Abstract
The evolution of peptide-specific CD4+ T-cell responses to acute viral infections of humans is poorly understood. We analyzed the response to parvovirus B19 (B19), a ubiquitous and clinically significant pathogen with a compact and conserved genome. The magnitude and breadth of the CD4+ T-cell response to the two B19 capsid proteins were investigated using a set of overlapping peptides and gamma interferon-specific enzyme-linked immunospot assays of peripheral blood mononuclear cells (PBMCs) from a cohort of acutely infected individuals who presented with acute arthropathy. These were compared to those for a cohort of B19-specific immunoglobulin M-negative (IgM−), IgG+ remotely infected individuals. Both cohorts of individuals were found to make broad CD4+ responses. However, while the responses following acute infection were detectable ex vivo, responses in remotely infected individuals were only detected after culture. One epitope (LASEESAFYVLEHSSFQLLG) was consistently targeted by both acutely (10/12) and remotely (6/7) infected individuals. This epitope was DRB1*1501 restricted, and a major histocompatibility complex peptide tetramer stained PBMCs from acutely infected individuals in the range of 0.003 to 0.042% of CD4+ T cells. Tetramer-positive populations were initially CD62Llo; unlike the case for B19-specific CD8+ T-cell responses, however, CD62L was reexpressed at later times, as responses remained stable or declined slowly. This first identification of B19 CD4+ T-cell epitopes, including a key immunodominant peptide, provides the tools to investigate the breadth, frequency, and functions of cellular responses to this virus in a range of specific clinical settings and gives an important reference point for analysis of peptide-specific CD4+ T cells during acute and persistent virus infections of humans.
Human parvovirus B19 (B19) is a ubiquitous, ∼5.6-kb DNA virus that causes erythema infectiosum, polyarthropathy, transient aplastic crisis, and fetal death. The genome is very stable and encodes only three major proteins. It is traditionally viewed as an acute but fully resolving human viral pathogen, although viral persistence in a number of cases (particularly in the immunocompromised) has been reported (18). The role of the cellular arm of the immune response to this virus has not been investigated extensively. We recently identified large CD8+ T-cell responses to B19 NS1 peptides, which in the first 2 years postinfection were sustained as mature “effector memory” populations (CD62Llo CCR7lo perforin+ CD57+) (15).
B19 has an icosahedral capsid consisting of two proteins, VP1 (83 kDa) and VP2 (58 kDa). The two proteins are identical except that VP1 has an additional 227 amino acids at its N terminus, known as the VP1 unique region (VP1U). VP2 is the major capsid protein and makes up approximately 95% of the 60 capsid protein units in the native virus (2). B19-specific CD4+ T-cell responses to recombinant B19 capsid proteins have been demonstrated in a number of studies but have not been defined at the peptide level (3, 5, 22). The role that B19-specific CD4+ T-cell responses play in protective immunity and/or immunity-mediated pathogenesis remains ill-defined. Increasing evidence points to a critical role of virus-specific CD4+ cells in protective immunity to a number of other viral infections, e.g., human immunodeficiency virus (HIV) (24), hepatitis C virus (12), and cytomegalovirus (CMV) (9, 10) infections. However, studies also suggest that CD4+ T cells can cause significant pathology upon overactivation, e.g., in human T-cell leukemia virus type 1 (HTLV-1) infection (13). A range of evidence indirectly supports the importance of CD4+ T cells in protection against B19 infection (14, 19), including, for example, the occurrence of pure red cell aplasia due to persistent B19 infection in patients with AIDS (8). On the other hand, immunity-mediated pathogenesis may cause a number of B19-related clinical symptoms, such as arthralgia. Thus, HLA-DR4-positive individuals are reported to be more susceptible to parvovirus arthritis (11, 16, 17).
To understand these issues in more depth, we set out to analyze the quantity and quality of CD4+ T-cell responses to parvovirus B19 infection at the T-cell epitope level. The response profile shows important differences from the CD8+ T-cell response and provides novel insights into the evolution of a normal, “successful” CD4+ T-cell response in humans.
MATERIALS AND METHODS
Study participants and sampling.
Thirteen previously healthy immunocompetent adults presenting to their general practitioners with symptoms of fever, arthralgia, fatigue, and rash were prospectively identified (B19 immunoglobulin M [IgM] positive) at the Department of Clinical Virology at the Oxford Radcliffe Hospitals, Oxford, United Kingdom. Patient information is displayed in Table 1. The timing of blood samples is given relative to the onset of symptoms. Eight healthy B19 IgG-positive, B19 IgM/DNA-negative laboratory volunteers were studied as a “remotely infected” cohort. Patients OR1 to -4 were included in a previous study of B19-specific CD8+ T-cell responses (15). They had no history suggestive of B19 infection and were likely infected in childhood. Four B19-seronegative individuals were used as control subjects. Peripheral blood mononuclear cells (PBMCs) were separated from heparinized blood samples within 8 h of sampling by gradient centrifugation on Ficoll-Paque (Amersham Biosciences, Uppsala, Sweden) or Lymphoprep (Fresenius Kabi Norge, Halden, Norway). Informed patient consent was obtained prior to study initiation. Ethical approval for the study was obtained from the Oxfordshire Clinical Research Ethics Committee (CO2.113).
TABLE 1.
