reported to calculate binding stoichiometry using two different methods as described in Materials and Methods. (a) Representative plot for the calculation of binding stoichiometry between trimeric N protein from Seoul virus and its S segment minipanhandle. (b) The results from panel a were verified by using a continuous-variation plot (see Materials and Methods for details). (c and d) Similar analysis for Puumala virus N protein.
DISCUSSION
Specific recognition of the vRNA genome by viral nucleocapsid protein is likely to be necessary for specific encapsidation of viral RNA and/or replication of viral genome. Recognition of vRNA by N requires the conserved higher order structure of the vRNA panhandle. We expressed and purified trimeric N protein from Andes, Prospect Hill, Puumala, Seoul, and Sin Nombre viruses and found that, as expected, N specifically recognizes the corresponding homologous viral RNA panhandles. Further, all N trimers except those of SNV recognize heterologous hantavirus panhandles but not panhandles derived from viruses of other genera in the family Bunyaviridae. Thus, both the higher-order structure of the hantavirus panhandle and the RNA binding domain of hantavirus N are highly functionally conserved. The ability of hantavirus N to discriminate between hantaviral panhandles and panhandles from other genera suggests that the primary sequence might be involved in specific recognition as the secondary structure of the vRNA panhandle is highly conserved throughout the Bunyaviridae. The primary sequence of the hantaviral RNA termini is highly conserved, with the first eight nucleotides in the panhandle being absolutely conserved. By using mutational analysis we found that N trimers from Andes, Puumala, Prospect Hill, and Seoul viruses required maintenance of the terminal conserved eight nucleotides of the panhandle. Additional nucleotide pairs adjacent to these terminal eight are also required for high-affinity binding. However, the exact sequence of these nucleotides pairs is not. For SNV the first 23 nucleotides are necessary for interaction of N protein with the panhandle. Andes virus and SNV panhandles are identical in sequence for the first 27 nucleotides. Thus, the terminal nucleotides of both panhandles probably interact with SNV N in identical ways.
The RNA binding domain of all N proteins, including that of SNV, is highly conserved and has been mapped to amino acids 175 to 217 in Hantaan virus N (39). Recently, the region spanning the amino acids from 194 to 204 was identified as the minimal region required for binding (36), and three amino acids, a glutamic acid, a tyrosine, and a serine in RNA binding domain have been suggested to be involved in RNA-N protein interaction. From fluorescence-quenching studies we have shown that N proteins from Andes, Puumala, Prospect Hill, Seoul, and Sin Nombre viruses assemble into trimeric forms with subtly different higher-order structures, as evidenced by their different Ksv values in the absence of RNA. This could be due to differential intermolecular protein-protein interactions between individual N molecules that comprise the trimer or to slightly different molecular environments surrounding the tryptophan residues. We suggest that for SNV N such a difference might be responsible for differential panhandle recognition compared with N from other hantaviruses.
We used two different methods to calculate the number of trimeric N molecules bound per panhandle. As reported earlier, we found that trimeric N molecules bind to individual vRNA panhandles with a stoichiometry of 1:1. If one assumes that a trimeric N molecule is globular in shape, then the average diameter for such a trimer will be around 80 to 100 Å. Double-stranded RNA typically assumes an A-type helix with a pitch of 28 Å, and the RNA panhandles of the various viruses are mostly double stranded. Thus, with an approximate linear length of about 650 Å there would enough space for multiple trimers on the panhandle. The specific association of a single trimer with the panhandle might be an initial interaction involved in either encapsidation or replication of the hantaviral genome. During encapsidation many N molecules are recruited on the viral genomic RNA. The specific binding of the N trimer at the panhandle might function in the preliminary discrimination of viral genomic RNA from nonviral RNA molecules, or the panhandle might serve as a nucleation site for further recruitment of N molecules during encapsidation. It is also likely that this specific interaction of N with the panhandle serves in conjunction with the viral RNA polymerase to recognize the viral RNA molecules during replication initiation (19, 20). Along these lines, it has been observed under in vitro and in vivo conditions that RdRp, along with N protein, is necessary for genome replication (3, 5, 12, 16).
In the Bunyaviridae family, the majority of the genomic RNAs, both those found intracellularly and those in virions, are found as ribonucleoproteins complexed with the viral N protein. Electron microscopic analysis indicated that the termini of Uukuniemi virus genomic RNAs interact to form circular structures that dissociate under denaturing conditions (11). Furthermore, the genome RNPs of other bunyaviruses have also been observed to be circular, indicating that intramolecular association of the vRNA termini forms in vivo and that the terminal panhandle ostensibly forms in the context of bound N protein (22, 24, 25). All members of the family Bunyaviridae contain terminal nucleotide sequences that could facilitate the association of termini through the formation of short panhandles, and for La Crosse virus, these termini have been shown to be base paired within the nucleocapsids, via psoralen cross-linking in situ. As with all minus-strand RNA viruses, these genome RNPs or nucleocapsids serve as templates for both mRNA and antigenome synthesis. Recent reverse genetic studies using Bunyamwera virus replicons has revealed that the ability of the genomic RNA termini to form base pairs is an important feature of bunyavirus replication (4). However, the way in which these termini form base pairs in the context of the nucleocapsid, and the precise role they play in viral RNA synthesis, is unclear.