Information on study participants
| Patient | Sexa | Age (yr) | HLA type
|
|||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | DRB1 | DRB3/4/5 | DQB1 | |||
| Remotely infected patients | ||||||||
| OR1 | M | 41 | 2,26 | 35,62 | 4 | 14,15 | 4,5 | 6,3 |
| OR2 | M | 43 | 2,29 | 44 | 7 | 4 | 2 | |
| OR3 | F | 24 | 2 | 40,14 | 4,6 | 15,13 | 5,3 | 6 |
| OR4 | M | 31 | 24,31 | 35 | 4,4 | 4,11 | 3,4 | 3 |
| OR6 | M | 37 | 3,11 | 55,49 | 3,7 | 1,4 | 4 | 5 |
| OR7 | M | 36 | 1,2 | 50,55 | 6,5 | 7,13 | 3,4 | 2,6 |
| OR8 | F | 29 | 1,2 | 8,57 | 6,7 | 3,11 | 3 | 2,3 |
| OR9 | F | 25 | 1,3 | 7,8 | 7 | 15,4 | 5,4 | 6 |
| Acutely infected patients | ||||||||
| O2 | F | 28 | 2,24 | 35,57 | 4,6 | 15,3 | 5,3 | 2,6 |
| O7 | F | 60 | 2 | 51,50 | 6,5 | 7,13 | 3,4 | 2,6 |
| O8 | F | 36 | 1,2 | 8,51 | 6 | 8,13 | 3 | 4,6 |
| O9 | F | 45 | 11,24 | 7,15 | 3,7 | 15,13 | 5,3 | 6 |
| O11 | F | 42 | 3,32 | 44,51 | 1,5 | 1 | 5 | |
| O12 | F | 40 | 2 | 40,51 | 3,14 | 15 | 5 | 6,3 |
| O13 | F | 51 | 1,2 | 7,44 | 3,7 | 15,4 | 5,4 | 6,3 |
| O15 | F | 49 | 11,25 | 7,35 | 7,4 | 15,14 | 5,3 | 5,6 |
| O19 | F | 43 | 1,29 | 8,52 | 7,12 | 15,3 | 5,3 | 2,6 |
| O20 | F | 31 | 2,3 | 7 | 7 | 1,15 | 5 | 5,6 |
| O21 | F | 37 | 2 | 7,40 | 7,3 | 16,4 | 5,4 | 5,3 |
| O22 | M | 49 | 1,24 | 8,40 | 2,7 | 15,3 | 5,3 | 2,6 |
| O23 | F | 48 | 3,31 | 8,7 | 7,7 | 15,51 | 6 | |
F, female; M, male.
Synthetic peptides.
A total of 77 20-mer peptides overlapping by 10 amino acids were synthesized by standard 9-fluorenylmethoxy carbonyl chemistry. These peptides covered the entire length of the VP1/2 proteins and were divided into 11 pools of seven peptides each. Peptides were used in both ex vivo and cultured gamma interferon (IFN-γ)-specific enzyme-linked immunospot (IFN-γ ELISPOT) assays and intracellular cytokine assays.
Ex vivo and cultured IFN-γ ELISPOT assays.
IFN-γ ELISPOT assays were performed as described previously (15). Briefly, 2.5 × 105 PBMCs were stimulated in triplicate with peptide pools/phytohemagglutinin (Sigma, St. Louis, Mo.). Synthetic peptides were used in pools of seven at a final concentration of 10 μM. IFN-γ responses were confirmed using individual peptides with optimal epitopes. Cultured ELISPOT assays involved the prior generation of short-term lines specific to VP1/2 peptide pools or individual 20-mer peptides. PBMCs were pulsed with 50 μM of the respective peptide and cultured at 2 × 106 cells/ml in 24-well plates for 12 to 18 days. On day 3, 10 units/ml of interleukin-2 (IL-2) was added. Half of the medium was replaced every third day with fresh medium containing 10 units/ml of IL-2. Lines were used in place of ex vivo PBMCs as described above (100,000 cells per well).
Generation and testing of T-cell lines.
Peptide-specific T-cell lines (TCLs) were generated as described for cultured ELISPOT assays and were maintained with IL-2. Intracellular cytokine staining (ICS) was used to carry out LASEESAFYVLEHSSFQLLG epitope-specific HLA restriction. This required the prepulsing of matched and mismatched PBMCs/B-cell lines/transfected L cells (a gift of L. Fugger) with peptide at 20 μM for 1 h at 37°C. Cells were washed and added to cytotoxic T lymphocytes at a 10:1 ratio for 1 h before the addition of brefeldin A. After another 5 h, cells were stained with anti-CD4 and anti-IFN-γ monoclonal antibodies (Becton Dickinson, Stockholm, Sweden).
Tetramer staining and enrichment.
Tetramer DRB1*1501 (LASEESAFYVLEHSSFQLLG) was purchased from Beckman Coulter and used according to the manufacturer's protocol. Due to the low frequencies of cells with positive staining for this tetramer, a magnetic bead-based tetramer enrichment technique was employed as described previously (1). Ten percent of the sample was not enriched, and this was used to calculate the input number of CD4+ T cells (by multiplying by 9). The frequencies of tetramer-positive cells in the initial samples were then calculated as follows: initial frequency = (measured output number/calculated input number) × 100.
This technique has been shown to have a high sensitivity and specificity as well as to maintain a good linear range at low frequencies (25).
RESULTS
Individuals acutely infected with B19 make broad CD4+ T-cell responses to VP1/2 peptides which are detectable by direct ex vivo ELISPOT assay.
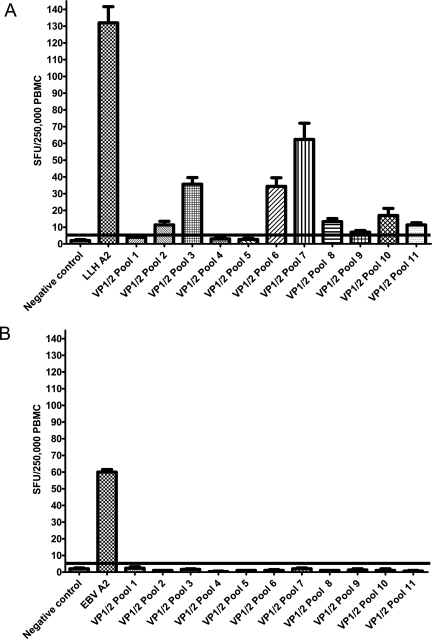
Figure 1A shows the results of a representative ex vivo IFN-γ ELISPOT assay carried out on PBMCs from a recently B19-infected (IgM+) individual (O22) 1 month after symptom development. Significant responses to 7 of 11 VP1/2 pools were seen, albeit at lower levels than the positive control (the CD8+ T-cell response to the HLA-A2-restricted B19 peptide LLHTDFEQV). Using this technique, ex vivo responses could be detected in seven of seven acutely infected individuals (1 to 5 months after symptom development) and were found for peptides located in both the VP1U and VP2 regions (Table 2). These responses were confirmed as CD4+ T-cell responses by magnetic bead depletion (data not shown). For the four B19-seronegative individuals tested, no responses were obtained in ex vivo assays. Pool 10 was the peptide pool targeted with the greatest frequency, with six of seven acutely infected individuals displaying an ex vivo response (Table 2). In contrast, no individuals made detectable ex vivo responses to pool 1. Individuals targeted two to eight (average, five pools) VP1/2 peptide pools. The levels of response ranged from 5 to 52 spot-forming units (SFU)/250,000 PBMCs, and the strongest responses (and greatest number of responses) were detected for peptides in the VP1/2 region. Due to restrictions in blood sample size, these responses were not dissected to identify the 20-mer peptides responsible. Three patients, however, were studied at sequential time points. Overall, they showed slight decreases in responses over time (Table 2). For example, the response of individual O7 to pool 10 dropped from 44 (at 1 month post-symptom development) to 21 (at 2 months post-symptom development) SFU/250,000 PBMCs. Similarly, the response of individual O12 to pool 6 dropped from 52 to 7 SFU/250,000 PBMCs between months 1 and 5 post-symptom development.
FIG. 1.
Responses to VP1/2 peptides can be detected by ex vivo IFN-γ ELISPOT assays for acute but not remote B19 infection. (A) Ex vivo ELISPOT assay carried out on PBMCs from patient O22 (HLA-A 1,24; HLA-B 8,40; HLA-C 2,7; DRB1 15,3; DRB5,3; DQB1 2,6) at 4 weeks post-symptom development. Cells were stimulated with VP1/2 peptide pools and a B19 CD8+ T-cell epitope HLA-A2 LLMTDFEQV. (B) Ex vivo ELISPOT assay of PBMCs from remotely infected individual OR1. Cells were stimulated with VP1/2 peptide pools and the EBV HLA-A2 GLCTLVAML peptide (as a positive control). Triplicate estimations are displayed. The cutoff for a positive result is the result for the negative control plus 2 standard deviations (horizontal line).
TABLE 2.
Data on frequency of ex vivo IFN-γ ELISPOT responses to VP1/2 peptide pools for seven acutely infected individuals
| Patient | Time (mo) post-symptom development | Response to peptide pool (SFU/250,000 PBMCs)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP1-specific pools
|
VP2-specific pools
|
|||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
| O22 | 1 | 12 | 46 | 38 | 24 | 16 | 29 | 13 | ||||
| O9 | 1 | 9 | 7 | |||||||||
| O7 | 1 | 8 | 11 | 16 | 5 | 44 | ||||||
| O7 | 2 | 6 | 11 | 12 | 6 | 21 | ||||||
| O8 | 1 | 8 | 32 | |||||||||
| O8 | 2 | |||||||||||
| O21 | 4 | 6 | 11 | 5 | 6 | 14 | ||||||
| O12 | 1 | 5 | 52 | 20 | 10 | 35 | ||||||
| O12 | 5 | 8 | 8 | 6 | 7 | 13 | 10 | |||||
| O11 | 2 | 11 | 15 | 16 | 16 | 10 | 11 | 9 | 17 | |||
A positive response (reported numbers) was taken as a response greater than that of the negative control (medium alone) plus 2 standard deviations.
Definition of an immunodominant B19 CD4+ T-cell epitope (LASEESAFYVLEHSSFQLLG).