Several attempts have been made to identify examples of intraspecific and interspecific segment reassortment as potentially contributory factors in the molecular evolution of members of the genus Hantavirus (10, 18, 28, 29). The ability of N protein and RdRp of one hantavirus to bind, replicate, or transcribe a genomic segment derived from a second hantavirus species could potentially represent a barrier that would need to be overcome to successfully reassort segments between viral species. Our results suggest that at least some such interspecific pairings could succeed, but the lower binding affinities exhibited between hantavirus N proteins and the panhandles of viruses in the family Bunyaviridae suggest that reassortment between such divergent viral species may be of limited efficiency.
Acknowledgments
This work was supported by the University of New Mexico School of Medicine Research Allocation Committee and research grant R21AI059330 from the NIH.
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Alfadhli, A., Z. Love, B. Arvidson, J. Seeds, J. Willey, and E. Barklis. 2001. Hantavirus nucleocapsid protein oligomerization. J. Virol. 75:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfadhli, A., E. Steel, L. Finlay, H. P. Bachinger, and E. Barklis. 2002. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 277:27103-27108. [DOI] [PubMed] [Google Scholar]
- 3.Barr, J. N., R. M. Elliott, E. F. Dunn, and G. W. Wertz. 2003. Segment-specific terminal sequences of Bunyamwera bunyavirus regulate genome replication. Virology 311:326-338. [DOI] [PubMed] [Google Scholar]
- 4.Barr, J. N., and G. W. Wertz. 2005. Role of the conserved nucleotide mismatch within 3′- and 5′-terminal regions of Bunyamwera virus in signaling transcription. J. Virol. 79:3586-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blakqori, G., G. Kochs, O. Haller, and F. Weber. 2003. Functional L polymerase of La Crosse virus allows in vivo reconstitution of recombinant nucleocapsids. J. Gen. Virol. 84:1207-1214. [DOI] [PubMed] [Google Scholar]
- 6.Das, S., and D. Dasgupta. 2005. Binding of (MTR)2Zn2+ complex to chromatin: a comparison with (MTR)2Mg2+ complex. J. Inorg. Biochem. 99:707-715. [DOI] [PubMed] [Google Scholar]
- 7.Gavrilovskaya, I. N., E. J. Brown, M. H. Ginsberg, and E. R. Mackow. 1999. Cellular entry of hantaviruses which cause hemorrhagic fever with renal syndrome is mediated by β3 integrins. J. Virol. 73:3951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gavrilovskaya, I. N., M. Shepley, R. Shaw, M. H. Ginsberg, and E. R. Mackow. 1998. β3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA 95:7074-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gott, P., R. Stohwasser, P. Schnitzler, G. Darai, and E. K. Bautz. 1993. RNA binding of recombinant nucleocapsid proteins of hantaviruses. Virology 194:332-337. [DOI] [PubMed] [Google Scholar]
- 10.Henderson, W. W., M. C. Monroe, S. C. St. Jeor, W. P. Thayer, J. E. Rowe, C. J. Peters, and S. T. Nichol. 1995. Naturally occurring Sin Nombre virus genetic reassortants. Virology 214:602-610. [DOI] [PubMed] [Google Scholar]
- 11.Hewlett, M. J., R. F. Pettersson, and D. Baltimore. 1977. Circular forms of Uukuniemi virion RNA: an electron microscopic study. J. Virol. 21:1085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikegami, T., C. J. Peters, and S. Makino. 2005. Rift valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J. Virol. 79:5606-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin, H., and R. M. Elliott. 1993. Characterization of Bunyamwera virus S RNA that is transcribed and replicated by the L protein expressed from recombinant vaccinia virus. J. Virol. 67:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaukinen, P., V. Koistinen, O. Vapalahti, A. Vaheri, and A. Plyusnin. 2001. Interaction between molecules of hantavirus nucleocapsid protein. J. Gen. Virol. 82:1845-1853. [DOI] [PubMed] [Google Scholar]
- 15.Kaukinen, P., A. Vaheri, and A. Plyusnin. 2003. Mapping of the regions involved in homotypic interactions of Tula hantavirus N protein. J. Virol. 77:10910-10916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohl, A., T. J. Hart, C. Noonan, E. Royall, L. O. Roberts, and R. M. Elliott. 2004. A Bunyamwera virus minireplicon system in mosquito cells. J. Virol. 78:5679-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakowicz, J. R. 1999. Principles of fluorescence spectroscopy, p. 237-265. Plenum Press, New York, N.Y.