In contrast to the data for acutely infected donors, responses to the VP1/2 pools were not detected for any of the remotely infected individuals in ex vivo assays. Figure 1B shows that for individual OR1, ex vivo IFN-γ responses could not be detected in response to stimulation with peptides from the capsid proteins VP1/2. However, B19-specific CD4+ T-cell peptide responses were readily detected in all seven remotely infected individuals (and zero of four B19-seronegative individuals) studied following in vitro culture of PBMCs with peptide pools for 14 days (Table 3). Fluorescence-activated cell sorting of short-term T-cell lines stimulated with peptide confirmed that they were CD4+ T-cell responses (Fig. 2A). Additionally, these responses were abrogated by CD4+ T-cell depletion (data not shown). Furthermore, in these cases, we were able to further define the pool responses to individual CD4+-restricted 20-mer peptides by repeated analysis of short-term T-cell lines (Table 3).
TABLE 3.
Summary of CD4+ T-cell VP1/2-specific 20-mer peptide sequences identified by cultured IFN-γ ELISPOT assays of cells from B19-seropositive remotely infected individuals (OR1-8)a
| B19 region | CD4+ T-cell-restricted peptide sequence | Peptide no. | Response of patient to peptide
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| OR1 | OR2 | OR3 | OR4 | OR6 | OR7 | OR8 | |||
| VP1U | MSKESGKWWESDDKFAKAVY | 1.1 | − | − | − | − | − | − | + |
| VP1U | LKDHYNISLDNPLENPSSLF | 1.5 | − | − | − | − | − | − | + |
| VP1U | DLVARIKNNLKNSPDLYSHH | 1.7 | − | − | − | − | + | + | − |
| VP1U | RIHDFRYSQLAKLGINPYTH | 3.2 | − | − | − | − | − | + | − |
| VP1U | WTVADEELLKNIKNETGFQA | 3.4 | + | − | − | − | − | − | − |
| VP1U | QVVKDYFTLKGAAAPVAHFQ | 3.6 | − | − | − | + | − | − | − |
| VP1/2 | GGGGSNPVKSMWSEGATFSA | 4.4 | − | − | + | − | − | − | − |
| VP1/2 | FLIPYDPEHHYKVFSPAASS | 4.7 | − | − | + | − | − | + | + |
| VP1/2 | YKVFSPAASSCHNASGKEAK | 5.1 | + | − | − | − | − | − | − |
| VP1/2 | STPWRYLDFNALNLFFSPLE | 5.4 | − | − | − | − | + | − | + |
| VP1/2 | ALNLFFSPLEFQHLIENYGS | 5.5 | + | + | − | − | − | − | − |
| VP1/2 | FQHLIENYGSIAPDALTVTI | 5.6 | − | − | + | − | − | − | − |
| VP1/2 | LPIWVYFPPQYAYLTVGDVN | 6.6 | + | + | − | − | − | − | − |
| VP1/2 | YAYLTVGDVNTQGISGDSKK | 6.7 | + | − | − | − | − | − | − |
| VP1/2 | LASEESAFYVLEHSSFQLLG | 7.2 | + | + | + | − | + | + | + |
| VP1/2 | LEHSSFQLLGTGGTATMYSK | 7.3 | − | + | − | − | − | − | + |
| VP1/2 | TGGTATMSYKFPPVPPENLE | 7.4 | + | − | − | − | − | − | − |
| VP1/2 | PDTLGGDPKFRSLTHEDHAI | 8.1 | − | − | − | − | − | − | + |
| VP1/2 | KYVPGINAISHGQTTYGNAE | 9.2 | − | − | − | − | − | − | + |
| VP1/2 | LQGLNMHTYFPNKGTQQYTD | 9.6 | − | − | − | − | − | − | + |
| VP1/2 | VWNRRALHYESQLWSKIPNL | 10.2 | − | + | − | + | − | − | − |
| VP1/2 | PQIFLKILPQSGPIGGIKSM | 10.6 | − | − | − | − | − | + | − |
| VP1/2 | GIMTVTMTFKLGPRKATGRW | 11.2 | − | − | − | − | − | − | + |
A positive response is taken as being greater than the value for the negative control (medium alone) plus 2 standard deviations. Positive responses were then confirmed by stimulating PBMCs with the identified 20-mer peptides and testing the T-cell lines in an ICS assay. Boldface type identifies the peptide sequence found to be restricted by HLA DRB1*1501.
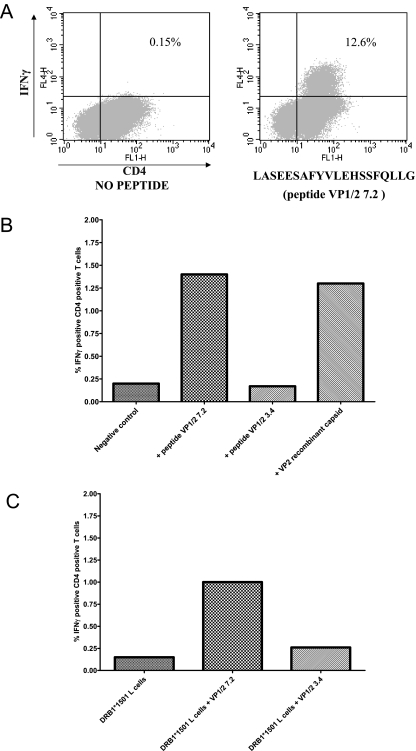
FIG. 2.
A peptide VP1/2 7.2-specific TCL can respond to stimulation with both peptide VP1/2 7.2 and VP2 recombinant capsid and can be shown to respond in a DRB1*1501-restricted manner. (A) ICS on 17-day-old peptide VP1/2 7.2-specific TCL (from individual OR3) stimulated with peptide VP1/2 7.2. Percentages shown are the numbers of IFN-γ-positive CD4+ T cells. (B) ICS on 17-day-old peptide VP1/2 7.2-specific TCL (from individual OR1) stimulated with peptide VP1/2 7.2 and VP2 recombinant capsid. Percentages shown are the numbers of IFN-γ-positive CD4+ T cells. (C) A peptide VP1/2 7.2-specific TCL from OR1 was rechallenged 17 days later with DRB1*1501-positive L cells (prepulsed with peptide VP1/2 7.2, VP1/2 3.4, or mock peptide) in an ICS assay. Percentages shown are the numbers of IFN-γ-positive CD4+ T cells.
In the above analysis, six of seven individuals were found to have clear responses to VP1/2 peptide 7.2 (LASEESAFYVLEHSSFQLLG). Positive 20-mer peptides were resynthesized to eliminate the potential existence of contaminants, and the responses were reproduced. Figure 2B shows that T-cell lines set up against these 20-mer peptides had the ability to react to B19 VP2 recombinant capsid protein, demonstrating that these cells recognize naturally processed epitopes. Similarly, VP2 capsid-derived T-cell lines responded to individual peptides (data not shown).
In individual OR1, the TCL response to peptide VP1/2 7.2 could be inhibited with an anti-HLA-DR monoclonal antibody but not with anti-HLA-DP and -DQ antibodies (data not shown). Intracellular cytokine staining using HLA-matched and mismatched PBMCs and B-cell lines as antigen-presenting cells was used to identify the DRB1*1501 restriction of this epitope in individual OR1. T-cell lines recognized LASEESAFYVLEHSSFQLLG presented by DRB1*1501-transfected L cells (Fig. 2C) but not by control B27-transfected L cells (data not shown). Peptide truncations showed the minimal binding region of the epitope to be an 11-mer peptide, FYVLEHSSFQL (data not shown), conforming to a published DRB1*1501 binding motif (21).
Targeting of the immunodominant epitope is common in acute infection.
Table 3 shows that six of seven (86%) remotely infected individuals responded to peptide VP1/2 7.2 in cultured IFN-γ ELISPOT assays, indicating that this epitope is not solely restricted by DRB1*1501. We therefore specifically analyzed responses to this peptide by using freeze-thawed samples from the acutely infected cohort (four non-DRB1*1501+ and eight DRB1*1501+ individuals). Seven of 12 acutely infected individuals responded to peptide VP1/2 in an ex vivo IFN-γ ELISPOT assay (data not shown). The frequencies of responses ranged from 10 to 115 SFU/million PBMCs. When the same individuals were tested by cultured IFN-γ ELISPOT assay, responses were detected in 10 of 12 (83%) individuals (3 non-DRB1*1501+ and 7 DRB1*1501+ individuals). For comparison, 9 of 12 acutely infected individuals were found to respond to VP2 recombinant capsid protein in ex vivo IFN-γ ELISPOT assays (data not shown). Responses ranged from approximately 50 to 200 SFU/million PBMCs.
Class II tetramer analysis of B19-specific CD4+ T-cell responses.
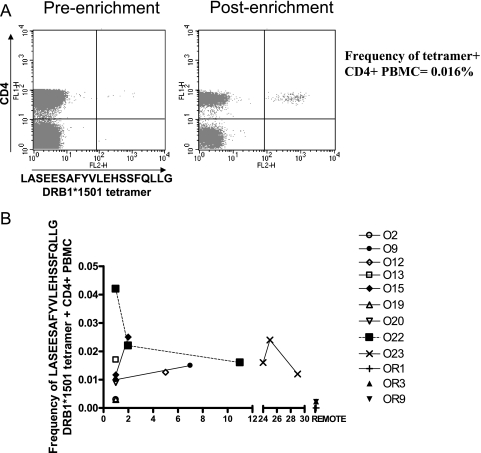
A DRB1*1501 LASEESAFYVLEHSSFQLLG tetramer was used to analyze the CD4+ T-cell responses ex vivo in nine of the acutely infected individuals who were DRB1*1501 positive. All nine DRB1*1501+ acutely infected individuals (at 1 to 29 months post-symptom development) had detectable responses, ranging from 0.003 to 0.042% CD4+ PBMCs (Fig. 3A). Clear and reproducible responses were observed only by using the enrichment technique, although a small population could be seen preenrichment, as shown in Fig. 3A. This high response rate supports the data obtained using the ex vivo ELISPOT assay.
FIG. 3.
DRB1*1501 LASEESAFYVLEHSSFQLLG tetramer-positive cells are present at high frequencies following B19 infection. (A) DRB1*1501 LASEESAFYVLEHSSFQLLG tetramer staining of cells from patient O23. A magnetic bead tetramer enrichment technique was required to clearly visualize the tetramer-positive populations. Preenrichment staining is shown on the left, while postenrichment staining is shown on the right. (B) Summary of the frequencies of DRB1*1501 LASEESAFYVLEHSSFQLLG tetramer-positive cells in nine DRB1*1501-positive acutely infected individuals and three remotely infected individuals.