- 18.Li, D., A. L. Schmaljohn, K. Anderson, and C. S. Schmaljohn. 1995. Complete nucleotide sequences of the M and S segments of two hantavirus isolates from California: evidence for reassortment in nature among viruses related to hantavirus pulmonary syndrome. Virology 206:973-983. [DOI] [PubMed] [Google Scholar]
- 19.Mir, M. A., and A. T. Panganiban. 2006. The bunyavirus nucleocapsid protein is an RNA chaperone: possible roles in viral RNA panhandle formation and genome replication. RNA 12:272-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mir, M. A., and A. T. Panganiban. 2005. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J. Virol. 79:1824-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mir, M. A., and A. T. Panganiban. 2004. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J. Virol. 78:8281-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obijeski, J. F., D. H. Bishop, F. A. Murphy, and E. L. Palmer. 1976. Structural proteins of La Crosse virus. J. Virol. 19:985-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osborne, J. C., and R. M. Elliott. 2000. RNA binding properties of Bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. J. Virol. 74:9946-9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersson, R. F., and C. H. von Bonsdorff. 1975. Ribonucleoproteins of Uukuniemi virus are circular. J. Virol. 15:386-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raju, R., and D. Kolakofsky. 1989. The ends of La Crosse virus genome and antigenome RNAs within nucleocapsids are base paired. J. Virol. 63:122-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rawlings, J. A., N. Torrez-Martinez, S. U. Neill, G. M. Moore, B. N. Hicks, S. Pichuantes, A. Nguyen, M. Bharadwaj, and B. Hjelle. 1996. Cocirculation of multiple hantaviruses in Texas, with characterization of the small (S) genome of a previously undescribed virus of cotton rats (Sigmodon hispidus). Am. J. Trop. Med. Hyg. 55:672-679. [DOI] [PubMed] [Google Scholar]
- 27.Richmond, K. E., K. Chenault, J. L. Sherwood, and T. L. German. 1998. Characterization of the nucleic acid binding properties of tomato spotted wilt virus nucleocapsid protein. Virology 248:6-11. [DOI] [PubMed] [Google Scholar]
- 28.Rizvanov, A. A., S. F. Khaiboullina, and S. St. Jeor. 2004. Development of reassortant viruses between pathogenic hantavirus strains. Virology 327:225-232. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez, L. L., J. H. Owens, C. J. Peters, and S. T. Nichol. 1998. Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology 242:99-106. [DOI] [PubMed] [Google Scholar]
- 30.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmaljohn, C. M. 1996. Molecular biology of hantaviruses. Plenum Press, New York, N.Y.
- 32.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1602. In K. A. Howley (ed.), Fields virology, vol. 2. Lippincott, Williams, and Wilkins, Philadelphia, Pa. [Google Scholar]
- 33.Schmaljohn, C. S., and C. B. Jonsson. 2001. Replication of hantaviruses, p. 15-32. In S. A. Nichol (ed.), Hantaviruses. Springer-Verlag, Berlin, Germany.
- 34.Schmaljohn, C. S., A. L. Schmaljohn, and J. M. Dalrymple. 1987. Hantaan virus M RNA: coding strategy, nucleotide sequence, and gene order. Virology 157:31-39. [DOI] [PubMed] [Google Scholar]
- 35.Severson, W., L. Partin, C. S. Schmaljohn, and C. B. Jonsson. 1999. Characterization of the Hantaan nucleocapsid protein-ribonucleic acid interaction. J. Biol. Chem. 274:33732-33739. [DOI] [PubMed] [Google Scholar]
- 36.Severson, W., X. Xu, M. Kuhn, N. Senutovitch, M. Thokala, F. Ferron, S. Longhi, B. Canard, and C. B. Jonsson. 2005. Essential amino acids of the Hantaan virus N protein in its interaction with RNA. J. Virol. 79:10032-10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Severson, W. E., X. Xu, and C. B. Jonsson. 2001. cis-acting signals in encapsidation of Hantaan virus S-segment viral genomic RNA by its N protein. J. Virol. 75:2646-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torrez-Martinez, N., M. Bharadwaj, D. Goade, J. Delury, P. Moran, B. Hicks, B. Nix, J. L. Davis, and B. Hjelle. 1998. Bayou virus-associated hantavirus pulmonary syndrome in Eastern Texas: identification of the rice rat, Oryzomys palustris, as reservoir host. Emerg. Infect. Dis. 4:105-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu, X., W. Severson, N. Villegas, C. S. Schmaljohn, and C. B. Jonsson. 2002. The RNA binding domain of the Hantaan virus N protein maps to a central, conserved region. J. Virol. 76:3301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]