For those individuals for whom longitudinal samples were available or from whom samples were taken beyond the acute phase (3 months or more), we analyzed the frequency of responses over time. Figure 3B reveals a relative stability of the responses, at around 0.02 to 0.03% of CD4+ T cells over the post-acute-phase period. Two remotely infected individuals studied (OR3 and OR9) had detectable tetramer-positive populations (following tetramer enrichment) at levels of 0.0023 and 0.0022% CD4+ PBMCs (Fig. 3B). Remotely infected individual OR1 did not display a detectable tetramer-positive population ex vivo, but cultured IFN-γ ELISPOT assays and ICS on a cultured T-cell line (Fig. 2) revealed that this individual had precursor CD4+ T cells that responded to this particular peptide. Tetramer-positive populations could not be detected in three B19-seropositive, DRB1*1501-negative individuals or in our B19-seronegative volunteers. Thus, responses appear to be maintained over a period of 1 to 2 years, with a decline at some point thereafter to the levels in remotely infected individuals, which are detectable ex vivo only after culture. These data are reminiscent of those obtained in studies of parvovirus B19-specific CD8+ T cells, although the sizes of the response were much greater in the latter case.
Phenotypic analysis of parvovirus B19-specific CD4+ T cells.
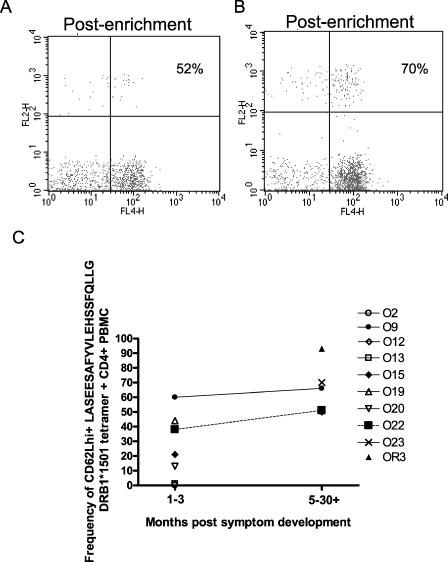
Since tetramer-positive populations were visible ex vivo during acute disease and for several months thereafter, we analyzed the phenotype of these populations with respect to “central” versus “effector” memory status (CD62L expression) (Fig. 4). In acute disease, CD62L+ tetramer-positive cells ranged from 0 to 60% of cells at time points within the first 3 months following symptom development (Fig. 4C). At time points from 5 to 30+ months (i.e., including one remotely infected individual), the range was 50 to 93%. This overall increase in CD62L expression was statistically significant (P = 0.01; Mann-Whitney test) and in marked contrast to data from the study of B19-specific CD8+ T-cell populations, where CD62L expression was maintained at very low levels over time.
FIG. 4.
DRB1*1501 LASEESAFYVLEHSSFQLLG tetramer-positive cells express high levels of CD62L following B19 infection. DRB1*1501 LASEESAFYVLEHSSFQLLG tetramer and CD62L staining results for patients O22 (A) and O23 (B) are shown. A magnetic bead tetramer enrichment technique was required to clearly visualize the tetramer-positive populations. (C) Summary of CD62L expression on DRB1*1501 LASEESAFYVLEHSSFQLLG tetramer-positive cells from nine DRB1*1501-positive acutely infected individuals and one remotely infected individual.
DISCUSSION
CD4+ T-cell responses play an important role in the control of both acute and persistent virus infections. However, in contrast to the case for CD8+ T cells, relatively little is known about the quantity and quality of CD4+ T-cell responses which emerge during acute infections of humans and how they evolve thereafter. Furthermore, although class II tetramers have been used to analyze CD4+ T-cell “memory” responses or for studies of persistent infection, there is little direct ex vivo data using these tools for acute disease. Since the B19 genome is small and conserved and since we had recently observed striking CD8+ T-cell responses to this virus, we aimed to dissect the CD4+ T-cell responses to this virus in detail.
CD4+ T-cell frequencies were generally greater in the acute than in the late or “remote” memory phase (as detected by ELISPOT assay and tetramer staining), and these responses declined over time. However, the kinetics of this decline requires further elucidation since the decline may be relatively slow, and indeed, responses were relatively stable over the first year or two after infection. This does, however, contrast with the case for CD8+ T cells regarding frequency after the resolution of clinical symptoms (15).
The phenotypes of CD4+ and CD8+ tetramer-positive cells also showed a difference, with a large proportion of tetramer-positive CD4+ T cells bearing the lymph node homing marker CD62L (even at times as early as 1 month post-symptom development). This phenotypic marker suggests that a significant number of these B19-specific cells are “central” memory cells. Only limited stains for CCR7 were performed (data not shown), but these confirmed a largely CCR7+ status. Thus, confronted with the same infection, CD4+ and CD8+ T-cell responses appear to follow different pathways after acute resolution. Indeed, CD8+ T-cell responses showed an increase in frequency and the acquisition of a “mature” phenotype (e.g., expression of perforin and CD57) as well as maintaining activation (CD38+) over many months after acute disease. Ultimately, the frequencies and activation status of the CD8+ T-cell responses do decline in “remote” infections, although they may remain CD62Llow (15).
In our study, there was no obvious preferential targeting of the VP2- or VP1U-specific capsid regions in either the acutely or remotely infected individuals. The remotely infected cohort responded to epitopes over the entire length of the VP1/2 sequence. The above findings are consistent with those of von Poblotzki and colleagues for recombinant antigens (VP1U, VP1, VP2, and NS1) (26), although there is some discrepancy on this point in the literature, possibly due to methodologic differences (6). The use of overlapping synthetic peptides is a highly sensitive method for investigating the fine detail of CD4+ T-cell responses to viral antigens.
Although our study sample was relatively small, the vast majority of individuals tested in both the acute and remote cohorts responded to epitope VP1/2 7.2. We propose that this epitope be considered as a candidate for any vaccine incorporating CD4-boosting responses. Although we defined this epitope as HLA-DR15 restricted, it is targeted by DR15-negative patients as well. Preliminary data suggest that at least one of the alternative HLA molecules capable of binding this epitope is DQ*06 (data not shown). The epitope may be equivalent to the immunodominant hemagglutinin epitope of influenza virus, which is commonly targeted and binds multiple HLA alleles, and the DR*1101/DR*0401-restricted NS3-derived peptide from hepatitis C virus, which has similar characteristics. Both of these have been studied ex vivo using class II tetramers (4, 20), and the memory responses elicited are of similar frequencies to those identified in the “remotely” infected individuals. Clearly, even though immunodominant and strong responses are observed during acute infection, the memory responses are at a level where detection ex vivo is very difficult, although the cells possess strong proliferative capacity, as confirmed in this as well as previous studies using recombinant antigens.
Overall, the exact role of B19-specific CD4+ T cells upon acute infection is unknown. In our study, symptom disappearance may be correlated with a decrease in CD4+ T-cell frequency, but limited data were available for these very early time points, and no asymptomatic seroconverters were studied. The possibility exists that the short-term arthropathy observed in this patient cohort could be attributed to a robust and overactive acute response. Previous studies have produced conflicting findings in this area and are based on small cohort sizes (7, 23). The new data on epitopes generated in this study should allow further definition of this issue. Hypothetically, B19-specific CD4+ T cells recognizing persisting B19 or cross-reactive self antigens could play a role in both viral elimination and the pathogenesis of autoimmune phenomena such as postviral arthritis. DR15 itself is linked to autoimmunity in the case of multiple sclerosis, but potential DR4-binding peptides may also be of most relevance in B19-associated arthropathy, given the previously described genetic link to this allele (11, 16).
In conclusion, this study is the first to identify CD4+ T-cell epitopes in B19, a prototype of a virus causing acute resolving infection. We observed an evolution of CD4+ T-cell responses which diverges substantially from that of CD8+ T-cell responses studied in the same clinical setting using similar techniques. Not only does this study provide an important reference point for other infections—particularly those where high levels of persistence occur—but it provides the tools to investigate the breadth, frequency, and functions of cellular responses to B19 in a range of specific clinical settings where the virus remains a significant problem.
Acknowledgments
We acknowledge funding from the Medical Research Council, the Wellcome Trust, and the James Martin School of the 21st Century (Oxford, United Kingdom).
We thank Tim Rostron (MRC HIU) for technical assistance with HLA typing and Gillian Harcourt for assistance with the CD4+ T-cell lines.
Footnotes
Published ahead of print on 30 August 2006.
REFERENCES
- 1.Barnes, E., S. M. Ward, V. O. Kasprowicz, G. Dusheiko, P. Klenerman, and M. Lucas. 2004. Ultra-sensitive class I tetramer analysis reveals previously undetectable populations of antiviral CD8+ T cells. Eur. J. Immunol. 34:1570-1577. [DOI] [PubMed] [Google Scholar]
- 2.Brown, K. E., and N. S. Young. 1997. Parvovirus B19 in human disease. Annu. Rev. Med. 48:59-67. [DOI] [PubMed] [Google Scholar]
- 3.Corcoran, A., S. Doyle, D. Waldron, A. Nicholson, and B. P. Mahon. 2000. Impaired gamma interferon responses against parvovirus B19 by recently infected children. J. Virol. 74:9903-9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day, C. L., N. P. Seth, M. Lucas, H. Appel, L. Gauthier, G. M. Lauer, G. K. Robbins, Z. M. Szczepiorkowski, D. R. Casson, R. T. Chung, S. Bell, G. Harcourt, B. D. Walker, P. Klenerman, and K. W. Wucherpfennig. 2003. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J. Clin. Investig. 112:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franssila, R., J. Auramo, S. Modrow, M. Mobs, C. Oker-Blom, P. Kapyla, M. Soderlund-Venermo, and K. Hedman. 2005. T helper cell-mediated interferon-gamma expression after human parvovirus B19 infection: persisting VP2-specific and transient VP1u-specific activity. Clin. Exp. Immunol. 142:53-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franssila, R., and K. Hedman. 2004. T-helper cell-mediated interferon-gamma, interleukin-10 and proliferation responses to a candidate recombinant vaccine for human parvovirus B19. Vaccine 22:3809-3815. [DOI] [PubMed] [Google Scholar]
- 7.Franssila, R., K. Hokynar, and K. Hedman. 2001. T helper cell-mediated in vitro responses of recently and remotely infected subjects to a candidate recombinant vaccine for human parvovirus B19. J. Infect. Dis. 183:805-809. [DOI] [PubMed] [Google Scholar]
- 8.Frickhofen, N., J. L. Abkowitz, M. Safford, J. M. Berry, J. Antunez-de-Mayolo, A. Astrow, R. Cohen, I. Halperin, L. King, D. Mintzer, et al. 1990. Persistent B19 parvovirus infection in patients infected with human immunodeficiency virus type 1 (HIV-1): a treatable cause of anemia in AIDS. Ann. Intern. Med. 113:926-933. [DOI] [PubMed] [Google Scholar]
- 9.Gamadia, L. E., E. B. Remmerswaal, J. F. Weel, F. Bemelman, R. A. van Lier, and I. J. Ten Berge. 2003. Primary immune responses to human CMV: a critical role for IFN-gamma-producing CD4+ T cells in protection against CMV disease. Blood 101:2686-2692. [DOI] [PubMed] [Google Scholar]
- 10.Gamadia, L. E., R. J. Rentenaar, R. A. van Lier, and I. J. ten Berge. 2004. Properties of CD4(+) T cells in human cytomegalovirus infection. Hum. Immunol. 65:486-492. [DOI] [PubMed] [Google Scholar]
- 11.Gendi, N. S., K. Gibson, and B. P. Wordsworth. 1996. Effect of HLA type and hypocomplementaemia on the expression of parvovirus arthritis: one year follow up of an outbreak. Ann. Rheum. Dis. 55:63-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlach, J. T., H. M. Diepolder, M. C. Jung, N. H. Gruener, W. W. Schraut, R. Zachoval, R. Hoffmann, C. A. Schirren, T. Santantonio, and G. R. Pape. 1999. Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C. Gastroenterology 117:933-941. [DOI] [PubMed] [Google Scholar]
- 13.Goon, P. K., E. Hanon, T. Igakura, Y. Tanaka, J. N. Weber, G. P. Taylor, and C. R. Bangham. 2002. High frequencies of Th1-type CD4(+) T cells specific to HTLV-1 Env and Tax proteins in patients with HTLV-1-associated myelopathy/tropical spastic paraparesis. Blood 99:3335-3341. [DOI] [PubMed] [Google Scholar]
- 14.Heegaard, E. D., and K. Schmiegelow. 2002. Serologic study on parvovirus B19 infection in childhood acute lymphoblastic leukemia during chemotherapy: clinical and hematologic implications. J. Pediatr. Hematol. Oncol. 24:368-373. [DOI] [PubMed] [Google Scholar]
- 15.Isa, A., V. Kasprowicz, O. Norbeck, A. Loughry, K. Jeffery, K. Broliden, P. Klenerman, T. Tolfvenstam, and P. Bowness. 2005. Prolonged activation of virus-specific CD8+ T cells after acute B19 infection. PLoS Med. 2:e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerr, J. R., D. L. Mattey, W. Thomson, K. V. Poulton, and W. E. Ollier. 2002. Association of symptomatic acute human parvovirus B19 infection with human leukocyte antigen class I and II alleles. J. Infect. Dis. 186:447-452. [DOI] [PubMed] [Google Scholar]
- 17.Klouda, P. T., S. A. Corbin, B. A. Bradley, B. J. Cohen, and A. D. Woolf. 1986. HLA and acute arthritis following human parvovirus infection. Tissue Antigens 28:318-319. [DOI] [PubMed] [Google Scholar]
- 18.Kurtzman, G. J., B. J. Cohen, A. M. Field, R. Oseas, R. M. Blaese, and N. S. Young. 1989. Immune response to B19 parvovirus and an antibody defect in persistent viral infection. J. Clin. Investig. 84:1114-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurtzman, G. J., B. Cohen, P. Meyers, A. Amunullah, and N. S. Young. 1988. Persistent B19 parvovirus infection as a cause of severe chronic anaemia in children with acute lymphocytic leukaemia. Lancet ii:1159-1162. [DOI] [PubMed] [Google Scholar]
- 20.Lucas, M., C. L. Day, J. R. Wyer, S. L. Cunliffe, A. Loughry, A. J. McMichael, and P. Klenerman. 2004. Ex vivo phenotype and frequency of influenza virus-specific CD4 memory T cells. J. Virol. 78:7284-7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marsh, S. G. E., P. Parham, and L. D. Barber. 2000. The HLA factsbook. Academic Press, San Diego, Calif.
- 22.Mitchell, L. A., R. Leong, and K. A. Rosenke. 2001. Lymphocyte recognition of human parvovirus B19 non-structural (NS1) protein: associations with occurrence of acute and chronic arthropathy? J. Med. Microbiol. 50:627-635. [DOI] [PubMed] [Google Scholar]
- 23.Murai, C., Y. Munakata, Y. Takahashi, T. Ishii, S. Shibata, T. Muryoi, T. Funato, M. Nakamura, K. Sugamura, and T. Sasaki. 1999. Rheumatoid arthritis after human parvovirus B19 infection. Ann. Rheum. Dis. 58:130-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberg, E. S., J. M. Billingsley, A. M. Caliendo, S. L. Boswell, P. E. Sax, S. A. Kalams, and B. D. Walker. 1997. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 278:1447-1450. [DOI] [PubMed] [Google Scholar]
- 25.Scriba, T. J., M. Purbhoo, C. L. Day, N. Robinson, S. Fidler, J. Fox, J. N. Weber, P. Klenerman, A. K. Sewell, and R. E. Phillips. 2005. Ultrasensitive detection and phenotyping of CD4+ T cells with optimised HLA class II tetramer staining. J. Immunol. 175:6334-6343. [DOI] [PubMed] [Google Scholar]
- 26.von Poblotzki, A., C. Gerdes, U. Reischl, H. Wolf, and S. Modrow. 1996. Lymphoproliferative responses after infection with human parvovirus B19. J. Virol. 70:7327-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]